Plasma Copy Number Alteration-Based Prognostic and Predictive Multi-Gene Risk Score in Metastatic Castration-Resistant Prostate Cancer
Abstract
Simple Summary
Abstract: Background
1. Introduction
2. Materials and Methods
2.1. Study Cohorts
2.2. cfDNA Extraction and Low-Pass Whole Genome Sequencing
2.3. Sequencing Data Processing and Gene-Specific Copy Number Calling
2.4. Copy Number Alteration Analysis
2.5. Survival Analysis
2.6. Multi-Gene Risk Scores Analysis
3. Results
3.1. Clinical Characteristics of Study Cohorts
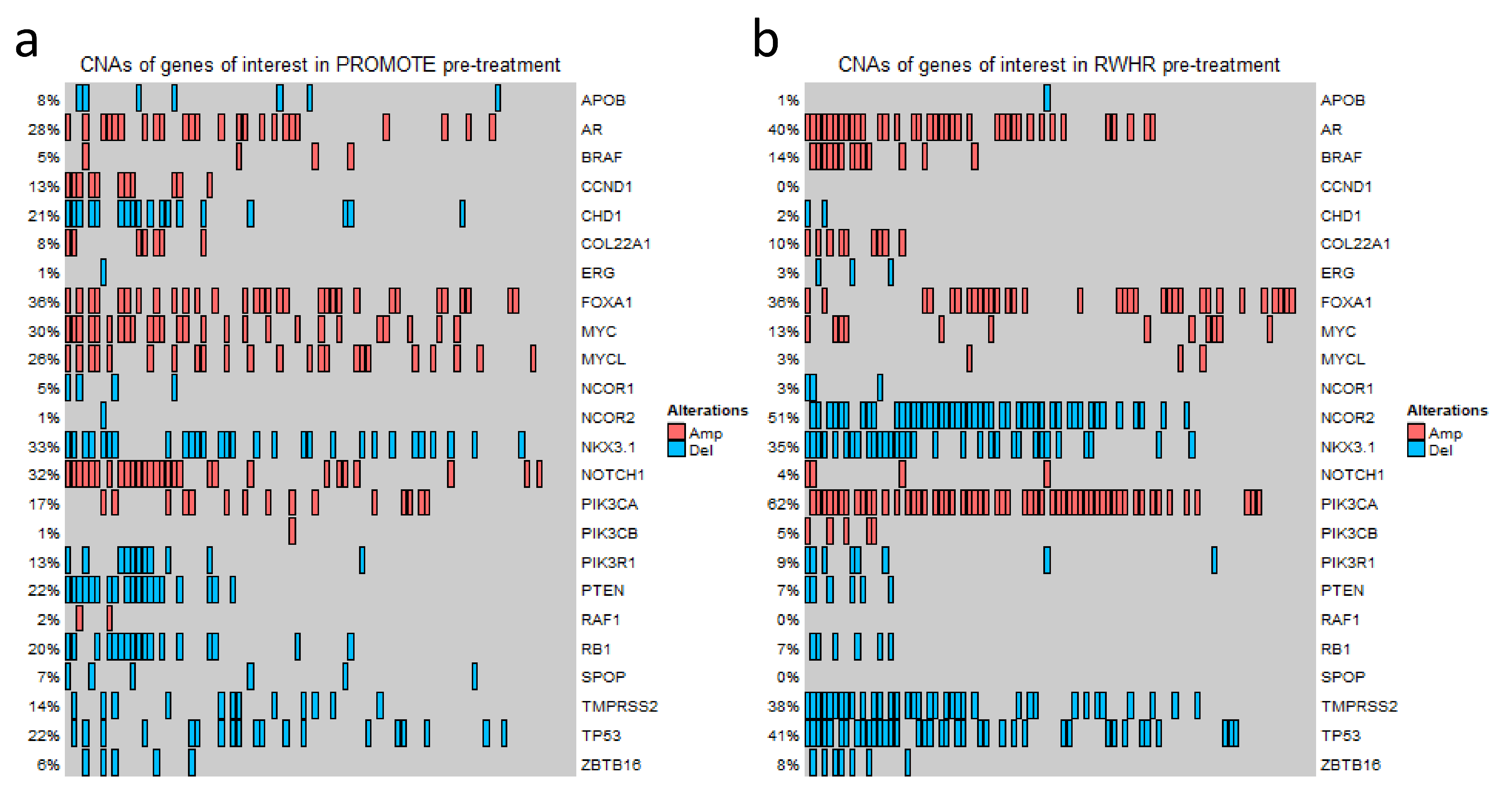
3.2. Broad Range of Detectable CNAs in cfDNA
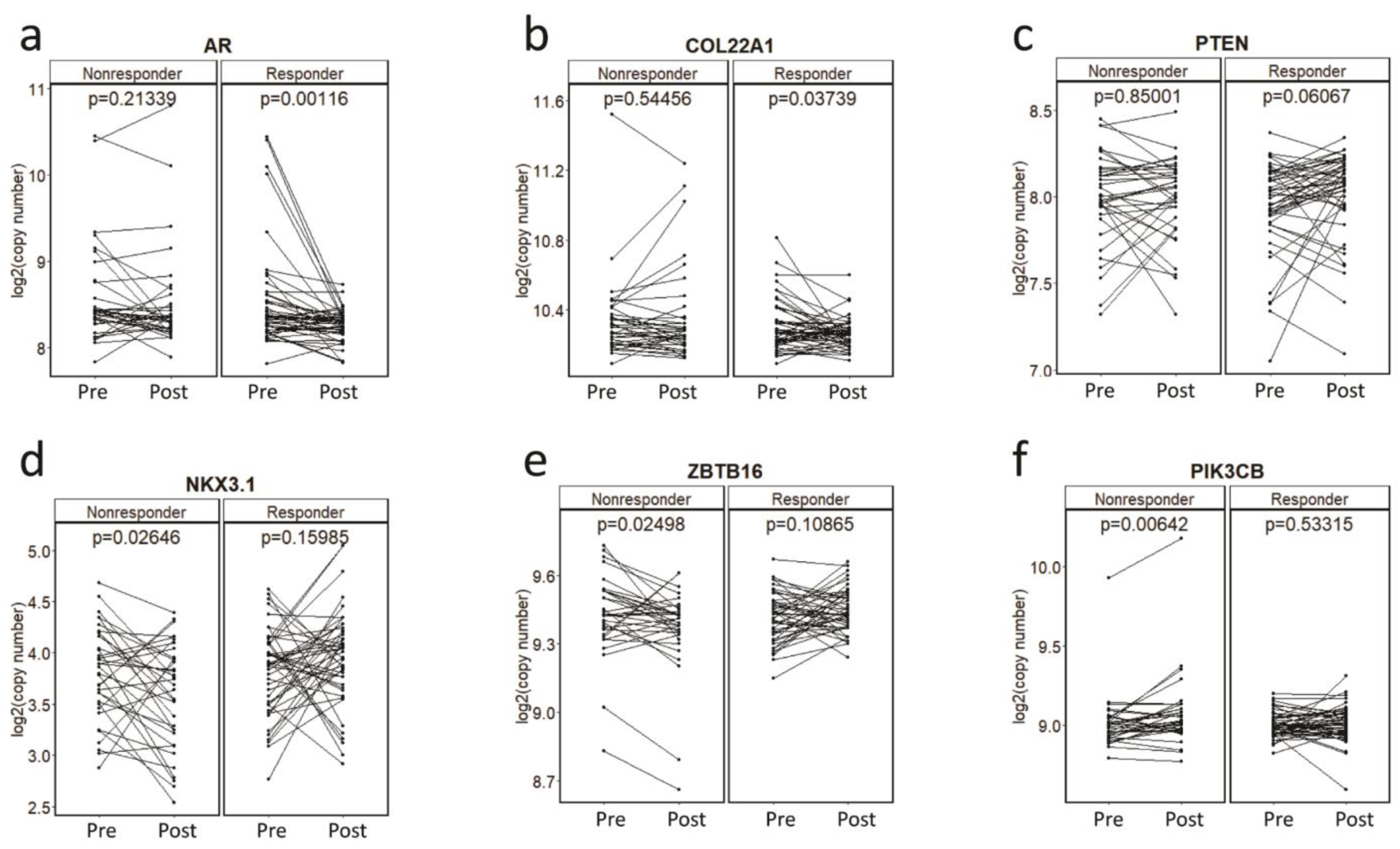
3.3. Gene-Specific and Pharmacodynamics Changes in CNAs for Primary Resistance to Abiraterone Acetate/Prednisone (AA/P) in the PROMOTE Cohort
3.4. Gene-Specific CNAs Predict Acquired Resistance and Survival
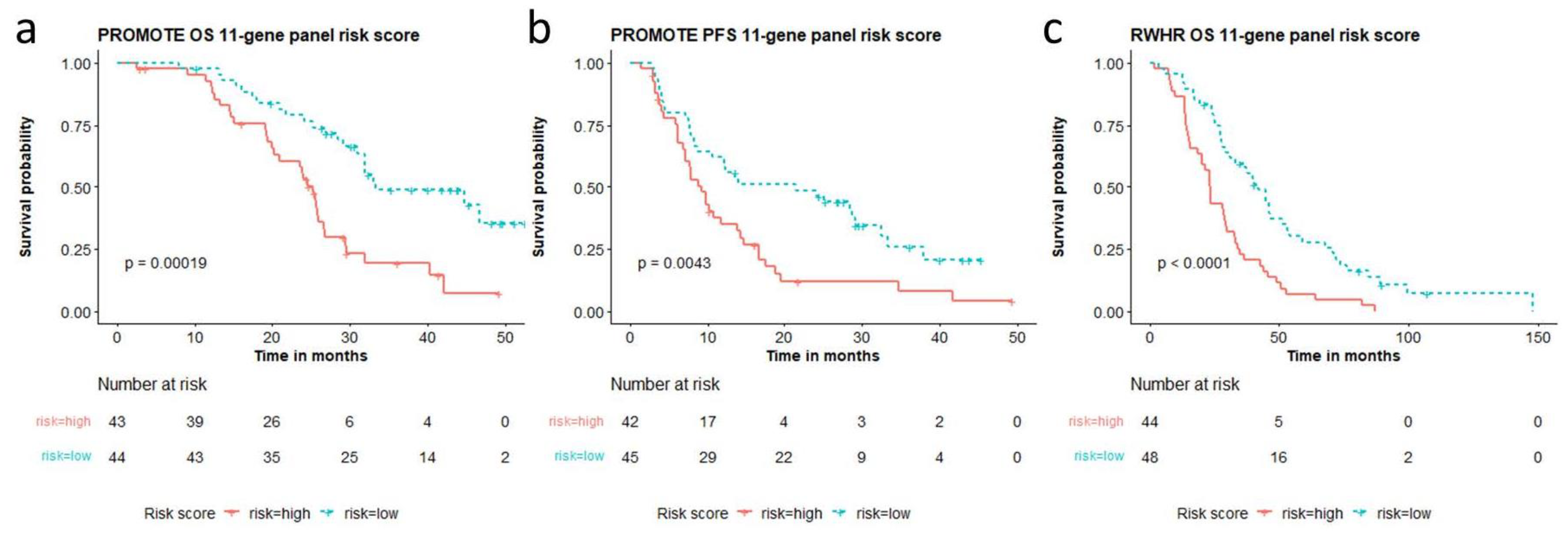
3.5. Multi-Gene CNAs-Based Risk Scores Are Predictive and Prognostic in Two Separate Cohorts
3.6. A Unified 11-Gene Risk Score Panel for Clinical Application
3.7. Multi-Gene Risk Score and Clinical Factor-Based Score Association with Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2.0.2.1. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; van den Bergh, R.C.N.; Briers, E.; Van den Broeck, T.; Cumberbatch, M.G.; De Santis, M.; Fanti, S.; Fossati, N.; Gandaglia, G.; Gillessen, S.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer-2020 Update. Part 1: Screening, Diagnosis, and Local Treatment with Curative Intent. Eur. Urol. 2021, 79, 243–262. [Google Scholar] [CrossRef]
- Tan, J.-L.; Sathianathen, N.; Geurts, N.; Nair, R.; Murphy, D.G.; Lamb, A.D. Androgen receptor targeted therapies in metastatic castration-resistant prostate cancer-The urologists’ perspective. Urol. Sci. 2017, 28, 190–196. [Google Scholar] [CrossRef]
- Wang, L.; Dehm, S.M.; Hillman, D.W.; Sicotte, H.; Tan, W.; Gormley, M.; Bhargava, V.; Jimenez, R.; Xie, F.; Yin, P.; et al. A prospective genome-wide study of prostate cancer metastases reveals association of wnt pathway activation and increased cell cycle proliferation with primary resistance to abiraterone acetate-prednisone. Ann. Oncol. 2018, 29, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Kohli, M.; Tan, W.; Zheng, T.; Wang, A.; Montesinos, C.; Wong, C.; Du, P.; Jia, S.; Yadav, S.; Horvath, L.G.; et al. Clinical and genomic insights into circulating tumor DNA-based alterations across the spectrum of metastatic hormone-sensitive and castrate-resistant prostate cancer. EBioMedicine 2020, 54, 102728. [Google Scholar] [CrossRef] [PubMed]
- Kwan, E.M.; Dai, C.; Fettke, H.; Hauser, C.; Docanto, M.M.; Bukczynska, P.; Ng, N.; Foroughi, S.; Graham, L.K.; Mahon, K.; et al. Plasma Cell-Free DNA Profiling of PTEN-PI3K-AKT Pathway Aberrations in Metastatic Castration-Resistant Prostate Cancer. JCO Precis. Oncol. 2021, 5, 622–637. [Google Scholar] [CrossRef] [PubMed]
- Fettke, H.; Kwan, E.M.; Docanto, M.M.; Bukczynska, P.; Ng, N.; Graham, L.K.; Mahon, K.; Hauser, C.; Tan, W.; Wang, X.H.; et al. Combined Cell-free DNA and RNA Profiling of the Androgen Receptor: Clinical Utility of a Novel Multianalyte Liquid Biopsy Assay for Metastatic Prostate Cancer. Eur. Urol. 2020, 78, 173–180. [Google Scholar] [CrossRef]
- Du, M.; Huang, C.C.; Tan, W.; Kohli, M.; Wang, L. Multiplex Digital PCR to Detect Amplifications of Specific Androgen Receptor Loci in Cell-Free DNA for Prognosis of Metastatic Castration-Resistant Prostate Cancer. Cancers 2020, 12, 2139. [Google Scholar] [CrossRef]
- Du, M.; Tian, Y.; Tan, W.; Wang, L.; Wang, L.; Kilari, D.; Huang, C.C.; Wang, L.; Kohli, M. Plasma cell-free DNA-based predictors of response to abiraterone acetate/prednisone and prognostic factors in metastatic castration-resistant prostate cancer. Prostate Cancer Prostatic Dis. 2020, 23, 705–713. [Google Scholar] [CrossRef]
- Vandekerkhove, G.; Lavoie, J.M.; Annala, M.; Murtha, A.J.; Sundahl, N.; Walz, S.; Sano, T.; Taavitsainen, S.; Ritch, E.; Fazli, L.; et al. Plasma ctDNA is a tumor tissue surrogate and enables clinical-genomic stratification of metastatic bladder cancer. Nat. Commun. 2021, 12, 184. [Google Scholar] [CrossRef] [PubMed]
- Jayaram, A.; Wingate, A.; Wetterskog, D.; Conteduca, V.; Khalaf, D.; Sharabiani, M.T.A.; Calabro, F.; Barwell, L.; Feyerabend, S.; Grande, E.; et al. Plasma Androgen Receptor Copy Number Status at Emergence of Metastatic Castration-Resistant Prostate Cancer: A Pooled Multicohort Analysis. JCO Precis. Oncol. 2019, 3, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Wyatt, A.W.; Annala, M.; Aggarwal, R.; Beja, K.; Feng, F.; Youngren, J.; Foye, A.; Lloyd, P.; Nykter, M.; Beer, T.M.; et al. Concordance of Circulating Tumor DNA and Matched Metastatic Tissue Biopsy in Prostate Cancer. J. Natl. Cancer Inst. 2017, 109, djx118. [Google Scholar] [CrossRef] [PubMed]
- Vandekerkhove, G.; Todenhofer, T.; Annala, M.; Struss, W.J.; Wong, A.; Beja, K.; Ritch, E.; Brahmbhatt, S.; Volik, S.V.; Hennenlotter, J.; et al. Circulating Tumor DNA Reveals Clinically Actionable Somatic Genome of Metastatic Bladder Cancer. Clin. Cancer Res. 2017, 23, 6487–6497. [Google Scholar] [CrossRef]
- Annala, M.; Vandekerkhove, G.; Khalaf, D.; Taavitsainen, S.; Beja, K.; Warner, E.W.; Sunderland, K.; Kollmannsberger, C.; Eigl, B.J.; Finch, D.; et al. Circulating Tumor DNA Genomics Correlate with Resistance to Abiraterone and Enzalutamide in Prostate Cancer. Cancer Discov. 2018, 8, 444–457. [Google Scholar] [CrossRef]
- Azad, A.A.; Volik, S.V.; Wyatt, A.W.; Haegert, A.; Le Bihan, S.; Bell, R.H.; Anderson, S.A.; McConeghy, B.; Shukin, R.; Bazov, J.; et al. Androgen Receptor Gene Aberrations in Circulating Cell-Free DNA: Biomarkers of Therapeutic Resistance in Castration-Resistant Prostate Cancer. Clin. Cancer Res. 2015, 21, 2315–2324. [Google Scholar] [CrossRef]
- Ryan, C.J.; Smith, M.R.; de Bono, J.S.; Molina, A.; Logothetis, C.J.; de Souza, P.; Fizazi, K.; Mainwaring, P.; Piulats, J.M.; Ng, S.; et al. Abiraterone in metastatic prostate cancer without previous chemotherapy. N. Engl. J. Med. 2013, 368, 138–148. [Google Scholar] [CrossRef]
- de Bono, J.S.; Logothetis, C.J.; Molina, A.; Fizazi, K.; North, S.; Chu, L.; Chi, K.N.; Jones, R.J.; Goodman, O.B.; Jr Saad, F.; et al. Abiraterone and increased survival in metastatic prostate cancer. N. Engl. J. Med. 2011, 364, 1995–2005. [Google Scholar] [CrossRef]
- Smith, M.R.; Hussain, M.; Saad, F.; Fizazi, K.; Sternberg, C.N.; Crawford, E.D.; Kopyltsov, E.; Park, C.H.; Alekseev, B.; Montesa-Pino, A.; et al. Darolutamide and Survival in Metastatic, Hormone-Sensitive Prostate Cancer. N. Engl. J. Med. 2022, 386, 1132–1142. [Google Scholar] [CrossRef]
- Chi, K.N.; Agarwal, N.; Bjartell, A.; Chung, B.H.; Pereira de Santana Gomes, A.J.; Given, R.; Juarez Soto, A.; Merseburger, A.S.; Ozguroglu, M.; Uemura, H.; et al. Apalutamide for Metastatic, Castration-Sensitive Prostate Cancer. N. Engl. J. Med. 2019, 381, 13–24. [Google Scholar] [CrossRef]
- Scher, H.I.; Halabi, S.; Tannock, I.; Morris, M.; Sternberg, C.N.; Carducci, M.A.; Eisenberger, M.A.; Higano, C.; Bubley, G.J.; Dreicer, R.; et al. Design and end points of clinical trials for patients with progressive prostate cancer and castrate levels of testosterone: Recommendations of the Prostate Cancer Clinical Trials Working Group. J. Clin. Oncol. 2008, 26, 1148–1159. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Yuan, T.; Liang, M.; Du, M.; Xia, S.; Dittmar, R.; Wang, D.; See, W.; Costello, B.A.; Quevedo, F.; et al. Exosomal miR-1290 and miR-375 as Prognostic Markers in Castration-resistant Prostate Cancer. Eur. Urol. 2015, 67, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. Genome Project Data Processing S: The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Robinson, D.; Van Allen, E.M.; Wu, Y.M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.M.; Montgomery, B.; Taplin, M.E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Grasso, C.S.; Wu, Y.M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The mutational landscape of lethal castration-resistant prostate cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef]
- Quigley, D.A.; Dang, H.X.; Zhao, S.G.; Lloyd, P.; Aggarwal, R.; Alumkal, J.J.; Foye, A.; Kothari, V.; Perry, M.D.; Bailey, A.M.; et al. Genomic Hallmarks and Structural Variation in Metastatic Prostate Cancer. Cell 2018, 174, 758–769.e759. [Google Scholar] [CrossRef]
- Halabi, S.; Lin, C.Y.; Kelly, W.K.; Fizazi, K.S.; Moul, J.W.; Kaplan, E.B.; Morris, M.J.; Small, E.J. Updated prognostic model for predicting overall survival in first-line chemotherapy for patients with metastatic castration-resistant prostate cancer. J. Clin. Oncol. 2014, 32, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Scher, H.I.; Jia, X.; de Bono, J.S.; Fleisher, M.; Pienta, K.J.; Raghavan, D.; Heller, G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: A reanalysis of IMMC38 trial data. Lancet Oncol. 2009, 10, 233–239. [Google Scholar] [CrossRef]
- Herberts, C.; Annala, M.; Sipola, J.; Ng, S.W.S.; Chen, X.E.; Nurminen, A.; Korhonen, O.V.; Munzur, A.D.; Beja, K.; Schonlau, E.; et al. Deep whole-genome ctDNA chronology of treatment-resistant prostate cancer. Nature 2022, 608, 199–208. [Google Scholar] [CrossRef]
- Carreira, S.; Romanel, A.; Goodall, J.; Grist, E.; Ferraldeschi, R.; Miranda, S.; Prandi, D.; Lorente, D.; Frenel, J.S.; Pezaro, C.; et al. Tumor clone dynamics in lethal prostate cancer. Sci. Transl. Med. 2014, 6, 254ra125. [Google Scholar] [CrossRef]
- Romanel, A.; Gasi Tandefelt, D.; Conteduca, V.; Jayaram, A.; Casiraghi, N.; Wetterskog, D.; Salvi, S.; Amadori, D.; Zafeiriou, Z.; Rescigno, P.; et al. Plasma AR and abiraterone-resistant prostate cancer. Sci. Transl. Med. 2015, 7, 312re310. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.Y.; Chang, C.W.; Chng, K.R.; Wansa, K.D.; Sung, W.K.; Cheung, E. Integration of regulatory networks by NKX3-1 promotes androgen-dependent prostate cancer survival. Mol. Cell. Biol. 2012, 32, 399–414. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Nenseth, H.Z.; Saatcioglu, F. Role of PLZF as a tumor suppressor in prostate cancer. Oncotarget 2017, 8, 71317–71324. [Google Scholar] [CrossRef]
- Ferraldeschi, R.; Welti, J.; Luo, J.; Attard, G.; de Bono, J.S. Targeting the androgen receptor pathway in castration-resistant prostate cancer: Progresses and prospects. Oncogene 2015, 34, 1745–1757. [Google Scholar] [CrossRef]
- Carver, B.S.; Chapinski, C.; Wongvipat, J.; Hieronymus, H.; Chen, Y.; Chandarlapaty, S.; Arora, V.K.; Le, C.; Koutcher, J.; Scher, H.; et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011, 19, 575–586. [Google Scholar] [CrossRef]
- Teroerde, M.; Nientiedt, C.; Duensing, A.; Hohenfellner, M.; Stenzinger, A.; Duensing, S. Revisiting the Role of p53 in Prostate Cancer. In Prostate Cancer; Bott, S.R.J., Ng, K.L., Eds.; Exon Publications: Brisbane, Australia, 2021. [Google Scholar]
- O’Cearbhaill, R.; Gannon, J.M.; Prichard, R.S.; Walshe, J.M.; McDermott, E.; Quinn, C.M. The American Joint Commission Cancer 8th Edition Prognostic Stage Including Oncotype DX(R) Recurrence Score: Impact on Staging of Early Breast Cancer. Pathobiology 2019, 86, 77–82. [Google Scholar] [CrossRef]
- Sparano, J.A.; Gray, R.J.; Makower, D.F.; Pritchard, K.I.; Albain, K.S.; Hayes, D.F.; Geyer, C.E.; Dees, E.C., Jr.; Goetz, M.P.; Olson, J.A., Jr.; et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N. Engl. J. Med. 2018, 379, 111–121. [Google Scholar] [CrossRef] [PubMed]

| Patient Cohorts | PROMOTE | RWHR | p Value of Differences | |
|---|---|---|---|---|
| Total | 88 | 92 | ||
| Age | 0.375 | |||
| Median | 72 | 73 | ||
| IQR | 66–78 | 67–78 | ||
| Range | 39–91 | 43–92 | ||
| Gleason score at initial diagnosis | 0.251 | |||
| 2–6 | 16 | 9 | ||
| 7 | 27 | 34 | ||
| 8–10 | 45 | 44 | ||
| Missing | 0 | 5 | ||
| Volume of metastatic disease | <0.0001 | |||
| High | 50 | 30 | ||
| Low | 38 | 62 | ||
| Site of metastases | 0.901 | |||
| Bone | 78 | 81 | ||
| Others | 10 | 11 | ||
| Baseline Prostate-Specific Antigen (ng/mL) | 0.276 | |||
| Median | 14.7 | 19.25 | ||
| IQR | 6.38–41.9 | 3.25–79.72 | ||
| Lactate Dehydrogenase (U/L) | 0.4535 | |||
| Median | 185.5 | 188 | ||
| IQR | 170.8–209.2 | 158.5–230.5 | ||
| Missing | 4 | 49 | ||
| Alkaline phosphatase (U/L) | 0.767 | |||
| Median | 109.5 | 96 | ||
| IQR | 83.25–178.75 | 68.25–137.50 | ||
| Missing | 6 | 6 | ||
| Time from ADT start to ADT fail (months) | 0.224 | |||
| Median | 24.87 | 19.97 | ||
| IQR | 11.55–53.8 | 7.88–44.92 | ||
| Time from date of enrollment to last follow-up (months) | 0.0001 | |||
| Median | 58.52 | 128.83 | ||
| IQR | 51.74–62.92 | 120.69–139.01 | ||
| Dead/alive during follow up | <0.0001 | |||
| Dead | 55 | 86 | ||
| Alive | 33 | 6 | ||
| Primary radical prostatectomy | 0.944 | |||
| Yes | 37 | 42 | ||
| No | 45 | 50 | ||
| Missing | 6 | 0 | ||
| Gene | CNA | PROMOTE PFS | PROMOTE OS | RWHR OS | |||
|---|---|---|---|---|---|---|---|
| HR | p Value | HR | p Value | HR | p Value | ||
| APOB | Del | 1.07 | 0.8812 | 0.69 | 0.5273 | 4.54 | 0.1061 |
| AR | Amp | 2.17 | 0.0042 | 1.98 | 0.0180 | 1.59 | 0.0377 |
| BRAF | Amp | 0.58 | 0.4452 | 0.38 | 0.3204 | 1.93 | 0.0293 |
| CCND1 | Amp | 1.32 | 0.4225 | 1.04 | 0.9120 | NA | 1.0000 |
| CHD1 | Del | 1.17 | 0.5946 | 1.02 | 0.9452 | 1.84 | 0.3899 |
| COL22A1 | Amp | 2.47 | 0.0216 | 1.86 | 0.1456 | 3.58 | 0.0002 |
| ERG | Del | 0.00 | 0.3336 | 0.00 | 0.6569 | 0.82 | 0.7421 |
| FOXA1 | Amp | 1.15 | 0.5805 | 0.78 | 0.3800 | 1.10 | 0.6601 |
| MYC | Amp | 1.57 | 0.0801 | 1.43 | 0.2088 | 0.55 | 0.0888 |
| MYCL | Amp | 0.71 | 0.2145 | 0.62 | 0.1294 | 0.89 | 0.8479 |
| NCOR1 | Del | 1.98 | 0.1816 | 4.00 | 0.0050 | 7.47 | 0.0002 |
| NCOR2 | Del | 0.00 | 0.3336 | 0.00 | 0.6569 | 1.53 | 0.0529 |
| NKX3.1 | Del | 0.69 | 0.1833 | 1.02 | 0.9585 | 1.80 | 0.0084 |
| NOTCH1 | Amp | 1.04 | 0.8705 | 1.02 | 0.9469 | 5.45 | 0.0005 |
| PIK3CA | Amp | 1.22 | 0.5408 | 2.16 | 0.0225 | 1.68 | 0.0247 |
| PIK3CB | Amp | 20.89 | 0.0001 | 3.61 | 0.1797 | 2.54 | 0.0391 |
| PIK3R1 | Del | 1.13 | 0.7427 | 1.79 | 0.1071 | 1.94 | 0.0902 |
| PTEN | Del | 1.10 | 0.7420 | 1.07 | 0.8421 | 1.70 | 0.2116 |
| RAF1 | Amp | 1.00 | 0.9964 | 3.06 | 0.1063 | NA | 1.0000 |
| RB1 | Del | 1.15 | 0.6240 | 1.02 | 0.9515 | 2.49 | 0.0278 |
| SPOP | Del | 1.44 | 0.4378 | 1.32 | 0.5539 | NA | 1.0000 |
| TMPRSS2 | Del | 0.93 | 0.8522 | 1.95 | 0.0965 | 1.73 | 0.0136 |
| TP53 | Del | 1.43 | 0.2357 | 2.07 | 0.0286 | 1.50 | 0.0655 |
| ZBTB16 | Del | 2.29 | 0.1547 | 6.63 | 0.0008 | 1.03 | 0.9405 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Du, M.; Soupir, A.; Wang, L.; Tan, W.; Kalari, K.R.; Kilari, D.; Park, J.; Huang, C.-C.; Kohli, M.; et al. Plasma Copy Number Alteration-Based Prognostic and Predictive Multi-Gene Risk Score in Metastatic Castration-Resistant Prostate Cancer. Cancers 2022, 14, 4714. https://doi.org/10.3390/cancers14194714
Huang J, Du M, Soupir A, Wang L, Tan W, Kalari KR, Kilari D, Park J, Huang C-C, Kohli M, et al. Plasma Copy Number Alteration-Based Prognostic and Predictive Multi-Gene Risk Score in Metastatic Castration-Resistant Prostate Cancer. Cancers. 2022; 14(19):4714. https://doi.org/10.3390/cancers14194714
Chicago/Turabian StyleHuang, Jinyong, Meijun Du, Alex Soupir, Liewei Wang, Winston Tan, Krishna R. Kalari, Deepak Kilari, Jong Park, Chiang-Ching Huang, Manish Kohli, and et al. 2022. "Plasma Copy Number Alteration-Based Prognostic and Predictive Multi-Gene Risk Score in Metastatic Castration-Resistant Prostate Cancer" Cancers 14, no. 19: 4714. https://doi.org/10.3390/cancers14194714
APA StyleHuang, J., Du, M., Soupir, A., Wang, L., Tan, W., Kalari, K. R., Kilari, D., Park, J., Huang, C.-C., Kohli, M., & Wang, L. (2022). Plasma Copy Number Alteration-Based Prognostic and Predictive Multi-Gene Risk Score in Metastatic Castration-Resistant Prostate Cancer. Cancers, 14(19), 4714. https://doi.org/10.3390/cancers14194714







