Expansion of Colorectal Cancer Biomarkers Based on Gut Bacteria and Viruses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Design
2.2. Metagenomic Sequence Data Processing
2.3. Sequence Taxonomic Annotation
2.4. Gene Prediction and Metabolic Pathway Functional Annotation
2.5. Microbial Ecological Analysis
2.6. Difference Analysis
2.7. Associations between Species and Pathways
2.8. Correlation Network
2.9. Microbial Classifier Selection
2.10. External Validation Data Acquisition and Analysis
3. Results
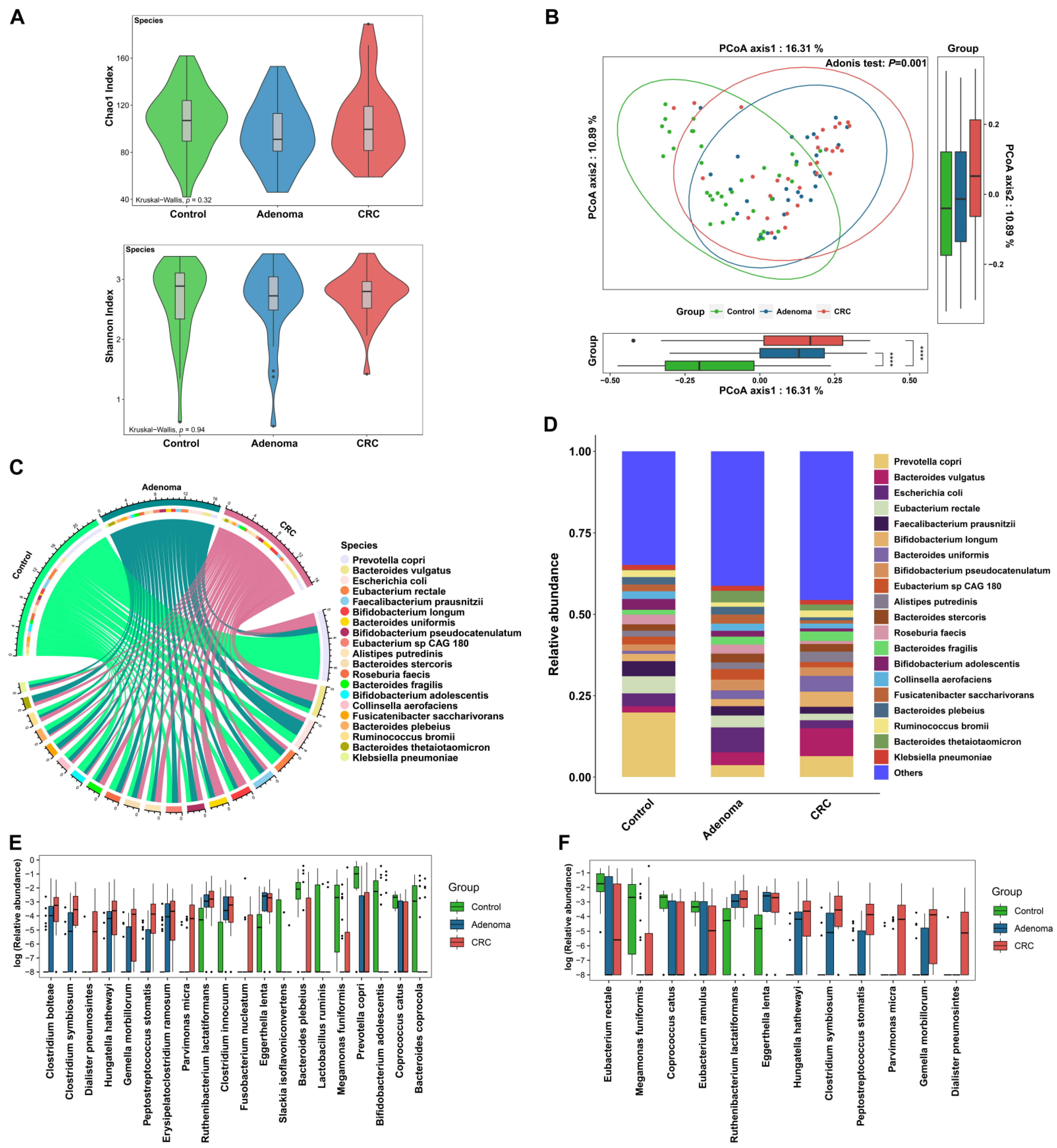
3.1. General Characteristics of Intestinal Microorganisms of the Participants
3.2. Bacteria Alteration across Groups
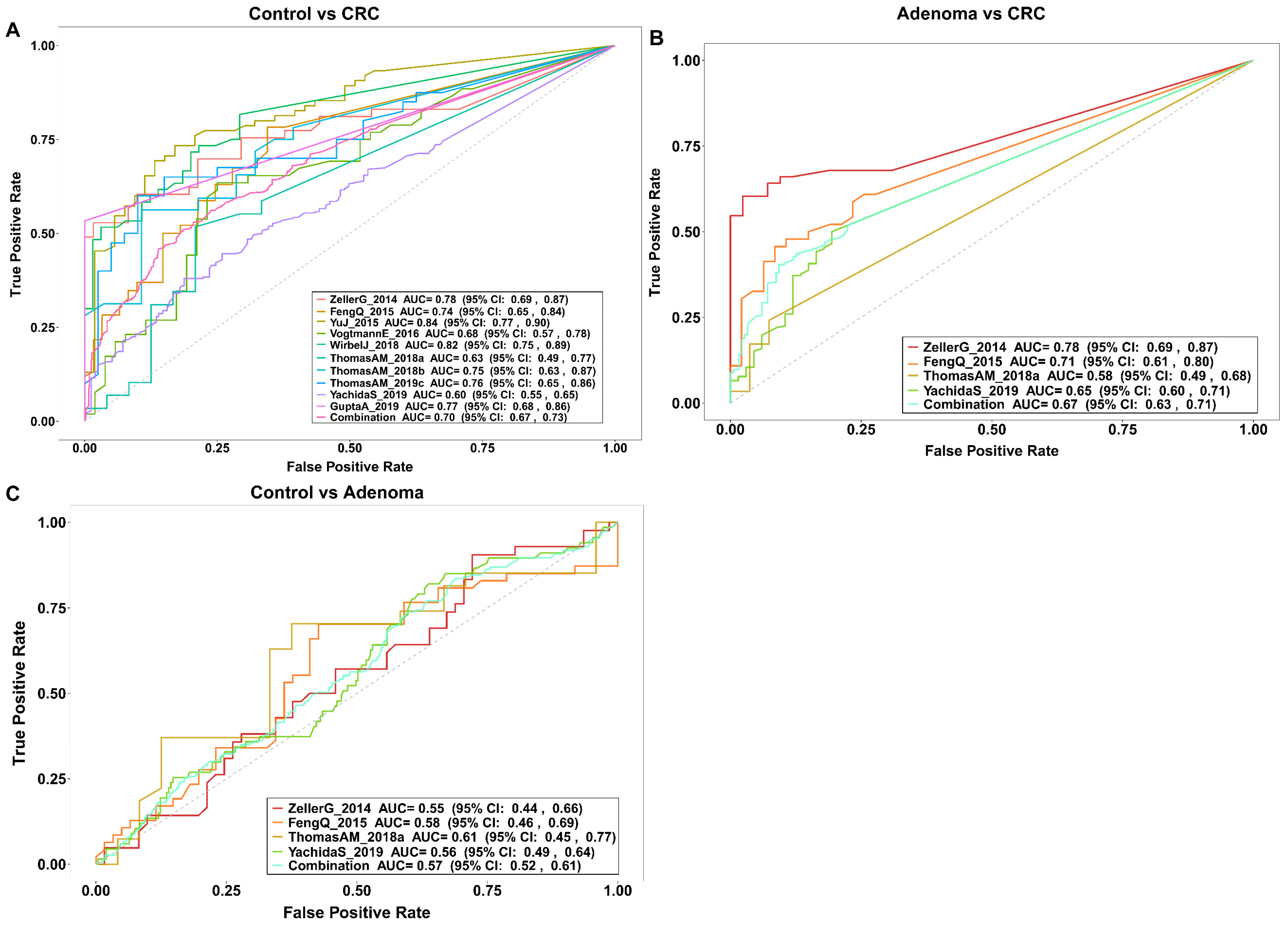
3.3. Gut Bacteria Distinguish Colorectal Tumor Patients from Controls
3.4. Metabolic Pathways Alteration across Groups and Associations between Bacteria and Metabolic Pathways
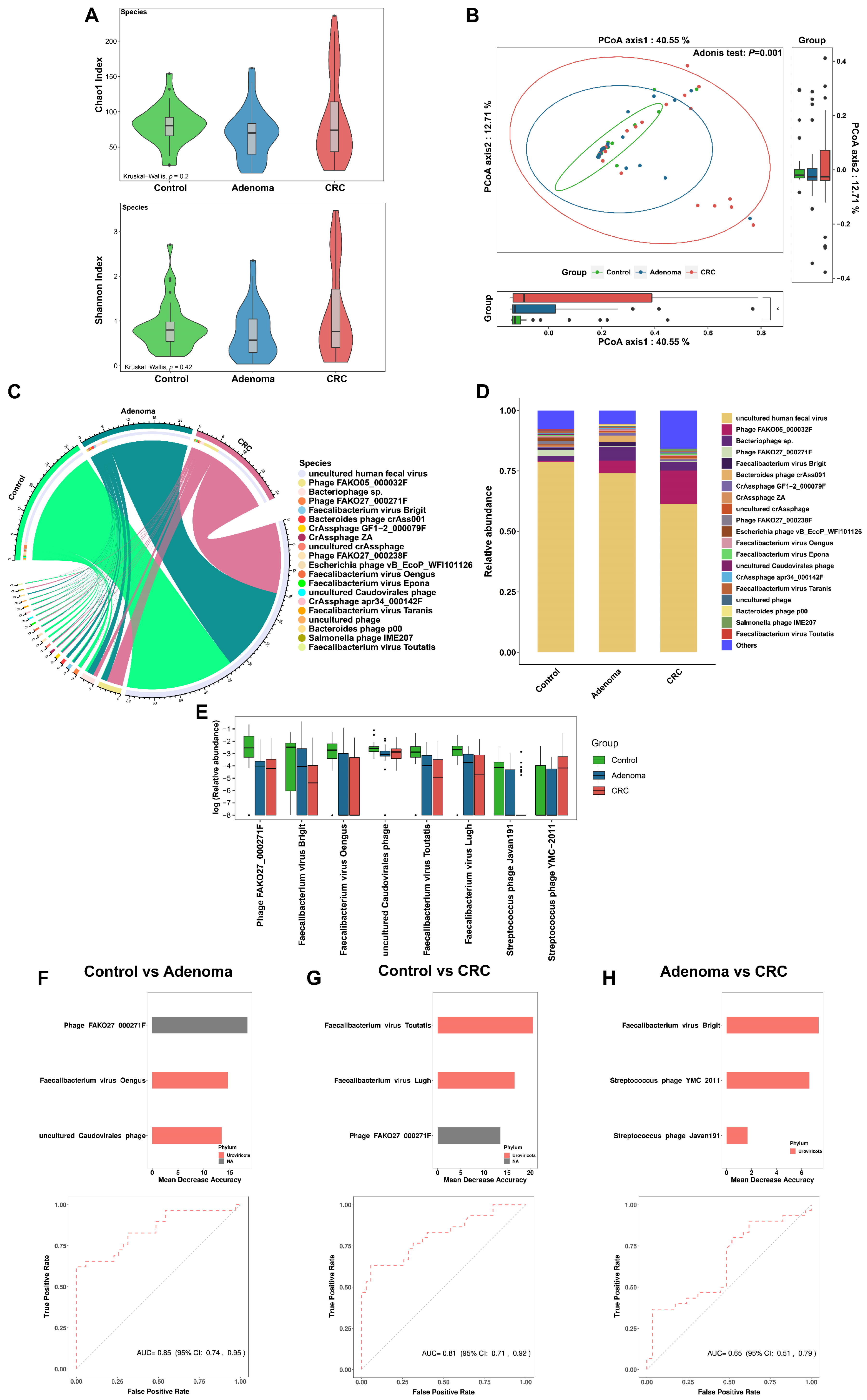
3.5. Gut Virome Alteration across Groups
3.6. Gut Virome Distinguishes Colorectal Tumor Patients from Controls
3.7. Associations across Microbiome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- International Agency for Research on Cancer. Global Cancer Observatory—Cancer Today. 2022. Available online: https://gco.iarc.fr/ (accessed on 2 May 2022).
- Fang, J.-Y. Chinese consensus on prevention of colorectal neoplasia (2021, Shanghai). J. Dig. Dis. 2022, 23, 58–90. [Google Scholar] [CrossRef]
- Keum, N.; Giovannucci, E. Global burden of colorectal cancer: Emerging trends, risk factors and prevention strategies. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 713–732. [Google Scholar] [CrossRef]
- Dekker, E.; Tanis, P.J.; Vleugels, J.L.A.; Kasi, P.M.; Wallace, M.B. Colorectal cancer. Lancet 2019, 394, 1467–1480. [Google Scholar] [CrossRef]
- Wang, T.; Cai, G.; Qiu, Y.; Fei, N.; Zhang, M.; Pang, X.; Jia, W.; Cai, S.; Zhao, L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 2012, 6, 320–329. [Google Scholar] [CrossRef]
- Yang, Y.; Misra, B.B.; Liang, L.; Bi, D.; Weng, W.; Wu, W.; Cai, S.; Qin, H.; Goel, A.; Li, X.; et al. Integrated microbiome and metabolome analysis reveals a novel interplay between commensal bacteria and metabolites in colorectal cancer. Theranostics 2019, 9, 4101–4114. [Google Scholar] [CrossRef]
- Wong, S.H.; Yu, J. Gut microbiota in colorectal cancer: Mechanisms of action and clinical applications. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 690–704. [Google Scholar] [CrossRef]
- Liang, Q.; Chiu, J.; Chen, Y.; Huang, Y.; Higashimori, A.; Fang, J.; Brim, H.; Ashktorab, H.; Ng, S.C.; Ng, S.S.M.; et al. Fecal Bacteria Act as Novel Biomarkers for Noninvasive Diagnosis of Colorectal Cancer. Clin. Cancer Res. 2017, 23, 2061–2070. [Google Scholar] [CrossRef]
- Dai, Z.; Coker, O.O.; Nakatsu, G.; Wu, W.K.K.; Zhao, L.; Chen, Z.; Chan, F.K.L.; Kristiansen, K.; Sung, J.J.Y.; Wong, S.H.; et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome 2018, 6, 70. [Google Scholar] [CrossRef]
- Wirbel, J.; Pyl, P.T.; Kartal, E.; Zych, K.; Kashani, A.; Milanese, A.; Fleck, J.S.; Voigt, A.Y.; Palleja, A.; Ponnudurai, R.; et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat. Med. 2019, 25, 679–689. [Google Scholar] [CrossRef]
- Coker, O.O.; Nakatsu, G.; Dai, R.Z.; Wu, W.K.K.; Wong, S.H.; Ng, S.C.; Chan, F.K.L.; Sung, J.J.Y.; Yu, J. Enteric fungal microbiota dysbiosis and ecological alterations in colorectal cancer. Gut 2019, 68, 654–662. [Google Scholar] [CrossRef]
- Hannigan, G.D.; Duhaime, M.B.; Ruffin, M.T.; Koumpouras, C.C.; Schloss, P.D. Diagnostic Potential and Interactive Dynamics of the Colorectal Cancer Virome. mBio 2018, 9, e02248-18. [Google Scholar] [CrossRef]
- Nakatsu, G.; Zhou, H.; Wu, W.K.K.; Wong, S.H.; Coker, O.O.; Dai, Z.; Li, X.; Szeto, C.H.; Sugimura, N.; Lam, T.Y.; et al. Alterations in Enteric Virome Are Associated With Colorectal Cancer and Survival Outcomes. Gastroenterology 2018, 155, 529–541.e5. [Google Scholar] [CrossRef]
- Shen, S.; Huo, D.; Ma, C.; Jiang, S.; Zhang, J. Expanding the Colorectal Cancer Biomarkers Based on the Human Gut Phageome. Microbiol. Spectr. 2021, 9, e0009021. [Google Scholar] [CrossRef]
- Beghini, F.; McIver, L.J.; Blanco-Míguez, A.; Dubois, L.; Asnicar, F.; Maharjan, S.; Mailyan, A.; Manghi, P.; Scholz, M.; Thomas, A.M.; et al. Integrating taxonomic, functional, and strain-level profiling of diverse microbial communities with bioBakery 3. ELife 2021, 10, e65088. [Google Scholar] [CrossRef]
- Wood, D.E.; Lu, J.; Langmead, B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019, 20, 257. [Google Scholar] [CrossRef]
- Friedman, J.; Alm, E.J. Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 2012, 8, e1002687. [Google Scholar] [CrossRef]
- Kim, H.; Lee, K.K.; Jeon, J.; Harris, W.A.; Lee, Y.-H. Domestication of Oryza species eco-evolutionarily shapes bacterial and fungal communities in rice seed. Microbiome 2020, 8, 20. [Google Scholar] [CrossRef]
- Shang, J.; Sun, Y. CHERRY: A Computational metHod for accuratE pRediction of virus-pRokarYotic interactions using a graph encoder-decoder model. Brief. Bioinform. 2022, 23, bbac182. [Google Scholar] [CrossRef]
- Pasolli, E.; Schiffer, L.; Manghi, P.; Renson, A.; Obenchain, V.; Truong, D.T.; Beghini, F.; Malik, F.; Ramos, M.; Dowd, J.B.; et al. Accessible, curated metagenomic data through ExperimentHub. Nat. Methods 2017, 14, 1023–1024. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Feng, Q.; Liang, S.; Jia, H.; Stadlmayr, A.; Tang, L.; Lan, Z.; Zhang, D.; Xia, H.; Xu, X.; Jie, Z.; et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat. Commun. 2015, 6, 6528. [Google Scholar] [CrossRef]
- Vogtmann, E.; Hua, X.; Zeller, G.; Sunagawa, S.; Voigt, A.Y.; Hercog, R.; Goedert, J.J.; Shi, J.; Bork, P.; Sinha, R. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. PLoS ONE 2016, 11, e0155362. [Google Scholar] [CrossRef]
- Yu, J.; Feng, Q.; Wong, S.H.; Zhang, D.; Liang, Q.Y.; Qin, Y.; Tang, L.; Zhao, H.; Stenvang, J.; Li, Y.; et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut 2017, 66, 70–78. [Google Scholar] [CrossRef]
- Thomas, A.M.; Manghi, P.; Asnicar, F.; Pasolli, E.; Armanini, F.; Zolfo, M.; Beghini, F.; Manara, S.; Karcher, N.; Pozzi, C.; et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nat. Med. 2019, 25, 667–678. [Google Scholar] [CrossRef]
- Yachida, S.; Mizutani, S.; Shiroma, H.; Shiba, S.; Nakajima, T.; Sakamoto, T.; Watanabe, H.; Masuda, K.; Nishimoto, Y.; Kubo, M.; et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat. Med. 2019, 25, 968–976. [Google Scholar] [CrossRef]
- Gupta, A.; Dhakan, D.B.; Maji, A.; Saxena, R.; PK, V.P.; Mahajan, S.; Pulikkan, J.; Kurian, J.; Gomez, A.M.; Scaria, J.; et al. Association of Flavonifractor plautii, a Flavonoid-Degrading Bacterium, with the Gut Microbiome of Colorectal Cancer Patients in India. MSystems 2019, 4, e00438-19. [Google Scholar] [CrossRef]
- Kwong, T.N.Y.; Wang, X.; Nakatsu, G.; Chow, T.C.; Tipoe, T.; Dai, R.Z.W.; Tsoi, K.K.K.; Wong, M.C.S.; Tse, G.; Chan, M.T.V.; et al. Association Between Bacteremia From Specific Microbes and Subsequent Diagnosis of Colorectal Cancer. Gastroenterology 2018, 155, 383–390.e8. [Google Scholar] [CrossRef]
- Drewes, J.L.; White, J.R.; Dejea, C.M.; Fathi, P.; Iyadorai, T.; Vadivelu, J.; Roslani, A.C.; Wick, E.C.; Mongodin, E.F.; Loke, M.F.; et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017, 3, 34. [Google Scholar] [CrossRef]
- Cao, Y.; Wang, Z.; Yan, Y.; Ji, L.; He, J.; Xuan, B.; Shen, C.; Ma, Y.; Jiang, S.; Ma, D.; et al. Enterotoxigenic Bacteroidesfragilis Promotes Intestinal Inflammation and Malignancy by Inhibiting Exosome-Packaged miR-149-3p. Gastroenterology 2021, 161, 1552–1566.e12. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.-H.; Gao, Q.-Y.; Cai, G.-X.; Sun, X.-M.; Sun, X.-M.; Zou, T.-H.; Chen, H.-M.; Yu, S.-Y.; Qiu, Y.-W.; Gu, W.-Q.; et al. Fecal Clostridium symbiosum for Noninvasive Detection of Early and Advanced Colorectal Cancer: Test and Validation Studies. EBioMedicine 2017, 25, 32–40. [Google Scholar] [CrossRef]
- Xia, X.; Wu, W.K.K.; Wong, S.H.; Liu, D.; Kwong, T.N.Y.; Nakatsu, G.; Yan, P.S.; Chuang, Y.-M.; Chan, M.W.-Y.; Coker, O.O.; et al. Bacteria pathogens drive host colonic epithelial cell promoter hypermethylation of tumor suppressor genes in colorectal cancer. Microbiome 2020, 8, 108. [Google Scholar] [CrossRef]
- Liang, J.Q.; Zeng, Y.; Kwok, G.; Cheung, C.P.; Suen, B.Y.; Ching, J.Y.L.; To, K.F.; Yu, J.; Chan, F.K.L.; Ng, S.C. Novel microbiome signatures for non-invasive diagnosis of adenoma recurrence after colonoscopic polypectomy. Aliment. Pharmacol. Ther. 2022, 55, 847–855. [Google Scholar] [CrossRef]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; DeStefano Shields, C.E.; Hechenbleikner, E.M.; Huso, D.L.; et al. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef]
- Bai, X.; Wei, H.; Liu, W.; Coker, O.O.; Gou, H.; Liu, C.; Zhao, L.; Li, C.; Zhou, Y.; Wang, G.; et al. Cigarette smoke promotes colorectal cancer through modulation of gut microbiota and related metabolites. Gut 2022, online ahead of print. [CrossRef]
- Gao, R.; Zhu, Y.; Kong, C.; Xia, K.; Li, H.; Zhu, Y.; Zhang, X.; Liu, Y.; Zhong, H.; Yang, R.; et al. Alterations, Interactions, and Diagnostic Potential of Gut Bacteria and Viruses in Colorectal Cancer. Front. Cell. Infect. Microbiol. 2021, 11, 657867. [Google Scholar] [CrossRef]
- Gregory, A.C.; Zablocki, O.; Zayed, A.A.; Howell, A.; Bolduc, B.; Sullivan, M.B. The Gut Virome Database Reveals Age-Dependent Patterns of Virome Diversity in the Human Gut. Cell Host Microbe 2020, 28, 724–740.e8. [Google Scholar] [CrossRef]
- Camarillo-Guerrero, L.F.; Almeida, A.; Rangel-Pineros, G.; Finn, R.D.; Lawley, T.D. Massive expansion of human gut bacteriophage diversity. Cell 2021, 184, 1098–1109.e9. [Google Scholar] [CrossRef]
- Zhao, L.; Shi, Y.; Lau, H.C.-H.; Liu, W.; Luo, G.; Wang, G.; Liu, C.; Pan, Y.; Zhou, Q.; Ding, Y.; et al. Uncovering 1058 novel human enteric DNA viruses through deep long-read third-generation sequencing and their clinical impact. Gastroenterology 2022, online ahead of print. [CrossRef]
- Liu, N.-N.; Jiao, N.; Tan, J.-C.; Wang, Z.; Wu, D.; Wang, A.-J.; Chen, J.; Tao, L.; Zhou, C.; Fang, W.; et al. Multi-kingdom microbiota analyses identify bacterial-fungal interactions and biomarkers of colorectal cancer across cohorts. Nat. Microbiol. 2022, 7, 238–250. [Google Scholar] [CrossRef]
- Li, Y.-K.; Xu, Q.; Sun, L.-P.; Gong, Y.-H.; Jing, J.-J.; Xing, C.-Z.; Yuan, Y. Nucleotide excision repair pathway gene polymorphisms are associated with risk and prognosis of colorectal cancer. World J. Gastroenterol. 2020, 26, 307–323. [Google Scholar] [CrossRef]
- Li, S.K.H.; Martin, A. Mismatch Repair and Colon Cancer: Mechanisms and Therapies Explored. Trends Mol. Med. 2016, 22, 274–289. [Google Scholar] [CrossRef]
- Hinnebusch, B.F.; Meng, S.; Wu, J.T.; Archer, S.Y.; Hodin, R.A. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J. Nutr. 2002, 132, 1012–1017. [Google Scholar] [CrossRef]
- Nguyen, N.K.; Deehan, E.C.; Zhang, Z.; Jin, M.; Baskota, N.; Perez-Muñoz, M.E.; Cole, J.; Tuncil, Y.E.; Seethaler, B.; Wang, T.; et al. Gut microbiota modulation with long-chain corn bran arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate. Microbiome 2020, 8, 118. [Google Scholar] [CrossRef]
- Yang, L.; Xue, R.; Pan, C. Prognostic and clinicopathological value of PD-L1 in colorectal cancer: A systematic review and meta-analysis. OncoTargets Ther. 2019, 12, 3671–3682. [Google Scholar] [CrossRef]
- Salek Farrokhi, A.; Darabi, N.; Yousefi, B.; Askandar, R.H.; Shariati, M.; Eslami, M. Is it true that gut microbiota is considered as panacea in cancer therapy? J. Cell. Physiol. 2019, 234, 14941–14950. [Google Scholar] [CrossRef]
- Vetizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P.M.; et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef]
- TIANGEN. TIANamp Stool DNA Kit: Rapid Extraction of High Quality Genomic DNA from Various Stool Samples. Available online: https://en.tiangen.com/content/details_43_4245.html (accessed on 6 May 2022).




| Characteristics | Control | Adenoma | CRC | Statistics * | p Value |
|---|---|---|---|---|---|
| (n = 35) | (n = 29) | (n = 30) | |||
| Age ( ± sd) | 57.53 ± 7.70 | 56.71 ± 8.21 | 57.82 ± 9.30 | 0.14 | 0.870 |
| Gender, n (%) | 0.69 | 0.710 | |||
| male | 20 (57.14) | 17 (58.62) | 20 (66.67) | ||
| female | 15 (42.86) | 12 (41.38) | 10 (33.33) | ||
| Hypertension, n (%) | 2.61 | 0.271 | |||
| without HTN | 27 (77.14) | 19 (65.52) | 25 (83.33) | ||
| with HTN | 8 (22.86) | 10 (34.48) | 5 (16.67) | ||
| Diabetes, n (%) | |||||
| without DM | 31 (88.57) | 25 (86.21) | 25 (83.33) | 0.06 | 0.930 |
| with DM | 4 (11.43) | 4 (13.79) | 5 (16.67) | ||
| BMI ( ± sd) | 23.09 ± 2.39 | 24.01 ± 2.60 | 22.51 ± 2.38 | 2.82 | 0.065 |
| Smoking, n (%) | 0.16 | 0.923 | |||
| smoking now | 10 (28.57) | 7 (24.14) | 8 (26.67) | ||
| no smoking now | 25 (71.43) | 22 (75.86) | 22 (73.33) | ||
| Drinking, n (%) | 0.19 | 0.907 | |||
| drinking now | 6 (17.14) | 6 (20.69) | 5 (16.67) | ||
| no drinking now | 29 (82.86) | 23 (79.31) | 25 (83.33) |
| Dataset | Group (N) | Gender_Male (%) | Age ± sd | Country | |
|---|---|---|---|---|---|
| ZellerG_2014 | Control | 61 | 45.90 | 60.57 ± 11.39 | FRA |
| Adenoma | 42 | 71.43 | 62.95 ± 9.11 | ||
| CRC | 53 | 54.72 | 66.81 ± 10.88 | ||
| FengQ_2015 | Control | 61 | 59.02 | 66.97 ± 6.45 | AUT |
| Adenoma | 47 | 48.94 | 66.49 ± 7.86 | ||
| CRC | 46 | 60.87 | 67.07 ± 10.91 | ||
| ThomasAM_2018a | Control | 24 | 54.17 | 67.92 ± 7.01 | ITA |
| Adenoma | 27 | 59.26 | 62.89 ± 8.67 | ||
| CRC | 29 | 79.31 | 71.45 ± 8.23 | ||
| YachidaS_2019 | Control | 251 | 54.18 | 60.81 ± 12.64 | JPN |
| Adenoma | 67 | 71.64 | 63.15 ± 9.12 | ||
| CRC | 258 | 62.40 | 62.72 ± 9.64 | ||
| YuJ_2015 | Control | 53 | 62.26 | 61.83 ± 5.70 | CHN |
| CRC | 75 | 64.00 | 65.93 ± 10.57 | ||
| VogtmannE_2016 | Control | 52 | 71.15 | 61.23 ± 11.03 | USA |
| CRC | 52 | 71.15 | 61.85 ± 13.58 | ||
| WirbelJ_2018 | Control | 65 | 56.92 | 55.97 ± 12.15 | DEU |
| CRC | 60 | 60.00 | 63.45 ± 12.64 | ||
| ThomasAM_2018b | Control | 28 | 57.14 | 57.81 ± 8.26 | ITA |
| CRC | 32 | 71.88 | 58.44 ± 8.39 | ||
| ThomasAM_2019c | Control | 40 | 60.00 | 63.23 ± 12.17 | JPN |
| CRC | 40 | 52.50 | 59.05 ± 12.83 | ||
| GuptaA_2019 | Control | 30 | 40.00 | 41.50 ± 16.90 | IND |
| CRC | 30 | 60.00 | 59.80 ± 7.81 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; He, Y.; Xia, L.; Yi, J.; Wang, Z.; Zhao, Y.; Song, X.; Li, J.; Liu, H.; Liang, X.; et al. Expansion of Colorectal Cancer Biomarkers Based on Gut Bacteria and Viruses. Cancers 2022, 14, 4662. https://doi.org/10.3390/cancers14194662
Zhang J, He Y, Xia L, Yi J, Wang Z, Zhao Y, Song X, Li J, Liu H, Liang X, et al. Expansion of Colorectal Cancer Biomarkers Based on Gut Bacteria and Viruses. Cancers. 2022; 14(19):4662. https://doi.org/10.3390/cancers14194662
Chicago/Turabian StyleZhang, Jia, Yangting He, Lu Xia, Jing Yi, Zhen Wang, Yingying Zhao, Xuemei Song, Jia Li, Hongli Liu, Xinjun Liang, and et al. 2022. "Expansion of Colorectal Cancer Biomarkers Based on Gut Bacteria and Viruses" Cancers 14, no. 19: 4662. https://doi.org/10.3390/cancers14194662
APA StyleZhang, J., He, Y., Xia, L., Yi, J., Wang, Z., Zhao, Y., Song, X., Li, J., Liu, H., Liang, X., Nie, S., & Liu, L. (2022). Expansion of Colorectal Cancer Biomarkers Based on Gut Bacteria and Viruses. Cancers, 14(19), 4662. https://doi.org/10.3390/cancers14194662






