The Drivers, Mechanisms, and Consequences of Genome Instability in HPV-Driven Cancers
Abstract
Simple Summary
Abstract
1. Introduction
2. The HPV Life Cycle and Oncogenesis
2.1. Expression of HPV Genes through the Viral Life Cycle
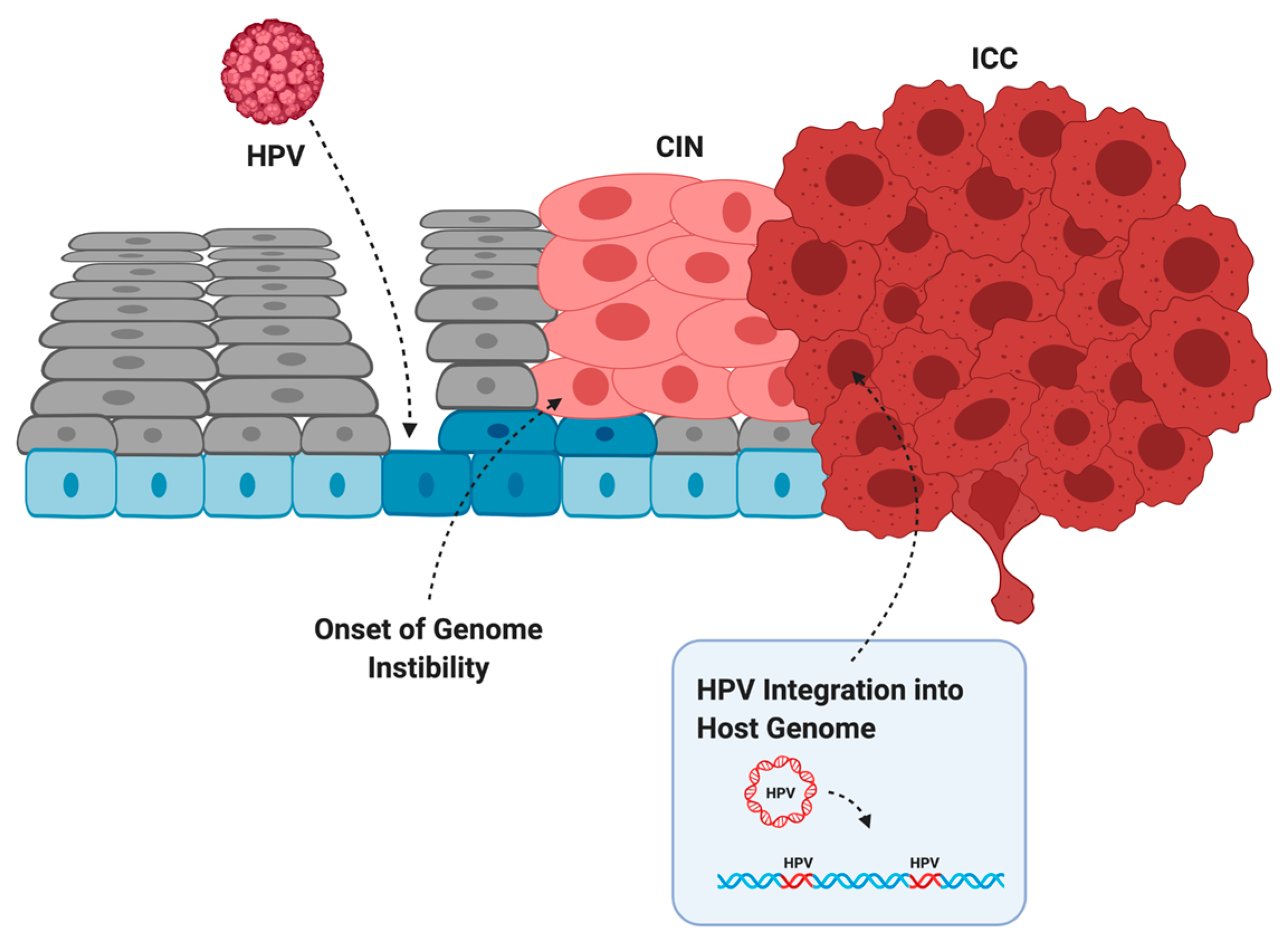
2.2. Transformation of Infected Cells into Cancer
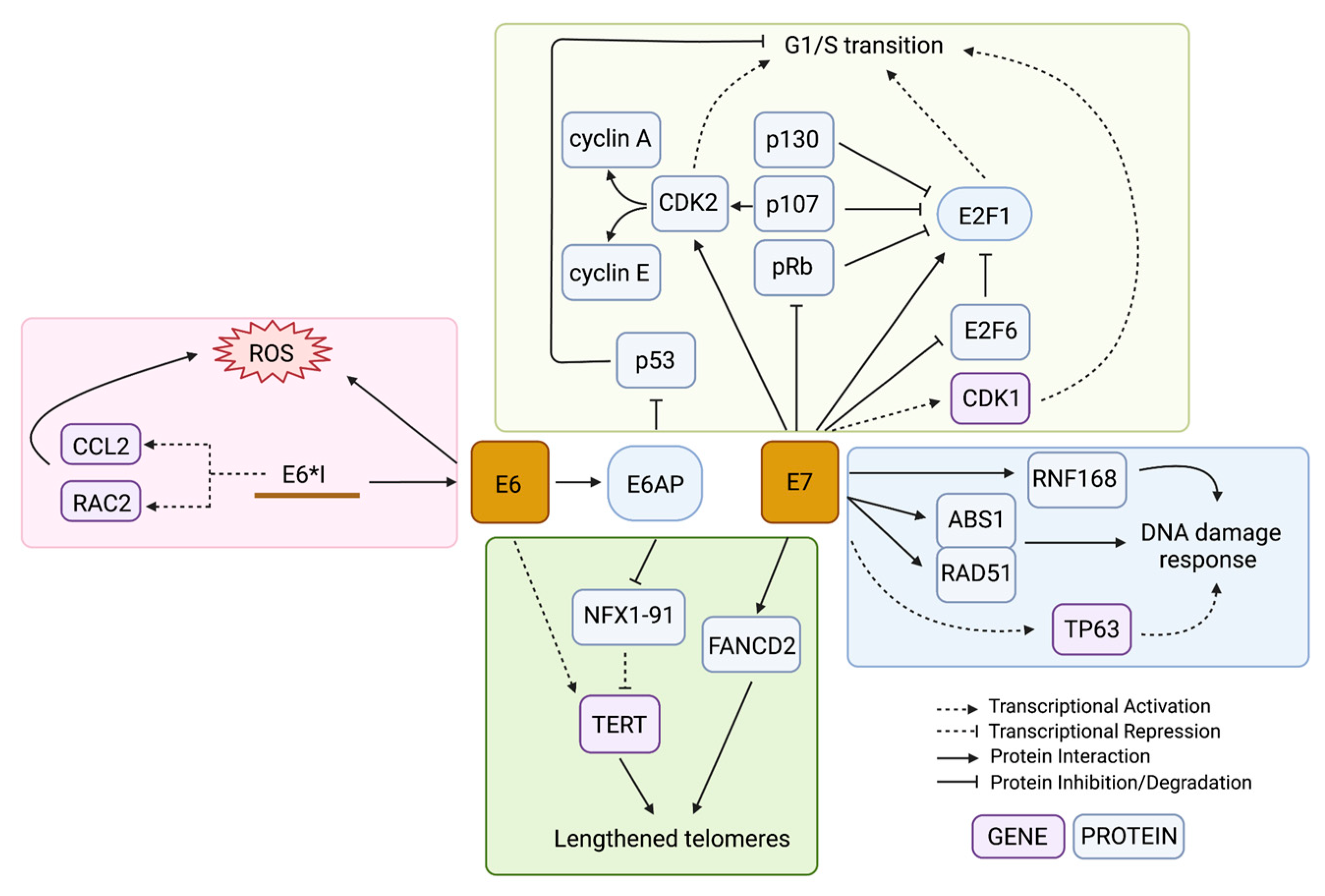
3. Direct Effects of the HPV Genes on Genome Instability
3.1. Cell Cycle Dysregulation Allows an Accumulation of Unchecked DNA Damage
3.2. Interactions with the Host DNA Damage Response
3.3. Generation of Oxidative Stress
3.4. Alteration of Telomeres
4. Indirect Effects of Viral Infection on Genome Instability
Genome Mutagenesis by APOBEC Enzymes
5. HPV-Independent Mutations That Cause Genome Instability
5.1. Mutation of DNA Repair Genes
5.2. Activating TERT Mutations
6. Genomic of the Integrated HPV Genome
6.1. Mechanisms of HPV Integration
6.2. Genome Instability at Regions of HPV Integration
7. Targeting Genome Instability in HPV-Driven Cancer Treatment
8. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Negrini, S.; Gorgoulis, V.G.; Halazonetis, T.D. Genomic instability—An evolving hallmark of cancer. Nat. Rev. Mol. Cell Biol. 2010, 11, 220–228. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A.; Warburton, A. The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathog. 2017, 13, e1006211. [Google Scholar] [CrossRef]
- Plummer, M.; de Martel, C.; Vignat, J.; Ferlay, J.; Bray, F.; Franceschi, S. Global burden of cancers attributable to infections in 2012: A synthetic analysis. Lancet Glob. Health 2016, 4, e609–e616. [Google Scholar] [CrossRef]
- Zapatka, M.; Pathogens, P.; Borozan, I.; Brewer, D.S.; Iskar, M.; Grundhoff, A.; Alawi, M.; Desai, N.; Sültmann, H.; Moch, H.; et al. The landscape of viral associations in human cancers. Nat. Genet. 2020, 52, 320–330. [Google Scholar] [CrossRef]
- de Sanjosé, S.; Serrano, B.; Tous, S.; Alejo, M.; Lloveras, B.; Quirós, B.; Clavero, O.; Vidal, A.; Ferrándiz-Pulido, C.; Pavón, M.A. Burden of human papillomavirus (HPV)-related cancers attributable to HPVs 6/11/16/18/31/33/45/52 and 58. JNCI Cancer Spectr. 2018, 2, ky045. [Google Scholar] [CrossRef]
- Berman, T.A.; Schiller, J.T. Human papillomavirus in cervical cancer and oropharyngeal cancer: One cause, two diseases. Cancer 2017, 123, 2219–2229. [Google Scholar] [CrossRef]
- Falcaro, M.; Castañon, A.; Ndlela, B.; Checchi, M.; Soldan, K.; Lopez-Bernal, J.; Elliss-Brookes, L.; Sasieni, P. The effects of the national HPV vaccination programme in England, UK, on cervical cancer and grade 3 cervical intraepithelial neoplasia incidence: A register-based observational study. Lancet 2021, 398, 2084–2092. [Google Scholar] [CrossRef]
- Bruni, L.; Diaz, M.; Barrionuevo-Rosas, L.; Herrero, R.; Bray, F.; Bosch, F.X.; de Sanjosé, S.; Castellsagué, X. Global estimates of human papillomavirus vaccination coverage by region and income level: A pooled analysis. Lancet Glob. Health 2016, 4, e453–e463. [Google Scholar] [CrossRef]
- Saraiya, M.; Unger, E.; Thompson, T.D.; Lynch, C.F.; Hernandez, B.Y.; Lyu, C.W.; Steinau, M.; Watson, M.; Wilkinson, E.J.; Hopenhayn, C.; et al. US Assessment of HPV Types in Cancers: Implications for Current and 9-Valent HPV Vaccines. JNCI J. Natl. Cancer Inst. 2015, 107, djv086. [Google Scholar] [CrossRef]
- De Martel, C.; Plummer, M.; Vignat, J.; Franceschi, S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. Int. J. Cancer 2017, 141, 664–670. [Google Scholar] [CrossRef]
- Jeggo, P.A.; Pearl, L.H.; Carr, A.M. DNA repair, genome stability and cancer: A historical perspective. Nat. Cancer 2015, 16, 35–42. [Google Scholar] [CrossRef]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and Molecular Landscape of DNA Damage Repair Deficiency across The Cancer Genome Atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef]
- Pleasance, E.; Titmuss, E.; Williamson, L.; Kwan, H.; Culibrk, L.; Zhao, E.Y.; Dixon, K.; Fan, K.; Bowlby, R.; Jones, M.R.; et al. Pan-cancer analysis of advanced patient tumors reveals interactions between therapy and genomic landscapes. Nat. Cancer 2020, 1, 452–468. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Ng, A.W.T.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.-L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef]
- Koh, G.; Degasperi, A.; Zou, X.; Momen, S.; Nik-Zainal, S. Mutational signatures: Emerging concepts, caveats and clinical applications. Nat. Cancer 2021, 21, 619–637. [Google Scholar] [CrossRef]
- Akagi, K.; Li, J.; Broutian, T.R.; Padilla-Nash, H.; Xiao, W.; Jiang, B.; Rocco, J.W.; Teknos, T.N.; Kumar, B.; Wangsa, D.; et al. Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Res. 2013, 24, 185–199. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Research Network; Albert Einstein College of Medicine; Analytical Biological Services; Barretos Cancer Hospital; Baylor College of Medicine; Beckman Research Institute of City of Hope; Buck Institute for Research on Aging; Canada’s Michael Smith Genome Sciences Centre; Harvard Medical School; Helen F. Graham Cancer Center & Research Institute at Christiana Care Health Services; et al. Integrated genomic and molecular characterization of cervical cancer. Nature 2017, 543, 378–384. [Google Scholar] [CrossRef]
- Parfenov, M.; Pedamallu, C.S.; Gehlenborg, N.; Freeman, S.; Danilova, L.; Bristow, C.A.; Lee, S.; Hadjipanayis, A.G.; Ivanova, E.V.; Wilkerson, M.D.; et al. Characterization of HPV and host genome interactions in primary head and neck cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 15544–15549. [Google Scholar] [CrossRef] [PubMed]
- Nulton, T.J.; Olex, A.L.; Dozmorov, M.; Morgan, I.M.; Windle, B. Analysis of The Cancer Genome Atlas sequencing data reveals novel properties of the human papillomavirus 16 genome in head and neck squamous cell carcinoma. Oncotarget 2017, 8, 17684–17699. [Google Scholar] [CrossRef] [PubMed]
- Egawa, N.; Egawa, K.; Griffin, H.M.; Doorbar, J. Human Papillomaviruses; Epithelial Tropisms, and the Development of Neoplasia. Viruses 2015, 7, 3863–3890. [Google Scholar] [CrossRef] [PubMed]
- D'Souza, G.; Burk, R.D.; Zhong, Y.; Minkoff, H.; Massad, L.S.; Xue, X.; Watts, D.H.; Anastos, K.; Palefsky, J.M.; Levine, A.M.; et al. Cervicovaginal human papillomavirus (HPV)-infection before and after hysterectomy: Evidence of different tissue tropism for oncogenic and nononcogenic HPV types in a cohort of HIV-positive and HIV-negative women. Int. J. Cancer 2011, 131, 1472–1478. [Google Scholar] [CrossRef]
- Mattox, A.K.; Roelands, J.; Saal, T.M.; Cheng, Y.; Rinchai, D.; Hendrickx, W.; Young, G.D.; Diefenbach, T.J.; Berger, A.E.; Westra, W.H.; et al. Myeloid Cells Are Enriched in Tonsillar Crypts, Providing Insight into the Viral Tropism of Human Papillomavirus. Am. J. Pathol. 2021, 191, 1774–1786. [Google Scholar] [CrossRef]
- Chen, Z.; Schiffman, M.; Herrero, R.; DeSalle, R.; Anastos, K.; Segondy, M.; Sahasrabuddhe, V.V.; Gravitt, P.E.; Hsing, A.W.; Burk, R.D. Evolution and taxonomic classification of alphapapillomavirus 7 complete genomes: HPV18, HPV39, HPV45, HPV59, HPV68 and HPV70. PLoS ONE 2013, 8, e72565. [Google Scholar] [CrossRef]
- Strati, K. Changing Stem Cell Dynamics during Papillomavirus Infection: Potential Roles for Cellular Plasticity in the Viral Lifecycle and Disease. Viruses 2017, 9, 221. [Google Scholar] [CrossRef]
- Hong, S.; Laimins, L.A. Regulation of the life cycle of HPVs by differentiation and the DNA damage response. Futur. Microbiol. 2013, 8, 1547–1557. [Google Scholar] [CrossRef]
- Parish, J.; Bean, A.M.; Park, R.B.; Androphy, E.J. ChlR1 Is Required for Loading Papillomavirus E2 onto Mitotic Chromosomes and Viral Genome Maintenance. Mol. Cell 2006, 24, 867–876. [Google Scholar] [CrossRef]
- King, L.E.; Fisk, J.C.; Dornan, E.S.; Donaldson, M.M.; Melendy, T.; Morgan, I.M. Human papillomavirus E1 and E2 mediated DNA replication is not arrested by DNA damage signalling. Virology 2010, 406, 95–102. [Google Scholar] [CrossRef]
- Sakakibara, N.; Chen, D.; McBride, A.A. Papillomaviruses Use Recombination-Dependent Replication to Vegetatively Amplify Their Genomes in Differentiated Cells. PLoS Pathog. 2013, 9, e1003321. [Google Scholar] [CrossRef] [PubMed]
- Estêvão, D.; Costa, N.R.; Gil da Costa, R.M.; Medeiros, R. Hallmarks of HPV carcinogenesis: The role of E6, E7 and E5 oncoproteins in cellular malignancy. Biochim. Biophys. Acta (BBA)-Gene Regul. Mech. 2019, 1862, 153–162. [Google Scholar] [CrossRef] [PubMed]
- Klingelhutz, A.J.; Roman, A. Cellular transformation by human papillomaviruses: Lessons learned by comparing high- and low-risk viruses. Virology 2012, 424, 77–98. [Google Scholar] [CrossRef] [PubMed]
- Chaiwongkot, A.; Vinokurova, S.; Pientong, C.; Ekalaksananan, T.; Kongyingyoes, B.; Kleebkaow, P.; Chumworathayi, B.; Patarapadungkit, N.; Reuschenbach, M.; von Knebel Doeberitz, M. Differential methylation of E2 binding sites in episomal and integrated HPV 16 genomes in preinvasive and invasive cervical lesions. Int. J. Cancer 2013, 132, 2087–2094. [Google Scholar] [CrossRef]
- Graham, S.V.; Faizo, A.A.A. Control of human papillomavirus gene expression by alternative splicing. Virus Res. 2016, 231, 83–95. [Google Scholar] [CrossRef]
- Zhou, J.; Sun, X.Y.; Stenzel, D.J.; Frazer, I. Expression of vaccinia recombinant HPV 16 L1 and L2 ORF proteins in epithelial cells is sufficient for assembly of HPV virion-like particles. Virology 1991, 185, 251–257. [Google Scholar] [CrossRef]
- Johansson, C.; Somberg, M.; Li, X.; Winquist, E.B.; Fay, J.; Ryan, F.; Pim, D.; Banks, L.; Schwartz, S. HPV-16 E2 contributes to induction of HPV-16 late gene expression by inhibiting early polyadenylation. EMBO J. 2012, 31, 3212–3227. [Google Scholar] [CrossRef]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25 (Suppl. 1), 2–23. [Google Scholar] [CrossRef]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine 2012, 30 (Suppl. 5), F55–F70. [Google Scholar] [CrossRef]
- Kreimer, A.R.; Chaturvedi, A.K. HPV-associated Oropharyngeal Cancers—Are They Preventable? Cancer Prev. Res. 2011, 4, 1346–1349. [Google Scholar] [CrossRef]
- Safaeian, M.; Solomon, D.; Castle, P.E. Cervical Cancer Prevention—Cervical Screening: Science in Evolution. Obstet. Gynecol. Clin. N. Am. 2007, 34, 739–760. [Google Scholar] [CrossRef]
- Wilting, S.; Steenbergen, R.D. Molecular events leading to HPV-induced high grade neoplasia. Papillomavirus Res. 2016, 2, 85–88. [Google Scholar] [CrossRef]
- Hu, Z.; Zhu, D.; Wang, W.; Li, W.; Jia, W.; Zeng, X.; Ding, W.; Yu, L.; Wang, X.; Wang, L.; et al. Genome-wide profiling of HPV integration in cervical cancer identifies clustered genomic hot spots and a potential microhomology-mediated integration mechanism. Nat. Genet. 2015, 47, 158–163. [Google Scholar] [CrossRef]
- Choo, K.-B.; Pan, C.-C.; Han, S.-H. Integration of human papillomavirus type 16 into cellular DNA of cervical carcinoma: Preferential deletion of the E2 gene and invariable retention of the long control region and the E6/E7 open reading frames. Virology 1987, 161, 259–261. [Google Scholar] [CrossRef]
- Scarth, J.A.; Patterson, M.R.; Morgan, E.L.; Macdonald, A. The human papillomavirus oncoproteins: A review of the host pathways targeted on the road to transformation. J. Gen. Virol. 2021, 102, 001540. [Google Scholar] [CrossRef]
- Kalliala, I.; Dyba, T.; Nieminen, P.; Hakulinen, T.; Anttila, A. Mortality in a long-term follow-up after treatment of CIN. Int. J. Cancer 2009, 126, 224–231. [Google Scholar] [CrossRef]
- Thomas, M.; Pim, D.; Banks, L. The role of the E6-p53 interaction in the molecular pathogenesis of HPV. Oncogene 1999, 18, 7690–7700. [Google Scholar] [CrossRef]
- Jones, D.; Thompson, D.; Munger, K. Destabilization of the RB Tumor Suppressor Protein and Stabilization of p53 Contribute to HPV Type 16 E7-Induced Apoptosis. Virology 1997, 239, 97–107. [Google Scholar] [CrossRef]
- Hawley-Nelson, P.; Vousden, K.; Hubbert, N.; Lowy, D.; Schiller, J. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989, 8, 3905–3910. [Google Scholar] [CrossRef]
- Hudson, J.B.; Bedell, M.A.; McCance, D.J.; Laiminis, L.A. Immortalization and altered differentiation of human keratinocytes in vitro by the E6 and E7 open reading frames of human papillomavirus type. J. Virol. 1990, 64, 519–526. [Google Scholar] [CrossRef]
- Sedman, S.A.; Barbosa, M.S.; Vass, W.C.; Hubbert, N.L.; Haas, J.A.; Lowy, D.R.; Schiller, J.T. The full-length E6 protein of human papillomavirus type 16 has transforming and trans-activating activities and cooperates with E7 to immortalize keratinocytes in culture. J. Virol. 1991, 65, 4860–4866. [Google Scholar] [CrossRef] [PubMed]
- Sashiyama, H.; Shino, Y.; Kawamata, Y.; Tomita, Y.; Ogawa, N.; Shimada, H.; Kobayashi, S.; Asano, T.; Ochiai, T.; Shirasawa, H. Immortalization of human esophageal keratinocytes by E6 and E7 of human papillomavirus type 16. Int. J. Oncol. 2001, 19, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Halbert, C.L.; Demers, G.W.; Galloway, D.A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J. Virol. 1991, 65, 473–478. [Google Scholar] [CrossRef]
- Reznikoff, C.A.; Belair, C.; Savelieva, E.; Zhai, Y.; Pfeifer, K.; Yeager, T.; Thompson, K.J.; De Vries, S.; Bindley, C.; Newton, M.A. Long-term genome stability and minimal genotypic and phenotypic alterations in HPV16 E7-, but not E6-, immortalized human uroepithelial cells. Genes Dev. 1994, 8, 2227–2240. [Google Scholar] [CrossRef] [PubMed]
- Smith, M.L.; Chen, I.T.; Zhan, Q.; O’Connor, P.M.; Fornace, A.J., Jr. Involvement of the p53 tumor suppressor in repair of u.v.-type DNA damage. Oncogene 1995, 10, 1053–1059. [Google Scholar]
- Beaudenon, S.; Huibregtse, J.M. HPV E6, E6AP and cervical cancer. BMC Biochem. 2008, 9, S4. [Google Scholar] [CrossRef]
- Martinez-Zapien, D.; Ruiz, F.X.; Poirson, J.; Mitschler, A.; Ramirez, J.; Forster, A.; Cousido-Siah, A.; Masson, M.; Pol, S.V.; Podjarny, A.; et al. Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53. Nature 2016, 529, 541–545. [Google Scholar] [CrossRef]
- Fu, L.; Van Doorslaer, K.; Chen, Z.; Ristriani, T.; Masson, M.; Travé, G.; Burk, R.D. Degradation of p53 by Human Alphapapillomavirus E6 Proteins Shows a Stronger Correlation with Phylogeny than Oncogenicity. PLoS ONE 2010, 5, e12816. [Google Scholar] [CrossRef]
- Willemsen, A.; Bravo, I.G. Origin and evolution of papillomavirus (onco)genes and genomes. Philos. Trans. R. Soc. B Biol. Sci. 2019, 374, 20180303. [Google Scholar] [CrossRef]
- Li, S.; Hong, X.; Wei, Z.; Xie, M.; Li, W.; Liu, G.; Guo, H.; Yang, J.; Wei, W.; Zhang, S. Ubiquitination of the HPV Oncoprotein E6 Is Critical for E6/E6AP-Mediated p53 Degradation. Front. Microbiol. 2019, 10, 2483. [Google Scholar] [CrossRef]
- Gu, Z.; Pim, D.; Labrecque, S.; Banks, L.; Matlashewski, G. DNA damage induced p53 mediated transcription is inhibited by human papillomavirus type 18 E6. Oncogene 1994, 9, 629–633. [Google Scholar]
- Zhou, G.; Liu, Z.; Myers, J.N. TP53 Mutations in Head and Neck Squamous Cell Carcinoma and Their Impact on Disease Progression and Treatment Response. J. Cell. Biochem. 2016, 117, 2682–2692. [Google Scholar] [CrossRef]
- Muller, P.A.J.; Vousden, K.H. p53 mutations in cancer. Nat. Cell Biol. 2013, 15, 2–8. [Google Scholar] [CrossRef]
- Barrow-Laing, L.; Chen, W.; Roman, A. Low- and high-risk human papillomavirus E7 proteins regulate p130 differently. Virology 2010, 400, 233–239. [Google Scholar] [CrossRef]
- Jones, D.L.; Alani, R.M.; Münger, K. The human papillomavirus E7 oncoprotein can uncouple cellular differentiation and proliferation in human keratinocytes by abrogating p21Cip1-mediated inhibition of cdk2. Genes Dev. 1997, 11, 2101–2111. [Google Scholar] [CrossRef]
- Hwang, S.G.; Lee, D.; Kim, J.; Seo, T.; Choe, J. Human Papillomavirus Type 16 E7 Binds to E2F1 and Activates E2F1-driven Transcription in a Retinoblastoma Protein-independent Manner. J. Biol. Chem. 2002, 277, 2923–2930. [Google Scholar] [CrossRef]
- McLaughlin-Drubin, M.E.; Huh, K.-W.; Münger, K. Human Papillomavirus Type 16 E7 Oncoprotein Associates with E2F6. J. Virol. 2008, 82, 8695–8705. [Google Scholar] [CrossRef]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A.M. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Zerfass, K.; Schulze, A.; Spitkovsky, D.; Friedman, V.; Henglein, B.; Jansen-Dürr, P. Sequential activation of cyclin E and cyclin A gene expression by human papillomavirus type 16 E7 through sequences necessary for transformation. J. Virol. 1995, 69, 6389–6399. [Google Scholar] [CrossRef]
- Martin, L.G.; Demers, G.W.; Galloway, D.A. Disruption of the G1/S transition in human papillomavirus type 16 E7-expressing human cells is associated with altered regulation of cyclin E. J. Virol. 1998, 72, 975–985. [Google Scholar] [CrossRef]
- He, W.; Staples, D.; Smith, C.; Fisher, C. Direct Activation of Cyclin-Dependent Kinase 2 by Human Papillomavirus E7. J. Virol. 2003, 77, 10566–10574. [Google Scholar] [CrossRef] [PubMed]
- McIntyre, M.C.; Ruesch, M.N.; Laimins, L.A. Human papillomavirus E7 oncoproteins bind a single form of cyclin E in a complex with cdk2 and p107. Virology 1996, 215, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Chen, J.J. Role of Cdk1 in DNA damage-induced G1 checkpoint abrogation by the human papillomavirus E7 oncogene. Cell Cycle 2014, 13, 3249–3259. [Google Scholar] [CrossRef] [PubMed]
- Qiao, L.; Zheng, J.; Tian, Y.; Zhang, Q.; Wang, X.; Chen, J.J.; Zhang, W. Regulator of chromatin condensation 1 abrogates the G1 cell cycle checkpoint via Cdk1 in human papillomavirus E7-expressing epithelium and cervical cancer cells. Cell Death Dis. 2018, 9, 583. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Qiao, L.; Liu, R.; Han, X.; Zhang, W. Phosphorylation of RCC1 on Serine 11 Facilitates G1/S Transition in HPV E7-Expressing Cells. Biomolecules 2021, 11, 995. [Google Scholar] [CrossRef]
- Stöppler, M.C.; Straight, S.W.; Tsao, G.; Schlegel, R.; Mccance, D.J. The E5 Gene of HPV-16 Enhances Keratinocyte Immortalization by Full-Length DNA. Virology 1996, 223, 251–254. Available online: https://www.sciencedirect.com/science/article/pii/S00426822 (accessed on 15 September 2022). [CrossRef]
- Maufort, J.P.; Shai, A.; Pitot, H.C.; Lambert, P.F. A Role for HPV16 E5 in Cervical CarcinogenesisHPV16 E5 in Cervical Cancer. Cancer Res. 2010, 70, 2924–2931. Available online: https://aacrjournals.org/cancerres/article-abstract/70/7/2924/561812 (accessed on 15 September 2022). [CrossRef]
- Straight, S.W.; Hinkle, P.M.; Jewers, R.J.; McCance, D.J. The E5 oncoprotein of human papillomavirus type 16 transforms fibroblasts and effects the downregulation of the epidermal growth factor receptor in keratinocytes. J. Virol. 1993, 67, 4521–4532. [Google Scholar] [CrossRef]
- Straight, S.W.; Herman, B.; McCance, D.J. The E5 oncoprotein of human papillomavirus type 16 inhibits the acidification of endosomes in human keratinocytes. J. Virol. 1995, 69, 3185–3192. [Google Scholar] [CrossRef]
- Deane, R.; Schäfer, W.; Zimmermann, H.P.; Mueller, L.; Görlich, D.; Prehn, S.; Ponstingl, H.; Bischoff, F.R. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-beta but interacts differently with RanBP1. Mol. Cell. Biol. 1997, 17, 5087–5096. [Google Scholar] [CrossRef]
- Crusius, K.; Rodriguez, I.; Alonso, A. The human papillomavirus type 16 E5 protein modulates ERK1/2 and p38 MAP kinase activation by an EGFR-independent process in stressed human keratinocytes. Virus Genes 2000, 20, 65–69. [Google Scholar] [CrossRef]
- Crusius, K.; Auvinen, E.; Alonso, A. Enhancement of EGF- and PMA-mediated MAP kinase activation in cells expressing the human papillomavirus type 16 E5 protein. Oncogene 1997, 15, 1437–1444. [Google Scholar] [CrossRef]
- Williams, S.M.G.; Disbrow, G.L.; Schlegel, R.; Lee, D.; Threadgill, D.W.; Lambert, P.F. Requirement of Epidermal Growth Factor Receptor for Hyperplasia Induced by E5, a High-Risk Human Papillomavirus Oncogene. Cancer Res. 2005, 65, 6534–6542. [Google Scholar] [CrossRef]
- Zhang, B.; Spandau, D.F.; Roman, A. E5 Protein of Human Papillomavirus Type 16 Protects Human Foreskin Keratinocytes from UV B-Irradiation-Induced Apoptosis. J. Virol. 2002, 76, 220–231. [Google Scholar] [CrossRef]
- Um, S.H.; Mundi, N.; Yoo, J.; Palma, D.A.; Fung, K.; MacNeil, D.; Wehrli, B.; Mymryk, J.S.; Barrett, J.W.; Nichols, A.C. Variable expression of the forgotten oncogene E5 in HPV-positive oropharyngeal cancer. J. Clin. Virol. 2014, 61, 94–100. [Google Scholar] [CrossRef]
- Chang, J.L.; Tsao, Y.P.; Liu, D.W.; Huang, S.J.; Lee, W.H.; Chen, S.L. The expression of HPV-16 E5 protein in squamous neoplastic changes in the uterine cervix. J. Biomed. Sci. 2001, 8, 206–213. [Google Scholar] [CrossRef]
- Duensing, S.; Münger, K. The human papillomavirus type 16 E6 and E7 oncoproteins independently induce numerical and structural chromosome instability. Cancer Res. 2002, 62, 7075–7082. [Google Scholar]
- Duensing, S.; Duensing, A.; Flores, E.R.; Do, A.; Lambert, P.F.; Münger, K. Centrosome Abnormalities and Genomic Instability by Episomal Expression of Human Papillomavirus Type 16 in Raft Cultures of Human Keratinocytes. J. Virol. 2001, 75, 7712–7716. [Google Scholar] [CrossRef]
- Krejci, L.; Altmannova, V.; Spirek, M.; Zhao, X. Homologous recombination and its regulation. Nucleic Acids Res. 2012, 40, 5795–5818. [Google Scholar] [CrossRef]
- Lavin, M.F.; Kozlov, S.; Gatei, M.; Kijas, A.W. ATM-Dependent Phosphorylation of All Three Members of the MRN Complex: From Sensor to Adaptor. Biomolecules 2015, 5, 2877–2902. [Google Scholar] [CrossRef]
- Cannavo, E.; Cejka, P. Sae2 promotes dsDNA endonuclease activity within Mre11-Rad50-Xrs2 to resect DNA breaks. Nature 2014, 514, 122–125. [Google Scholar] [CrossRef] [PubMed]
- López-Contreras, A.J.; Fernandez-Capetillo, O. The ATR barrier to replication-born DNA damage. DNA Repair 2010, 9, 1249–1255. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.-H.; Lindsey-Boltz, L.A.; Kemp, M.; Mason, A.C.; Wold, M.S.; Sancar, A. Reconstitution of RPA-covered single-stranded DNA-activated ATR-Chk1 signaling. Proc. Natl. Acad. Sci. USA 2010, 107, 13660–13665. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, C.C.; Laimins, L.A. Human Papillomavirus and the DNA Damage Response: Exploiting Host Repair Pathways for Viral Replication. Viruses 2017, 9, 232. [Google Scholar] [CrossRef] [PubMed]
- Spriggs, C.C.; Blanco, L.Z.; Maniar, K.P.; Laimins, L.A. Expression of HPV-induced DNA Damage Repair Factors Correlates with CIN Progression. Int. J. Gynecol. Pathol. 2019, 38, 1–10. [Google Scholar] [CrossRef]
- Hong, S.; Cheng, S.; Iovane, A.; Laimins, L.A. STAT-5 Regulates Transcription of the Topoisomerase IIβ-Binding Protein 1 (TopBP1) Gene to Activate the ATR Pathway and Promote Human Papillomavirus Replication. mBio 2015, 6, e02006-15. [Google Scholar] [CrossRef]
- Kono, T.; Hoover, P.; Poropatich, K.; Paunesku, T.; Mittal, B.B.; Samant, S.; Laimins, L.A. Activation of DNA damage repair factors in HPV positive oropharyngeal cancers. Virology 2020, 547, 27–34. [Google Scholar] [CrossRef]
- Holcomb, A.J.; Brown, L.; Tawfik, O.; Madan, R.; Shnayder, Y.; Thomas, S.M.; Wallace, N.A. DNA repair gene expression is increased in HPV positive head and neck squamous cell carcinomas. Virology 2020, 548, 174–181. [Google Scholar] [CrossRef]
- Moody, C.A.; Laimins, L.A. Human Papillomaviruses Activate the ATM DNA Damage Pathway for Viral Genome Amplification upon Differentiation. PLoS Pathog. 2009, 5, e1000605. [Google Scholar] [CrossRef]
- Anacker, D.C.; Gautam, D.; Gillespie, K.A.; Chappell, W.H.; Moody, C.A. Productive Replication of Human Papillomavirus 31 Requires DNA Repair Factor Nbs1. J. Virol. 2014, 88, 8528–8544. [Google Scholar] [CrossRef]
- Sakakibara, N.; Mitra, R.; McBride, A.A. The Papillomavirus E1 Helicase Activates a Cellular DNA Damage Response in Viral Replication Foci. J. Virol. 2011, 85, 8981–8995. [Google Scholar] [CrossRef]
- Das, D.; Bristol, M.L.; Smith, N.W.; James, C.D.; Wang, X.; Pichierri, P.; Morgan, I.M. Werner Helicase Control of Human Papillomavirus 16 E1-E2 DNA Replication Is Regulated by SIRT1 Deacetylation. mBio 2019, 10, e00263-19. [Google Scholar] [CrossRef]
- James, C.D.; Das, D.; Morgan, E.L.; Otoa, R.; Macdonald, A.; Morgan, I.M. Werner Syndrome Protein (WRN) Regulates Cell Proliferation and the Human Papillomavirus 16 Life Cycle during Epithelial Differentiation. mSphere 2020, 5, e00858-20. [Google Scholar] [CrossRef]
- Wallace, N.A.; Khanal, S.; Robinson, K.L.; Wendel, S.O.; Messer, J.J.; Galloway, D.A. High-Risk Alphapapillomavirus Oncogenes Impair the Homologous Recombination Pathway. J. Virol. 2017, 91, e01084-17. [Google Scholar] [CrossRef]
- Johnson, B.A.; Aloor, H.L.; Moody, C.A. The Rb binding domain of HPV31 E7 is required to maintain high levels of DNA repair factors in infected cells. Virology 2017, 500, 22–34. [Google Scholar] [CrossRef]
- Eldakhakhny, S.; Zhou, Q.; Crosbie, E.; Sayan, B.S. Human papillomavirus E7 induces p63 expression to modulate DNA damage response. Cell Death Dis. 2018, 9, 127. [Google Scholar] [CrossRef]
- Wallace, N.A. Catching HPV in the Homologous Recombination Cookie Jar. Trends Microbiol. 2019, 28, 191–201. [Google Scholar] [CrossRef]
- Gusho, E.; Laimins, L. Human Papillomaviruses Target the DNA Damage Repair and Innate Immune Response Pathways to Allow for Persistent Infection. Viruses 2021, 13, 1390. Available online: https://www.mdpi.com/1999-4915/13/7/1390 (accessed on 15 September 2022). [CrossRef]
- Harberts, E.; Gaspari, A.A. TLR Signaling and DNA Repair: Are They Associated? J. Investig. Dermatol. 2013, 133, 296–302. Available online: https://www.sciencedirect.com/science/article/pii/S0022202X1536108X (accessed on 15 September 2022). [CrossRef]
- Li, T.; Chen, Z.J. The cGAS–cGAMP–STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 2018, 215, 1287–1299. Available online: https://rupress.org/jem/article-abstract/215/5/1287/42581 (accessed on 15 September 2022). [CrossRef]
- Saulters, E.; Woolley, J.F.; Varadarajan, S.; Jones, T.M.; Dahal, L.N. STINGing Viral Tumors: What We Know from Head and Neck Cancers. Cancer Res. 2021, 81, 3945–3952. Available online: https://aacrjournals.org/cancerres/article-abstract/81/15/3945/670234 (accessed on 15 September 2022). [CrossRef] [PubMed]
- Yang, X.; Cheng, Y.; Li, C. The role of TLRs in cervical cancer with HPV infection: A review. Signal Transduct. Target. Ther. 2017, 2, 17055. Available online: https://www.nature.com/articles/sigtrans201755 (accessed on 15 September 2022). [CrossRef] [PubMed]
- Gagliardi, A.; Porter, V.L.; Zong, Z.; Bowlby, R.; Titmuss, E.; Namirembe, C.; Griner, N.B.; Petrello, H.; Bowen, J.; Chan, S.K.; et al. Analysis of Ugandan cervical carcinomas identifies human papillomavirus clade–specific epigenome and transcriptome landscapes. Nat. Genet. 2020, 52, 800–810. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Ma, L.; Jones, T.; Palomero, L.; Pujana, M.A.; Martinez-Ruiz, H.; Ha, P.K.; Murnane, J.; Cuartas, I.; Seoane, J.; et al. Subjugation of TGFβ Signaling by Human Papilloma Virus in Head and Neck Squamous Cell Carcinoma Shifts DNA Repair from Homologous Recombination to Alternative End Joining. Clin. Cancer Res. 2018, 24, 6001–6014. [Google Scholar] [CrossRef] [PubMed]
- Mehta, K.; Laimins, L. Human Papillomaviruses Preferentially Recruit DNA Repair Factors to Viral Genomes for Rapid Repair and Amplification. mBio 2018, 9, e00064-18. [Google Scholar] [CrossRef]
- Sitz, J.; Blanchet, S.A.; Gameiro, S.F.; Biquand, E.; Morgan, T.M.; Galloy, M.; Dessapt, J.; Lavoie, E.G.; Blondeau, A.; Smith, B.C.; et al. Human papillomavirus E7 oncoprotein targets RNF168 to hijack the host DNA damage response. Proc. Natl. Acad. Sci. USA 2019, 116, 19552–19562. [Google Scholar] [CrossRef]
- Moloney, J.N.; Cotter, T.G. ROS signalling in the biology of cancer. Semin. Cell Dev. Biol. 2018, 80, 50–64. [Google Scholar] [CrossRef]
- Thanan, R.; Oikawa, S.; Hiraku, Y.; Ohnishi, S.; Mariko, M.; Pinlaor, S.; Yongvanit, P.; Kawanishi, S.; Murata, M. Oxidative Stress and Its Significant Roles in Neurodegenerative Diseases and Cancer. Int. J. Mol. Sci. 2014, 16, 193–217. [Google Scholar] [CrossRef]
- Kawanishi, S.; Ohnishi, S.; Ma, N.; Hiraku, Y.; Oikawa, S.; Murata, M. Nitrative and oxidative DNA damage in infection-related carcinogenesis in relation to cancer stem cells. Genes Environ. 2016, 38, 26. [Google Scholar] [CrossRef]
- Kuchino, Y.; Mori, F.; Kasai, H.; Inoue, H.; Iwai, S.; Miura, K.; Ohtsuka, E.; Nishimura, S. Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature 1987, 327, 77–79. [Google Scholar] [CrossRef]
- Barciszewska, A.-M.; Giel-Pietraszuk, M.; Perrigue, P.M.; Naskręt-Barciszewska, M. Total DNA Methylation Changes Reflect Random Oxidative DNA Damage in Gliomas. Cells 2019, 8, 1065. [Google Scholar] [CrossRef]
- Hiraku, Y.; Tabata, T.; Ma, N.; Murata, M.; Ding, X.; Kawanishi, S. Nitrative and oxidative DNA damage in cervical intraepithelial neoplasia associated with human papilloma virus infection. Cancer Sci. 2007, 98, 964–972. [Google Scholar] [CrossRef]
- Visalli, G.; Riso, R.; Facciolà, A.; Mondello, P.; Caruso, C.; Picerno, I.; Di Pietro, A.; Spataro, P.; Bertuccio, M.P. Higher levels of oxidative DNA damage in cervical cells are correlated with the grade of dysplasia and HPV infection. J. Med. Virol. 2015, 88, 336–344. [Google Scholar] [CrossRef]
- Cruz-Gregorio, A.; Manzo-Merino, J.; Gonzaléz-García, M.C.; Pedraza-Chaverri, J.; Medina-Campos, O.N.; Valverde, M.; Rojas, E.; Rodríguez-Sastre, M.A.; Garcia-Cuellar, C.M.; Lizano, M. Human Papillomavirus Types 16 and 18 Early-expressed Proteins Differentially Modulate the Cellular Redox State and DNA Damage. Int. J. Biol. Sci. 2018, 14, 21–35. [Google Scholar] [CrossRef]
- Paget-Bailly, P.; Meznad, K.; Bruyère, D.; Perrard, J.; Herfs, M.; Jung, A.C.; Mougin, C.; Prétet, J.-L.; Baguet, A. Comparative RNA sequencing reveals that HPV16 E6 abrogates the effect of E6*I on ROS metabolism. Sci. Rep. 2019, 9, 5938. [Google Scholar] [CrossRef]
- Williams, V.M.; Filippova, M.; Filippov, V.; Payne, K.; Duerksen-Hughes, P. Human Papillomavirus Type 16 E6* Induces Oxidative Stress and DNA Damage. J. Virol. 2014, 88, 6751–6761. [Google Scholar] [CrossRef]
- Olmedo-Nieva, L.; Muñoz-Bello, J.O.; Contreras-Paredes, A.; Lizano, M. The Role of E6 Spliced Isoforms (E6*) in Human Papillomavirus-Induced Carcinogenesis. Viruses 2018, 10, 45. [Google Scholar] [CrossRef]
- Cornelissen, M.T.; Smits, H.L.; Briët, M.A.; van den Tweel, J.G.; Struyk, A.P.; van der Noordaa, J.; ter Schegget, J. Uniformity of the splicing pattern of the E6/E7 transcripts in human papillomavirus type 16-transformed human fibroblasts, human cervical premalignant lesions and carcinomas. J. Gen. Virol. 1990, 71 Pt 5, 1243–1246. [Google Scholar] [CrossRef]
- Tang, S.; Tao, M.; McCoy, J.P., Jr.; Zheng, Z.-M. The E7 Oncoprotein Is Translated from Spliced E6*I Transcripts in High-Risk Human Papillomavirus Type 16- or Type 18-Positive Cervical Cancer Cell Lines via Translation Reinitiation. J. Virol. 2006, 80, 4249–4263. [Google Scholar] [CrossRef]
- Pim, D.; Massimi, P.; Banks, L. Alternatively spliced HPV-18 E6* protein inhibits E6 mediated degradation of p53 and suppresses transformed cell growth. Oncogene 1997, 15, 257–264. [Google Scholar] [CrossRef]
- Marullo, R.; Werner, E.; Zhang, H.; Chen, G.Z.; Shin, D.M.; Doetsch, P.W. HPV16 E6 and E7 proteins induce a chronic oxidative stress response via NOX2 that causes genomic instability and increased susceptibility to DNA damage in head and neck cancer cells. Carcinogenesis 2015, 36, 1397–1406. [Google Scholar] [CrossRef]
- Aubert, G.; Lansdorp, P.M. Telomeres and aging. Physiol. Rev. 2008, 88, 557–579. [Google Scholar] [CrossRef]
- Vinagre, J.; Almeida, A.; Pópulo, H.; Batista, R.; Lyra, J.; Pinto, V.; Coelho, R.; Celestino, R.; Prazeres, H.; Lima, L.; et al. Frequency of TERT promoter mutations in human cancers. Nat. Commun. 2013, 4, 2185. [Google Scholar] [CrossRef]
- Shay, J.W.; Wright, W.E. Senescence and immortalization: Role of telomeres and telomerase. Carcinogenesis 2005, 26, 867–874. [Google Scholar] [CrossRef]
- Cheung, A.L.M.; Deng, W. Telomere dysfunction, genome instability and cancer. Front. Biosci. 2008, 13, 2075–2090. [Google Scholar] [CrossRef] [PubMed]
- Plug-DeMaggio, A.W.; Sundsvold, T.; Wurscher, M.A.; Koop, J.I.; Klingelhutz, A.J.; McDougall, J.K. Telomere erosion and chromosomal instability in cells expressing the HPV oncogene 16E. Oncogene 2004, 23, 3561–3571. [Google Scholar] [CrossRef] [PubMed]
- Spardy, N.; Duensing, A.; Hoskins, E.E.; Wells, S.I.; Duensing, S. HPV-16 E7 Reveals a Link between DNA Replication Stress, Fanconi Anemia D2 Protein, and Alternative Lengthening of Telomere–Associated Promyelocytic Leukemia Bodies. Cancer Res. 2008, 68, 9954–9963. [Google Scholar] [CrossRef] [PubMed]
- Widschwendter, A.; Müller, H.M.; Hubalek, M.M.; Wiedemair, A.; Fiegl, H.; Goebel, G.; Mueller-Holzner, E.; Marth, C.; Widschwendter, M. Methylation status and expression of human telomerase reverse transcriptase in ovarian and cervical cancer. Gynecol. Oncol. 2004, 93, 407–416. [Google Scholar] [CrossRef]
- McMurray, H.; McCance, D.J. Human Papillomavirus Type 16 E6 Activates TERT Gene Transcription through Induction of c-Myc and Release of USF-Mediated Repression. J. Virol. 2003, 77, 9852–9861. [Google Scholar] [CrossRef]
- Gewin, L.; Galloway, D.A. E Box-dependent activation of telomerase by human papillomavirus type 16 E6 does not require induction of c-myc. J. Virol. 2001, 75, 7198–7201. [Google Scholar] [CrossRef]
- Veldman, T.; Horikawa, I.; Barrett, J.C.; Schlegel, R. Transcriptional Activation of the Telomerase hTERT Gene by Human Papillomavirus Type 16 E6 Oncoprotein. J. Virol. 2001, 75, 4467–4472. [Google Scholar] [CrossRef] [PubMed]
- Gewin, L.; Myers, H.; Kiyono, T.; Galloway, D.A. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes Dev. 2004, 18, 2269–2282. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Zhao, L.-J.; Zhao, C.; Zhang, G.; Zhao, Y.; Li, J.-R.; Li, X.-P.; Wei, L.-H. Hypomethylated CpG around the transcription start site enables TERT expression and HPV16 E6 regulates TERT methylation in cervical cancer cells. Gynecol. Oncol. 2012, 124, 534–541. [Google Scholar] [CrossRef] [PubMed]
- Rogeri, C.D.; Silveira, H.C.S.; Causin, R.L.; Villa, L.L.; Stein, M.D.; de Carvalho, A.C.; Arantes, L.M.R.B.; Scapulatempo-Neto, C.; Possati-Resende, J.C.; Antoniazzi, M.; et al. Methylation of the hsa-miR-124, SOX1, TERT, and LMX1A genes as biomarkers for precursor lesions in cervical cancer. Gynecol. Oncol. 2018, 150, 545–551. [Google Scholar] [CrossRef]
- Schütze, D.M.; Kooter, J.M.; Wilting, S.M.; Meijer, C.J.; Quint, W.; Snijders, P.J.; Steenbergen, R.D. Longitudinal assessment of DNA methylation changes during HPVE6E7-induced immortalization of primary keratinocytes. Epigenetics 2015, 10, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Molano, M.; Moreno-Acosta, P.; Morales, N.; Burgos, M.; Buitrago, L.; Gamboa, O.; Alvarez, R.; Garland, S.M.; Tabrizi, S.N.; Steenbergen, R.D.; et al. Association Between Type-specific HPV Infections and hTERT DNA Methylation in Patients with Invasive Cervical Cancer. Cancer Genom. Proteom. 2016, 13, 483–492. [Google Scholar] [CrossRef]
- James, M.A.; Lee, J.H.; Klingelhutz, A.J. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int. J. Cancer 2006, 119, 1878–1885. [Google Scholar] [CrossRef]
- Durzyńska, J.; Leśniewicz, K.; Poreba, E. Human papillomaviruses in epigenetic regulations. Mutat. Res. Mutat. Res. 2017, 772, 36–50. [Google Scholar] [CrossRef]
- Roberts, S.A.; Lawrence, M.S.; Klimczak, L.J.; AGrimm, S.A.; Fargo, D.; Stojanov, P.; Kiezun, A.; Kryukov, G.; Carter, S.L.; Saksena, G.; et al. An APOBEC cytidine deaminase mutagenesis pattern is widespread in human cancers. Nat. Genet. 2013, 45, 970–976. [Google Scholar] [CrossRef]
- Cannataro, V.L.; Gaffney, S.G.; Sasaki, T.; Issaeva, N.; Grewal, N.; Grandis, J.R.; Yarbrough, W.G.; Burtness, B.; Anderson, K.S.; Townsend, J.P. APOBEC-induced mutations and their cancer effect size in head and neck squamous cell carcinoma. Oncogene 2019, 38, 3475–3487. [Google Scholar] [CrossRef]
- Revathidevi, S.; Murugan, A.K.; Nakaoka, H.; Inoue, I.; Munirajan, A.K. APOBEC: A molecular driver in cervical cancer pathogenesis. Cancer Lett. 2020, 496, 104–116. [Google Scholar] [CrossRef]
- Feber, A.; Worth, D.C.; Chakravarthy, A.; de Winter, P.; Shah, K.; Arya, M.; Saqib, M.; Nigam, R.; Malone, P.R.; Tan, W.S.; et al. CSN1 Somatic Mutations in Penile Squamous Cell Carcinoma. Cancer Res. 2016, 76, 4720–4727. [Google Scholar] [CrossRef]
- Guerra, G.R.; Kong, J.C.; Millen, R.M.; Read, M.; Liu, D.S.; Roth, S.; Sampurno, S.; Sia, J.; Bernardi, M.-P.; Chittleborough, T.J.; et al. Molecular and genomic characterisation of a panel of human anal cancer cell lines. Cell Death Dis. 2021, 12, 959. [Google Scholar] [CrossRef]
- Williams, E.A.; Werth, A.J.; Sharaf, R.; Montesion, M.; Sokol, E.S.; Pavlick, D.C.; McLaughlin-Drubin, M.; Erlich, R.; Toma, H.; Williams, K.J.; et al. Vulvar Squamous Cell Carcinoma: Comprehensive Genomic Profiling of HPV+ Versus HPV– Forms Reveals Distinct Sets of Potentially Actionable Molecular Targets. JCO Precis. Oncol. 2020, 4, 647–661. [Google Scholar] [CrossRef]
- Shi, K.; Carpenter, M.A.; Banerjee, S.; Shaban, N.M.; Kurahashi, K.; Salamango, D.J.; McCann, J.L.; Starrett, G.J.; Duffy, J.V.; Demir, Ö.; et al. Structural basis for targeted DNA cytosine deamination and mutagenesis by APOBEC3A and APOBEC3B. Nat. Struct. Mol. Biol. 2016, 24, 131–139. [Google Scholar] [CrossRef]
- Kazanov, M.; Roberts, S.A.; Polak, P.; Stamatoyannopoulos, J.; Klimczak, L.J.; Gordenin, D.A.; Sunyaev, S.R. APOBEC-Induced Cancer Mutations Are Uniquely Enriched in Early-Replicating, Gene-Dense, and Active Chromatin Regions. Cell Rep. 2015, 13, 1103–1109. [Google Scholar] [CrossRef]
- Rebhandl, S.; Huemer, M.; Greil, R.; Geisberger, R. AID/APOBEC deaminases and cancer. Oncoscience 2015, 2, 320–333. [Google Scholar] [CrossRef]
- Harris, R.S.; Liddament, M.T. Retroviral restriction by APOBEC proteins. Nat. Rev. Immunol. 2004, 4, 868–877. [Google Scholar] [CrossRef]
- Imahashi, M.; Nakashima, M.; Iwatani, Y. Antiviral Mechanism and Biochemical Basis of the Human APOBEC3 Family. Front. Microbiol. 2012, 3, 250. [Google Scholar] [CrossRef]
- Nik-Zainal, S.; Alexandrov, L.B.; Wedge, D.C.; van Loo, P.; Greenman, C.D.; Raine, K.; Jones, D.; Hinton, J.; Marshall, J.; Stebbings, L.A.; et al. Breast Cancer Working Group of the International Cancer Genome Consortium, Mutational processes molding the genomes of 21 breast cancers. Cell 2012, 149, 979–993. [Google Scholar] [CrossRef]
- Zhu, B.; Xiao, Y.; Yeager, M.; Clifford, G.; Wentzensen, N.; Cullen, M.; Boland, J.F.; Bass, S.; Steinberg, M.K.; Raine-Bennett, T.; et al. Mutations in the HPV16 genome induced by APOBEC3 are associated with viral clearance. Nat. Commun. 2020, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Warren, C.J.; Xu, T.; Guo, K.; Griffin, L.M.; Westrich, J.A.; Lee, D.; Lambert, P.F.; Santiago, M.L.; Pyeon, D. APOBEC3A Functions as a Restriction Factor of Human Papillomavirus. J. Virol. 2015, 89, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Ahasan, M.; Wakae, K.; Wang, Z.; Kitamura, K.; Liu, G.; Koura, M.; Imayasu, M.; Sakamoto, N.; Hanaoka, K.; Nakamura, M.; et al. APOBEC3A and 3C decrease human papillomavirus 16 pseudovirion infectivity. Biochem. Biophys. Res. Commun. 2015, 457, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, J.-P.; Guétard, D.; Henry, M.; Wain-Hobson, S. Evidence for Editing of Human Papillomavirus DNA by APOBEC3 in Benign and Precancerous Lesions. Science 2008, 320, 230–233. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Onuki, M.; Tenjimbayashi, Y.; Mori, S.; Ishii, Y.; Takeuchi, T.; Tasaka, N.; Satoh, T.; Morisada, T.; Iwata, T.; et al. Within-Host Variations of Human Papillomavirus Reveal APOBEC Signature Mutagenesis in the Viral Genome. J. Virol. 2018, 92, e00017-18. [Google Scholar] [CrossRef]
- Warren, C.; Van Doorslaer, K.; Pandey, A.; Espinosa, J.M.; Pyeon, D. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus Evol. 2015, 1, vev015. [Google Scholar] [CrossRef] [PubMed]
- Lagström, S.; Løvestad, A.H.; Umu, S.U.; Ambur, O.H.; Nygård, M.; Rounge, T.B.; Christiansen, I.K. HPV16 and HPV18 type-specific APOBEC3 and integration profiles in different diagnostic categories of cervical samples. Tumour Virus Res. 2021, 12, 200221. [Google Scholar] [CrossRef]
- Vieira, V.C.; Leonard, B.; White, E.A.; Starrett, G.J.; Temiz, N.A.; Lorenz, L.D.; Lee, D.; Soares, M.A.; Lambert, P.F.; Howley, P.; et al. Human Papillomavirus E6 Triggers Upregulation of the Antiviral and Cancer Genomic DNA Deaminase APOBEC3B. mBio 2014, 5, e02234-14. [Google Scholar] [CrossRef]
- Periyasamy, M.; Singh, A.K.; Gemma, C.; Kranjec, C.; Farzan, R.; Leach, D.A.; Navaratnam, N.; Pálinkás, H.L.; Vértessy, B.G.; Fenton, T.R.; et al. p53 controls expression of the DNA deaminase APOBEC3B to limit its potential mutagenic activity in cancer cells. Nucleic Acids Res. 2017, 45, 11056–11069. [Google Scholar] [CrossRef]
- Chen, S.; Li, X.; Qin, J.; Chen, Y.; Liu, L.; Zhang, D.; Wang, M.; Wang, M.; Zhang, D. APOBEC3A possesses anticancer and antiviral effects by differential inhibition of HPV E6 and E7 expression on cervical cancer. Int. J. Clin. Exp. Med. 2015, 8, 10548–10557. [Google Scholar]
- Halle, M.K.; Sundaresan, A.; Zhang, J.; Pedamallu, C.S.; Srinivasasainagendra, V.; Blair, J.; Brooke, D.; Bertelsen, B.I.; Woie, K.; Shrestha, S.; et al. Genomic alterations associated with mutational signatures, DNA damage repair and chromatin remodeling pathways in cervical carcinoma. NPJ Genom. Med. 2021, 6, 82. [Google Scholar] [CrossRef]
- Kondo, S.; Wakae, K.; Wakisaka, N.; Nakanishi, Y.; Ishikawa, K.; Komori, T.; Moriyama-Kita, M.; Endo, K.; Murono, S.; Wang, Z.; et al. APOBEC3A associates with human papillomavirus genome integration in oropharyngeal cancers. Oncogene 2016, 36, 1687–1697. [Google Scholar] [CrossRef]
- Henderson, S.; Chakravarthy, A.; Su, X.; Boshoff, C.; Fenton, T.R. APOBEC-Mediated Cytosine Deamination Links PIK3CA Helical Domain Mutations to Human Papillomavirus-Driven Tumor Development. Cell Rep. 2014, 7, 1833–1841. [Google Scholar] [CrossRef]
- Wang, S.S.; Bratti, M.C.; Rodríguez, A.C.; Herrero, R.; Burk, R.D.; Porras, C.; González, P.; Sherman, M.E.; Wacholder, S.; Lan, Z.E.; et al. Common variants in immune and DNA repair genes and risk for human papillomavirus persistence and progression to cervical cancer. J. Infect. Dis. 2009, 199, 20–30. [Google Scholar] [CrossRef]
- Dylawerska, A.; Barczak, W.; Wegner, A.; Golusinski, W.; Suchorska, W.M. Association of DNA repair genes polymorphisms and mutations with increased risk of head and neck cancer: A review. Med. Oncol. 2017, 34, 197. [Google Scholar] [CrossRef]
- Burcher, K.; Faucheux, A.; Lantz, J.; Wilson, H.; Abreu, A.; Salafian, K.; Patel, M.; Song, A.; Petro, R.; Lycan, T.; et al. Prevalence of DNA Repair Gene Mutations in Blood and Tumor Tissue and Impact on Prognosis and Treatment in HNSCC. Cancers 2021, 13, 3118. [Google Scholar] [CrossRef]
- Seiwert, T.Y.; Zuo, Z.; Keck, M.K.; Khattri, A.; Pedamallu, C.S.; Stricker, T.; Brown, C.; Pugh, T.J.; Stojanov, P.; Cho, J.; et al. Integrative and Comparative Genomic Analysis of HPV-Positive and HPV-Negative Head and Neck Squamous Cell Carcinomas. Clin. Cancer Res. 2015, 21, 632–641. [Google Scholar] [CrossRef]
- Vinothkumar, V.; Arun, K.; Arunkumar, G.; Revathidevi, S.; Ramani, R.; Bhaskar, L.V.; Murugan, A.K.; Munirajan, A.K. Association between functional TERT promoter polymorphism rs2853669 and cervical cancer risk in South Indian women. Mol. Clin. Oncol. 2020, 12, 485–494. [Google Scholar] [CrossRef]
- Annunziata, C.; Pezzuto, F.; Greggi, S.; Ionna, F.; Losito, S.; Botti, G.; Buonaguro, L.; Buonaguro, F.M.; Tornesello, M.L. Distinct profiles of TERT promoter mutations and telomerase expression in head and neck cancer and cervical carcinoma. Int. J. Cancer 2018, 143, 1153–1161. [Google Scholar] [CrossRef]
- Vinothkumar, V.; Arunkumar, G.; Revathidevi, S.; Arun, K.; Manikandan, M.; Rao, A.K.D.M.; Rajkumar, K.S.; Ajay, C.; Rajaraman, R.; Ramani, R.; et al. TERT promoter hot spot mutations are frequent in Indian cervical and oral squamous cell carcinomas. Tumor Biol. 2015, 37, 7907–7913. [Google Scholar] [CrossRef]
- Yilmaz, I.; Erkul, E.; Sari, S.O.; Issin, G.; Tural, E.; Terzi, N.T.K.; Karatay, H.; Celik, M.; Ulusan, M.; Bilgic, B. Promoter region mutations of the telomerase reverse transcriptase (TERT) gene in head and neck squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2020, 130, 63–70. [Google Scholar] [CrossRef]
- Salama, A.M.; Momeni-Boroujeni, A.; Vanderbilt, C.; Ladanyi, M.; Soslow, R. Molecular landscape of vulvovaginal squamous cell carcinoma: New insights into molecular mechanisms of HPV-associated and HPV-independent squamous cell carcinoma. Mod. Pathol. 2021, 35, 274–282. [Google Scholar] [CrossRef]
- Kim, S.K.; Kim, J.-H.; Han, J.H.; Cho, N.H.; Kim, S.J.; Kim, S.I.; Choo, S.H.; Kim, J.S.; Park, B.; Kwon, J.E. TERT promoter mutations in penile squamous cell carcinoma: High frequency in non-HPV-related type and association with favorable clinicopathologic features. J. Cancer Res. Clin. Oncol. 2021, 147, 1125–1135. [Google Scholar] [CrossRef]
- Bhattacharjee, B.; Sengupta, S. CpG methylation of HPV 16 LCR at E2 binding site proximal to P97 is associated with cervical cancer in presence of intact E2. Virology 2006, 354, 280–285. [Google Scholar] [CrossRef]
- Amaro-Filho, S.M.; Chaves, C.B.P.; Felix, S.P.; Basto, D.L.; de Almeida, L.M.; Moreira, M.A.M. HPV DNA methylation at the early promoter and E1/E2 integrity: A comparison between HPV16, HPV18 and HPV45 in cervical cancer. Papillomavirus Res. 2018, 5, 172–179. [Google Scholar] [CrossRef]
- Fernandez, A.F.; Esteller, M. Viral epigenomes in human tumorigenesis. Oncogene 2010, 29, 1405–1420. [Google Scholar] [CrossRef]
- Vinokurova, S.; Wentzensen, N.; Kraus, I.; Klaes, R.; Driesch, C.; Melsheimer, P.; Kisseljov, F.; Dürst, M.; Schneider, A.; von Knebel Doeberitz, M. Type-Dependent Integration Frequency of Human Papillomavirus Genomes in Cervical Lesions. Cancer Res. 2008, 68, 307–313. [Google Scholar] [CrossRef]
- Egawa, N.; Doorbar, J. The low-risk papillomaviruses. Virus Res. 2017, 231, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Donne, A.; Hampson, L.; Homer, J.; Hampson, I. The role of HPV type in Recurrent Respiratory Papillomatosis. Int. J. Pediatr. Otorhinolaryngol. 2009, 74, 7–14. [Google Scholar] [CrossRef]
- Reidy, P.M.; Dedo, H.H.; Rabah, R.; Field, J.B.; Mathog, R.H.; Gregoire, L.; Lancaster, W.D. Integration of Human Papillomavirus Type 11 in Recurrent Respiratory Papilloma-Associated Cancer. Laryngoscope 2004, 114, 1906–1909. [Google Scholar] [CrossRef]
- Iden, M.; Tsaih, S.-W.; Huang, Y.-W.; Liu, P.; Xiao, M.; Flister, M.J.; Rader, J.S. Multi-omics mapping of human papillomavirus integration sites illuminates novel cervical cancer target genes. Br. J. Cancer 2021, 125, 1408–1419. [Google Scholar] [CrossRef]
- Bodelon, C.; Untereiner, M.E.; Machiela, M.J.; Vinokurova, S.; Wentzensen, N. Genomic characterization of viral integration sites in HPV-related cancers. Int. J. Cancer 2016, 139, 2001–2011. [Google Scholar] [CrossRef] [PubMed]
- Garza-Rodríguez, M.; Oyervides-Muñoz, M.; Pérez-Maya, A.; Sánchez-Domínguez, C.; Berlanga-Garza, A.; Antonio-Macedo, M.; Valdés-Chapa, L.; Vidal-Torres, D.; Vidal-Gutiérrez, O.; Pérez-Ibave, D.; et al. Analysis of HPV Integrations in Mexican Pre-Tumoral Cervical Lesions Reveal Centromere-Enriched Breakpoints and Abundant Unspecific HPV Regions. Int. J. Mol. Sci. 2021, 22, 3242. [Google Scholar] [CrossRef] [PubMed]
- Oyervides-Muñoz, M.A.; Pérez-Maya, A.A.; Rodríguez-Gutiérrez, H.F.; Gómez-Macias, G.S.; Fajardo, O.R.; Trevino, V.; Barrera-Saldaña, H.A.; Garza-Rodríguez, M.L. Understanding the HPV integration and its progression to cervical cancer. Infect. Genet. Evol. 2018, 61, 134–144. [Google Scholar] [CrossRef]
- Sfeir, A.; Symington, L.S. Microhomology-Mediated End Joining: A Back-up Survival Mechanism or Dedicated Pathway? Trends Biochem. Sci. 2015, 40, 701–714. [Google Scholar] [CrossRef]
- Wongworawat, Y.C.; Filippova, M.; Williams, V.M.; Filippov, V.; Duerksen-Hughes, P.J. Chronic oxidative stress increases the integration frequency of foreign DNA and human papillomavirus 16 in human keratinocytes. Am. J. Cancer Res. 2016, 6, 764. [Google Scholar]
- Williams, V.M.; Filippova, M.; Soto, U.; Duerksen-Hughes, P.J. HPV-DNA integration and carcinogenesis: Putative roles for inflammation and oxidative stress. Futur. Virol. 2011, 6, 45–57. [Google Scholar] [CrossRef]
- Adey, A.; Burton, J.; Kitzman, J.O.; Hiatt, J.B.; Lewis, A.P.; Martin, B.K.; Qiu, R.; Lee, C.; Shendure, J. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature 2013, 500, 207–211. [Google Scholar] [CrossRef]
- Peter, M.; Stransky, N.; Couturier, J.; Hupé, P.; Barillot, E.; de Cremoux, P.; Cottu, P.; Radvanyi, F.; Sastre-Garau, X. Frequent genomic structural alterations at HPV insertion sites in cervical carcinoma. J. Pathol. 2010, 221, 320–330. [Google Scholar] [CrossRef]
- Deshpande, V.; Luebeck, J.; Nguyen, N.-P.; Bakhtiari, M.; Turner, K.M.; Schwab, R.; Carter, H.; Mischel, P.S.; Bafna, V. Exploring the landscape of focal amplifications in cancer using AmpliconArchitect. Nat. Commun. 2019, 10, 392. [Google Scholar] [CrossRef]
- Koche, R.P.; Rodriguez-Fos, E.; Helmsauer, K.; Burkert, M.; MacArthur, I.C.; Maag, J.; Chamorro, R.; Munoz-Perez, N.; Puiggròs, M.; Garcia, H.D.; et al. Extrachromosomal circular DNA drives oncogenic genome remodeling in neuroblastoma. Nat. Genet. 2019, 52, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Nguyen, N.-P.; Turner, K.; Wu, S.; Gujar, A.D.; Luebeck, J.; Liu, J.; Deshpande, V.; Rajkumar, U.; Namburi, S.; et al. Extrachromosomal DNA is associated with oncogene amplification and poor outcome across multiple cancers. Nat. Genet. 2020, 52, 891–897. [Google Scholar] [CrossRef] [PubMed]
- Kadaja, M.; Isok-Paas, H.; Laos, T.; Ustav, E.; Ustav, M. Mechanism of Genomic Instability in Cells Infected with the High-Risk Human Papillomaviruses. PLoS Pathog. 2009, 5, e1000397. Available online: https://www.ncbi.nlm.nih.gov/pmc/articles/pmc2666264/ (accessed on 15 September 2022). [CrossRef]
- Rossi, N.M.; Dai, J.; Xie, Y.; Lou, H.; Boland, J.F.; Yeager, M.; Orozco, R.; Alvirez, E.; Mirabello, L.; Gharzouzi, E.; et al. Extrachromosomal Amplification of Human Papillomavirus Episomes as a Mechanism of Cervical Carcinogenesis. bioRxiv 2021, bioRxiv:2021.10.22.465367. [Google Scholar] [CrossRef]
- Banerjee, N.S.; Moore, D.; Parker, C.J.; Broker, T.R.; Chow, L.T. Targeting DNA Damage Response as a Strategy to Treat HPV Infections. Int. J. Mol. Sci. 2019, 20, 5455. [Google Scholar] [CrossRef]
- Chen, Q.; Cai, D.; Li, M.; Wu, X. The homologous recombination protein RAD51 is a promising therapeutic target for cervical carcinoma. Oncol. Rep. 2017, 38, 767–774. [Google Scholar] [CrossRef]
- Dok, R.; Bamps, M.; Glorieux, M.; Zhao, P.; Sablina, A.; Nuyts, S. Radiosensitization approaches for HPV-positive and HPV-negative head and neck squamous carcinomas. Int. J. Cancer 2019, 146, 1075–1085. [Google Scholar] [CrossRef]
- Alsahafi, E.N.; Thavaraj, S.; Sarvestani, N.; Novoplansky, O.; Elkabets, M.; Ayaz, B.; Tavassoli, M.; Legends, M.F. EGFR overexpression increases radiotherapy response in HPV-positive head and neck cancer through inhibition of DNA damage repair and HPV E6 downregulation. Cancer Lett. 2020, 498, 80–97. [Google Scholar] [CrossRef]
- De Lorenzo, S.; Patel, A.; Hurley, R.M.; Kaufmann, S.H.M. The Elephant and the Blind Men: Making Sense of PARP Inhibitors in Homologous Recombination Deficient Tumor Cells. Front. Oncol. 2013, 3, 228. [Google Scholar] [CrossRef]
- Molkentine, J.M.; Molkentine, D.P.; Bridges, K.A.; Xie, T.; Yang, L.; Sheth, A.; Heffernan, T.P.; Clump, D.A.; Faust, A.Z.; Ferris, R.L.; et al. Targeting DNA damage response in head and neck cancers through abrogation of cell cycle checkpoints. Int. J. Radiat. Biol. 2020, 97, 1121–1128. [Google Scholar] [CrossRef]
- Pirotte, E.F.; Holzhauser, S.; Owens, D.; Quine, S.; Al-Hussaini, A.; Christian, A.D.; Giles, P.J.; Man, S.T.; Evans, M.; Powell, N.G. Sensitivity to inhibition of DNA repair by Olaparib in novel oropharyngeal cancer cell lines infected with Human Papillomavirus. PLoS ONE 2018, 13, e0207934. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Gao, J.Z.; Gomendoza, S.M.T.; Li, J.W.; Yang, S. Recent Advances of WEE1 Inhibitors and Statins in Cancers with p53 Mutations. Front. Med. 2021, 8, 737951. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.-H.; Min, A.; Kim, S.; Jang, H.; Kim, S.H.; Kim, H.-J.; Ryu, H.S.; Ku, J.-L.; Lee, K.-H.; Im, S.-A. Antitumor effect of a WEE1 inhibitor and potentiation of olaparib sensitivity by DNA damage response modulation in triple-negative breast cancer. Sci. Rep. 2020, 10, 9930. [Google Scholar] [CrossRef]
- Osman, A.A.; Monroe, M.M.; Alves, M.V.O.; Patel, A.A.; Katsonis, P.; Fitzgerald, A.L.; Neskey, D.M.; Frederick, M.J.; Woo, S.H.; Caulin, C.; et al. Wee-1 Kinase Inhibition Overcomes Cisplatin Resistance Associated with High-Risk TP53 Mutations in Head and Neck Cancer through Mitotic Arrest Followed by SenescenceTargeting Wee-1 Kinase in mutp53 HNSCC. Available online: https://aacrjournals.org/mct/article-abstract/14/2/608/175773 (accessed on 15 September 2022).
- Friedman, J.; Morisada, M.; Sun, L.; Moore, E.C.; Padget, M.; Hodge, J.W.; Schlom, J.; Gameiro, S.; Allen, C.T. Inhibition of WEE1 kinase and cell cycle checkpoint activation sensitizes head and neck cancers to natural killer cell therapies. J. Immunother. Cancer 2018, 6, 59. [Google Scholar] [CrossRef]
- Diab, A.; Gem, H.; Swanger, J.; Kim, H.Y.; Smith, K.; Zou, G.; Raju, S.; Kao, M.; Fitzgibbon, M.; Loeb, K.R.; et al. FOXM1 drives HPV+ HNSCC sensitivity to WEE1 inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 28287–28296. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porter, V.L.; Marra, M.A. The Drivers, Mechanisms, and Consequences of Genome Instability in HPV-Driven Cancers. Cancers 2022, 14, 4623. https://doi.org/10.3390/cancers14194623
Porter VL, Marra MA. The Drivers, Mechanisms, and Consequences of Genome Instability in HPV-Driven Cancers. Cancers. 2022; 14(19):4623. https://doi.org/10.3390/cancers14194623
Chicago/Turabian StylePorter, Vanessa L., and Marco A. Marra. 2022. "The Drivers, Mechanisms, and Consequences of Genome Instability in HPV-Driven Cancers" Cancers 14, no. 19: 4623. https://doi.org/10.3390/cancers14194623
APA StylePorter, V. L., & Marra, M. A. (2022). The Drivers, Mechanisms, and Consequences of Genome Instability in HPV-Driven Cancers. Cancers, 14(19), 4623. https://doi.org/10.3390/cancers14194623





