Insight into LncRNA- and CircRNA-Mediated CeRNAs: Regulatory Network and Implications in Nasopharyngeal Carcinoma—A Narrative Literature Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Methods
3. Biological Functions of LncRNA/CircRNA-Mediated CeRNA Networks in NPC
3.1. Regulation of NPC Cell Proliferation
3.2. Regulation of NPC Cell Apoptosis
3.3. Modulating NPC Chemosensitivity
3.4. Modulating NPC Metastasis
4. Implications of LncRNA/CircRNA-Associated ceRNAs as Diagnostic Markers or Therapeutic Targets in NPC
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Chen, Y.-P.; Chan, A.T.C.; Le, Q.-T.; Blanchard, P.; Sun, Y.; Ma, J. Nasopharyngeal carcinoma. Lancet 2019, 394, 64–80. [Google Scholar] [CrossRef]
- Tang, L.-L.; Chen, W.-Q.; Xue, W.-Q.; He, Y.-Q.; Zheng, R.-S.; Zeng, Y.-X.; Jia, W.-H. Global trends in incidence and mortality of nasopharyngeal carcinoma. Cancer Lett. 2016, 374, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.C.W.; Hui, E.P.; Lo, K.-W.; Lam, W.K.J.; Johnson, D.; Li, L.; Tao, Q.; Chan, K.C.A.; To, K.-F.; King, A.D.; et al. Nasopharyngeal carcinoma: An evolving paradigm. Nat. Rev. Clin. Oncol. 2021, 18, 679–695. [Google Scholar] [CrossRef]
- Jia, W.-H.; Qin, H.-D. Non-viral environmental risk factors for nasopharyngeal carcinoma: A systematic review. Semin. Cancer Biol. 2012, 22, 117–126. [Google Scholar] [CrossRef]
- Tsao, S.W.; Yip, Y.L.; Tsang, C.M.; Pang, P.S.; Lau, V.M.Y.; Zhang, G.; Lo, K.W. Etiological factors of nasopharyngeal carcinoma. Oral Oncol. 2014, 50, 330–338. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jing, B.; Ke, L.; Li, B.; Xia, W.; He, C.; Qian, C.; Zhao, C.; Mai, H.; Chen, M.; et al. Development and validation of an endoscopic images-based deep learning model for detection with nasopharyngeal malignancies. Cancer Commun. 2018, 38, 59. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-S.; Li, X.-Y.; Chen, Q.-Y.; Tang, L.-Q.; Mai, H.-Q. Future of Radiotherapy in Nasopharyngeal Carcinoma. Br. J. Radiol. 2019, 92, 20190209. [Google Scholar] [CrossRef] [PubMed]
- Tan, Z.; Xiao, L.; Tang, M.; Bai, F.; Li, J.; Li, L.; Shi, F.; Li, N.; Li, Y.; Du, Q.; et al. Targeting CPT1A-mediated fatty acid oxidation sensitizes nasopharyngeal carcinoma to radiation therapy. Theranostics 2018, 8, 2329–2347. [Google Scholar] [CrossRef]
- Guan, S.; Wei, J.; Huang, L.; Wu, L. Chemotherapy and chemo-resistance in nasopharyngeal carcinoma. Eur. J. Med. Chem. 2020, 207, 112758. [Google Scholar] [CrossRef]
- Liu, S.-L.; Sun, X.-S.; Li, X.-Y.; Chen, Q.-Y.; Lin, H.-X.; Wen, Y.-F.; Guo, S.-S.; Liu, L.-T.; Xie, H.-J.; Tang, Q.-N.; et al. Liposomal paclitaxel versus docetaxel in induction chemotherapy using Taxanes, cisplatin and 5-fluorouracil for locally advanced nasopharyngeal carcinoma. BMC Cancer 2018, 18, 1279. [Google Scholar] [CrossRef]
- Liu, H.; Qi, B.; Guo, X.; Tang, L.-Q.; Chen, Q.-Y.; Zhang, L.; Guo, L.; Luo, D.-H.; Huang, P.-Y.; Mo, H.-Y.; et al. Genetic variations in radiation and chemotherapy drug action pathways and survival in locoregionally advanced nasopharyngeal carcinoma treated with chemoradiotherapy. PLoS ONE 2013, 8, e82750. [Google Scholar] [CrossRef] [PubMed]
- Bruce, J.P.; To, K.-F.; Lui, V.W.Y.; Chung, G.T.Y.; Chan, Y.-Y.; Tsang, C.M.; Yip, K.Y.; Ma, B.B.Y.; Woo, J.K.S.; Hui, E.P.; et al. Whole-genome profiling of nasopharyngeal carcinoma reveals viral-host co-operation in inflammatory NF-κB activation and immune escape. Nat. Commun. 2021, 12, 4193. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Luo, R.; Liu, Y.; Gao, L.; Fu, Z.; Fu, Q.; Luo, X.; Chen, Y.; Deng, X.; Liang, Z.; et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat. Commun. 2016, 7, 11309. [Google Scholar] [CrossRef]
- Qing, X.; Tan, G.-L.; Liu, H.-W.; Li, W.; Ai, J.-G.; Xiong, S.-S.; Yang, M.-Q.; Wang, T.-S. LINC00669 insulates the JAK/STAT suppressor SOCS1 to promote nasopharyngeal cancer cell proliferation and invasion. J. Exp. Clin. Cancer Res. 2020, 39, 166. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: LncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015, 11, 474–485. [Google Scholar] [CrossRef]
- Holdt, L.M.; Hoffmann, S.; Sass, K.; Langenberger, D.; Scholz, M.; Krohn, K.; Finstermeier, K.; Stahringer, A.; Wilfert, W.; Beutner, F.; et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 2013, 9, e1003588. [Google Scholar] [CrossRef]
- Isoda, T.; Moore, A.J.; He, Z.; Chandra, V.; Aida, M.; Denholtz, M.; Piet van Hamburg, J.; Fisch, K.M.; Chang, A.N.; Fahl, S.P.; et al. Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell 2017, 171, 103–119.e18. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Denzler, R.; Agarwal, V.; Stefano, J.; Bartel, D.P.; Stoffel, M. Assessing the ceRNA hypothesis with quantitative measurements of miRNA and target abundance. Mol. Cell 2014, 54, 766–776. [Google Scholar] [CrossRef]
- Kristensen, L.S.; Andersen, M.S.; Stagsted, L.V.W.; Ebbesen, K.K.; Hansen, T.B.; Kjems, J. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 2019, 20, 675–691. [Google Scholar] [CrossRef]
- Chen, L.; Wang, C.; Sun, H.; Wang, J.; Liang, Y.; Wang, Y.; Wong, G. The bioinformatics toolbox for circRNA discovery and analysis. Brief. Bioinform. 2021, 22, 1706–1728. [Google Scholar] [CrossRef] [PubMed]
- Obi, P.; Chen, Y.G. The design and synthesis of circular RNAs. Methods 2021, 196, 85–103. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.-L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Tang, L.; Xiong, W.; Zhang, L.; Wang, D.; Wang, Y.; Wu, Y.; Wei, F.; Mo, Y.; Hou, X.; Shi, L.; et al. circSETD3 regulates MAPRE1 through miR-615-5p and miR-1538 sponges to promote migration and invasion in nasopharyngeal carcinoma. Oncogene 2021, 40, 307–321. [Google Scholar] [CrossRef]
- Chen, L.; Zhou, H.; Guan, Z. CircRNA_000543 knockdown sensitizes nasopharyngeal carcinoma to irradiation by targeting miR-9/platelet-derived growth factor receptor B axis. Biochem. Biophys. Res. Commun. 2019, 512, 786–792. [Google Scholar] [CrossRef]
- Lin, J.; Qin, H.; Han, Y.; Li, X.; Zhao, Y.; Zhai, G. CircNRIP1 Modulates the miR-515-5p/IL-25 Axis to Control 5-Fu and Cisplatin Resistance in Nasopharyngeal Carcinoma. Drug Des. Dev. Ther. 2021, 15, 323–330. [Google Scholar] [CrossRef]
- Pushparaj, P.N.; Aarthi, J.J.; Manikandan, J.; Kumar, S.D. siRNA, miRNA, and shRNA: In vivo applications. J. Dent. Res. 2008, 87, 992–1003. [Google Scholar] [CrossRef]
- Kilikevicius, A.; Meister, G.; Corey, D.R. Reexamining assumptions about miRNA-guided gene silencing. Nucleic Acids Res. 2022, 50, 617–634. [Google Scholar] [CrossRef]
- Iwakawa, H.-O.; Tomari, Y. Life of RISC: Formation, action, and degradation of RNA-induced silencing complex. Mol. Cell 2022, 82, 30–43. [Google Scholar] [CrossRef]
- Kawamata, T.; Tomari, Y. Making RISC. Trends Biochem. Sci. 2010, 35, 368–376. [Google Scholar] [CrossRef] [PubMed]
- Lewis, B.P.; Burge, C.B.; Bartel, D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 2005, 120, 15–20. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, X.; Qin, Z.; Wei, L.; Lu, Y.; Peng, Q.; Gao, Y.; Zhang, X.; Zhang, X.; Li, Z.; et al. Epstein-Barr virus miR-BART3-3p promotes tumorigenesis by regulating the senescence pathway in gastric cancer. J. Biol. Chem. 2019, 294, 4854–4866. [Google Scholar] [CrossRef]
- Zuo, L.; Xie, Y.; Tang, J.; Xin, S.; Liu, L.; Zhang, S.; Yan, Q.; Zhu, F.; Lu, J. Targeting Exosomal EBV-LMP1 Transfer and miR-203 Expression via the NF-κB Pathway: The Therapeutic Role of Aspirin in NPC. Mol. Ther. Nucleic Acids 2019, 17, 175–184. [Google Scholar] [CrossRef]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Kong, X.; Duan, Y.; Sang, Y.; Li, Y.; Zhang, H.; Liang, Y.; Liu, Y.; Zhang, N.; Yang, Q. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J. Cell Physiol. 2019, 234, 9105–9117. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huo, X.; Yang, X.-R.; He, J.; Cheng, L.; Wang, N.; Deng, X.; Jin, H.; Wang, N.; Wang, C.; et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 2017, 16, 136. [Google Scholar] [CrossRef]
- Hong, X.; Liu, N.; Liang, Y.; He, Q.; Yang, X.; Lei, Y.; Zhang, P.; Zhao, Y.; He, S.; Wang, Y.; et al. Circular RNA CRIM1 functions as a ceRNA to promote nasopharyngeal carcinoma metastasis and docetaxel chemoresistance through upregulating FOXQ1. Mol. Cancer 2020, 19, 33. [Google Scholar] [CrossRef]
- Peng, J.; Liu, F.; Zheng, H.; Wu, Q.; Liu, S. IncRNA ZFAS1 contributes to the radioresistance of nasopharyngeal carcinoma cells by sponging hsa-miR-7-5p to upregulate ENO2. Cell Cycle 2021, 20, 126–141. [Google Scholar] [CrossRef]
- Liu, D.; Wang, Y.; Zhao, Y.; Gu, X. LncRNA SNHG5 promotes nasopharyngeal carcinoma progression by regulating miR-1179/HMGB3 axis. BMC Cancer 2020, 20, 178. [Google Scholar] [CrossRef]
- Hu, W.; Li, H.; Wang, S. LncRNA SNHG7 promotes the proliferation of nasopharyngeal carcinoma by miR-514a-5p/ELAVL1 axis. BMC Cancer 2020, 20, 376. [Google Scholar] [CrossRef] [PubMed]
- Liao, B.; Wang, Z.; Zhu, Y.; Wang, M.; Liu, Y. Long noncoding RNA DRAIC acts as a microRNA-122 sponge to facilitate nasopharyngeal carcinoma cell proliferation, migration and invasion via regulating SATB1. Artif. Cells Nanomed. Biotechnol. 2019, 47, 3585–3597. [Google Scholar] [CrossRef] [PubMed]
- Zhang, E.; Li, X. LncRNA SOX2-OT regulates proliferation and metastasis of nasopharyngeal carcinoma cells through miR-146b-5p/HNRNPA2B1 pathway. J. Cell Biochem. 2019, 120, 16575–16588. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Tan, S.; Song, L.; Song, L.; Wang, Y. LncRNA XIST knockdown suppresses the malignancy of human nasopharyngeal carcinoma through XIST/miRNA-148a-3p/ADAM17 pathway in vitro and in vivo. Biomed. Pharmacother. 2020, 121, 109620. [Google Scholar] [CrossRef]
- Zheng, Z.-Q.; Li, Z.-X.; Zhou, G.-Q.; Lin, L.; Zhang, L.-L.; Lv, J.-W.; Huang, X.-D.; Liu, R.-Q.; Chen, F.; He, X.-J.; et al. Long Noncoding RNA FAM225A Promotes Nasopharyngeal Carcinoma Tumorigenesis and Metastasis by Acting as ceRNA to Sponge miR-590-3p/miR-1275 and Upregulate ITGB3. Cancer Res. 2019, 79, 4612–4626. [Google Scholar] [CrossRef]
- Chen, W.; Du, M.; Hu, X.; Ma, H.; Zhang, E.; Wang, T.; Yin, L.; He, X.; Hu, Z. Long noncoding RNA cytoskeleton regulator RNA promotes cell invasion and metastasis by titrating miR-613 to regulate ANXA2 in nasopharyngeal carcinoma. Cancer Med. 2020, 9, 1209–1219. [Google Scholar] [CrossRef]
- Liu, F.; Wei, J.; Hao, Y.; Lan, J.; Li, W.; Weng, J.; Li, M.; Su, C.; Li, B.; Mo, M.; et al. Long intergenic non-protein coding RNA 02570 promotes nasopharyngeal carcinoma progression by adsorbing microRNA miR-4649-3p thereby upregulating both sterol regulatory element binding protein 1, and fatty acid synthase. Bioengineered 2021, 12, 7119–7130. [Google Scholar] [CrossRef]
- Gao, C.; Lu, W.; Lou, W.; Wang, L.; Xu, Q. Long noncoding RNA HOXC13-AS positively affects cell proliferation and invasion in nasopharyngeal carcinoma via modulating miR-383-3p/HMGA2 axis. J. Cell Physiol. 2019, 234, 12809–12820. [Google Scholar] [CrossRef]
- Zheng, Y.-J.; Zhao, J.-Y.; Liang, T.-S.; Wang, P.; Wang, J.; Yang, D.-K.; Liu, Z.-S. Long noncoding RNA SMAD5-AS1 acts as a microRNA-106a-5p sponge to promote epithelial mesenchymal transition in nasopharyngeal carcinoma. FASEB J. 2019, 33, 12915–12928. [Google Scholar] [CrossRef]
- Yi, L.; Ouyang, L.; Wang, S.; Li, S.-S.; Yang, X.-M. Long noncoding RNA PTPRG-AS1 acts as a microRNA-194-3p sponge to regulate radiosensitivity and metastasis of nasopharyngeal carcinoma cells via PRC1. J. Cell Physiol. 2019, 234, 19088–19102. [Google Scholar] [CrossRef]
- Shen, Z.; Wu, Y.; He, G. Long non-coding RNA PTPRG-AS1/microRNA-124-3p regulates radiosensitivity of nasopharyngeal carcinoma via the LIM Homeobox 2-dependent Notch pathway through competitive endogenous RNA mechanism. Bioengineered 2022, 13, 8208–8225. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Pan, J.; Luo, Z.; Duan, Q.; Wang, D. Long non-coding RNA FOXD3-AS1 silencing exerts tumor suppressive effects in nasopharyngeal carcinoma by downregulating FOXD3 expression via microRNA-185-3p upregulation. Cancer Gene Ther. 2021, 28, 602–618. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Liu, X.; Lv, B. Long non-coding RNA MEG3 promotes autophagy and apoptosis of nasopharyngeal carcinoma cells via PTEN up-regulation by binding to microRNA-21. J. Cell Mol. Med. 2021, 25, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Cheng, Y.; Xu, L.; Tian, C.; Jiao, H.; Wang, R.; Gao, X. LncRNA NEAT1/miR-129/Bcl-2 signaling axis contributes to HDAC inhibitor tolerance in nasopharyngeal cancer. Aging 2020, 12, 14174–14188. [Google Scholar] [CrossRef]
- Li, H.; You, J.; Xue, H.; Tan, X.; Chao, C. CircCTDP1 promotes nasopharyngeal carcinoma progression via a microRNA-320b/HOXA10/TGFβ2 pathway. Int. J. Mol. Med. 2020, 45, 836–846. [Google Scholar] [CrossRef]
- Ke, Z.; Xie, F.; Zheng, C.; Chen, D. CircHIPK3 promotes proliferation and invasion in nasopharyngeal carcinoma by abrogating miR-4288-induced ELF3 inhibition. J. Cell Physiol. 2019, 234, 1699–1706. [Google Scholar] [CrossRef]
- Li, W.; Lu, H.; Wang, H.; Ning, X.; Liu, Q.; Zhang, H.; Liu, Z.; Wang, J.; Zhao, W.; Gu, Y.; et al. Circular RNA TGFBR2 acts as a ceRNA to suppress nasopharyngeal carcinoma progression by sponging miR-107. Cancer Lett. 2021, 499, 301–313. [Google Scholar] [CrossRef]
- Wang, L.; Sang, J.; Zhang, Y.; Gao, L.; Zhao, D.; Cao, H. Circular RNA ITCH attenuates the progression of nasopharyngeal carcinoma by inducing PTEN upregulation via miR-214. J. Gene Med. 2022, 24, e3391. [Google Scholar] [CrossRef]
- Lo, A.K.-F.; Lung, R.W.-M.; Dawson, C.W.; Young, L.S.; Ko, C.-W.; Yeung, W.W.; Kang, W.; To, K.-F.; Lo, K.-W. Activation of sterol regulatory element-binding protein 1 (SREBP1)-mediated lipogenesis by the Epstein-Barr virus-encoded latent membrane protein 1 (LMP1) promotes cell proliferation and progression of nasopharyngeal carcinoma. J. Pathol. 2018, 246, 180–190. [Google Scholar] [CrossRef]
- Li, H.-L.; Deng, N.-H.; He, X.-S.; Li, Y.-H. Small biomarkers with massive impacts: PI3K/AKT/mTOR signalling and microRNA crosstalk regulate nasopharyngeal carcinoma. Biomark. Res. 2022, 10, 52. [Google Scholar] [CrossRef]
- Wu, G.; Huang, W.; Xu, J.; Li, W.; Wu, Y.; Yang, Q.; Liu, K.; Zhu, M.; Balasubramanian, P.S.; Li, M. Dynamic contrast-enhanced MRI predicts PTEN protein expression which can function as a prognostic measure of progression-free survival in NPC patients. J. Cancer Res. Clin. Oncol. 2022, 148, 1771–1780. [Google Scholar] [CrossRef] [PubMed]
- Lin, F.-J.; Lin, X.-D.; Xu, L.-Y.; Zhu, S.-Q. Long Noncoding RNA HOXA11-AS Modulates the Resistance of Nasopharyngeal Carcinoma Cells to Cisplatin via miR-454-3p/c-Met. Mol. Cells 2020, 43, 856–869. [Google Scholar] [CrossRef] [PubMed]
- Cao, C.; Zhou, S.; Hu, J. Long noncoding RNA MAGI2-AS3/miR-218-5p/GDPD5/SEC61A1 axis drives cellular proliferation and migration and confers cisplatin resistance in nasopharyngeal carcinoma. Int. Forum Allergy Rhinol. 2020, 10, 1012–1023. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, S.; Wang, H.; Cao, J.; Huang, X.; Chen, Z.; Xu, P.; Sun, G.; Xu, J.; Lv, J.; et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol. Cancer 2019, 18, 20. [Google Scholar] [CrossRef] [PubMed]
- D’Ambrosi, S.; Visser, A.; Antunes-Ferreira, M.; Poutsma, A.; Giannoukakos, S.; Sol, N.; Sabrkhany, S.; Bahce, I.; Kuijpers, M.J.E.; Oude Egbrink, M.G.A.; et al. The Analysis of Platelet-Derived circRNA Repertoire as Potential Diagnostic Biomarker for Non-Small Cell Lung Cancer. Cancers 2021, 13, 4644. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, W.; Hao, S. LncRNA CCAT1 modulates the sensitivity of paclitaxel in nasopharynx cancers cells via miR-181a/CPEB2 axis. Cell Cycle 2017, 16, 795–801. [Google Scholar] [CrossRef]
- Zhao, C.H.; Bai, X.F.; Hu, X.H. Knockdown of lncRNA XIST inhibits hypoxia-induced glycolysis, migration and invasion through regulating miR-381-3p/NEK5 axis in nasopharyngeal carcinoma. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 2505–2517. [Google Scholar] [CrossRef]
- D’Ambrogio, A.; Nagaoka, K.; Richter, J.D. Translational control of cell growth and malignancy by the CPEBs. Nat. Rev. Cancer 2013, 13, 283–290. [Google Scholar] [CrossRef]
- Tordjman, J.; Majumder, M.; Amiri, M.; Hasan, A.; Hess, D.; Lala, P.K. Tumor suppressor role of cytoplasmic polyadenylation element binding protein 2 (CPEB2) in human mammary epithelial cells. BMC Cancer 2019, 19, 561. [Google Scholar] [CrossRef]
- Lin, M.; Zhang, X.-L.; You, R.; Yang, Q.; Zou, X.; Yu, K.; Liu, Y.-P.; Zou, R.-H.; Hua, Y.-J.; Huang, P.-Y.; et al. Neoantigen landscape in metastatic nasopharyngeal carcinoma. Theranostics 2021, 11, 6427–6444. [Google Scholar] [CrossRef]
- Lee, V.; Kwong, D.; Leung, T.-W.; Lam, K.-O.; Tong, C.-C.; Lee, A. Palliative systemic therapy for recurrent or metastatic nasopharyngeal carcinoma—How far have we achieved? Crit. Rev. Oncol. Hematol. 2017, 114, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Tang, T.; Yang, L.; Cao, Y.; Wang, M.; Zhang, S.; Gong, Z.; Xiong, F.; He, Y.; Zhou, Y.; Liao, Q.; et al. LncRNA AATBC regulates Pinin to promote metastasis in nasopharyngeal carcinoma. Mol. Oncol. 2020, 14, 2251–2270. [Google Scholar] [CrossRef] [PubMed]
- Lian, Y.; Xiong, F.; Yang, L.; Bo, H.; Gong, Z.; Wang, Y.; Wei, F.; Tang, Y.; Li, X.; Liao, Q.; et al. Long noncoding RNA AFAP1-AS1 acts as a competing endogenous RNA of miR-423-5p to facilitate nasopharyngeal carcinoma metastasis through regulating the Rho/Rac pathway. J. Exp. Clin. Cancer Res. 2018, 37, 253. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Chen, J.; Ma, C.; Pei, S.; Du, M.; Zhang, Y.; Feng, Y.; Yin, R.; Bian, X.; He, X.; et al. Hsa_circ_0046263 functions as a ceRNA to promote nasopharyngeal carcinoma progression by upregulating IGFBP3. Cell Death Dis. 2020, 11, 562. [Google Scholar] [CrossRef]
- Liu, Q.; Shuai, M.; Xia, Y. Knockdown of EBV-encoded circRNA circRPMS1 suppresses nasopharyngeal carcinoma cell proliferation and metastasis through sponging multiple miRNAs. Cancer Manag. Res. 2019, 11, 8023–8031. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Pastushenko, I.; Blanpain, C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol. 2019, 29, 212–226. [Google Scholar] [CrossRef]
- Zhang, J.; Tian, X.-J.; Xing, J. Signal Transduction Pathways of EMT Induced by TGF-β, SHH, and WNT and Their Crosstalks. J. Clin. Med. 2016, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, S.; Bajdak, K.; Meidhof, S.; Burk, U.; Niedermann, G.; Firat, E.; Wellner, U.; Dimmler, A.; Faller, G.; Schubert, J.; et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011, 30, 770–782. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Feng, X.; Hao, X.; Wang, P.; Zhang, Y.; Zheng, X.; Li, L.; Ren, S.; Zhang, M.; Xu, M. CircSETD3 (Hsa_circ_0000567) acts as a sponge for microRNA-421 inhibiting hepatocellular carcinoma growth. J. Exp. Clin. Cancer Res. 2019, 38, 98. [Google Scholar] [CrossRef]
- Li, C.; Hu, J.; Hao, J.; Zhao, B.; Wu, B.; Sun, L.; Peng, S.; Gao, G.F.; Meng, S. Competitive virus and host RNAs: The interplay of a hidden virus and host interaction. Protein Cell 2014, 5, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Gunasekharan, V.; Laimins, L.A. Human papillomaviruses modulate microRNA 145 expression to directly control genome amplification. J. Virol. 2013, 87, 6037–6043. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Wang, Y.; Wang, S.; Wu, B.; Hao, J.; Fan, H.; Ju, Y.; Ding, Y.; Chen, L.; Chu, X.; et al. Hepatitis B virus mRNA-mediated miR-122 inhibition upregulates PTTG1-binding protein, which promotes hepatocellular carcinoma tumor growth and cell invasion. J. Virol. 2013, 87, 2193–2205. [Google Scholar] [CrossRef] [PubMed]
- Au, K.H.; Ngan, R.K.C.; Ng, A.W.Y.; Poon, D.M.C.; Ng, W.T.; Yuen, K.T.; Lee, V.H.F.; Tung, S.Y.; Chan, A.T.C.; Sze, H.C.K.; et al. Treatment outcomes of nasopharyngeal carcinoma in modern era after intensity modulated radiotherapy (IMRT) in Hong Kong: A report of 3328 patients (HKNPCSG 1301 study). Oral Oncol. 2018, 77, 16–21. [Google Scholar] [CrossRef]
- Samaridou, E.; Heyes, J.; Lutwyche, P. Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliv. Rev. 2020, 154–155, 37–63. [Google Scholar] [CrossRef]
- Mukherjee, A.; Waters, A.K.; Kalyan, P.; Achrol, A.S.; Kesari, S.; Yenugonda, V.M. Lipid-polymer hybrid nanoparticles as a next-generation drug delivery platform: State of the art, emerging technologies, and perspectives. Int. J. Nanomed. 2019, 14, 1937–1952. [Google Scholar] [CrossRef]
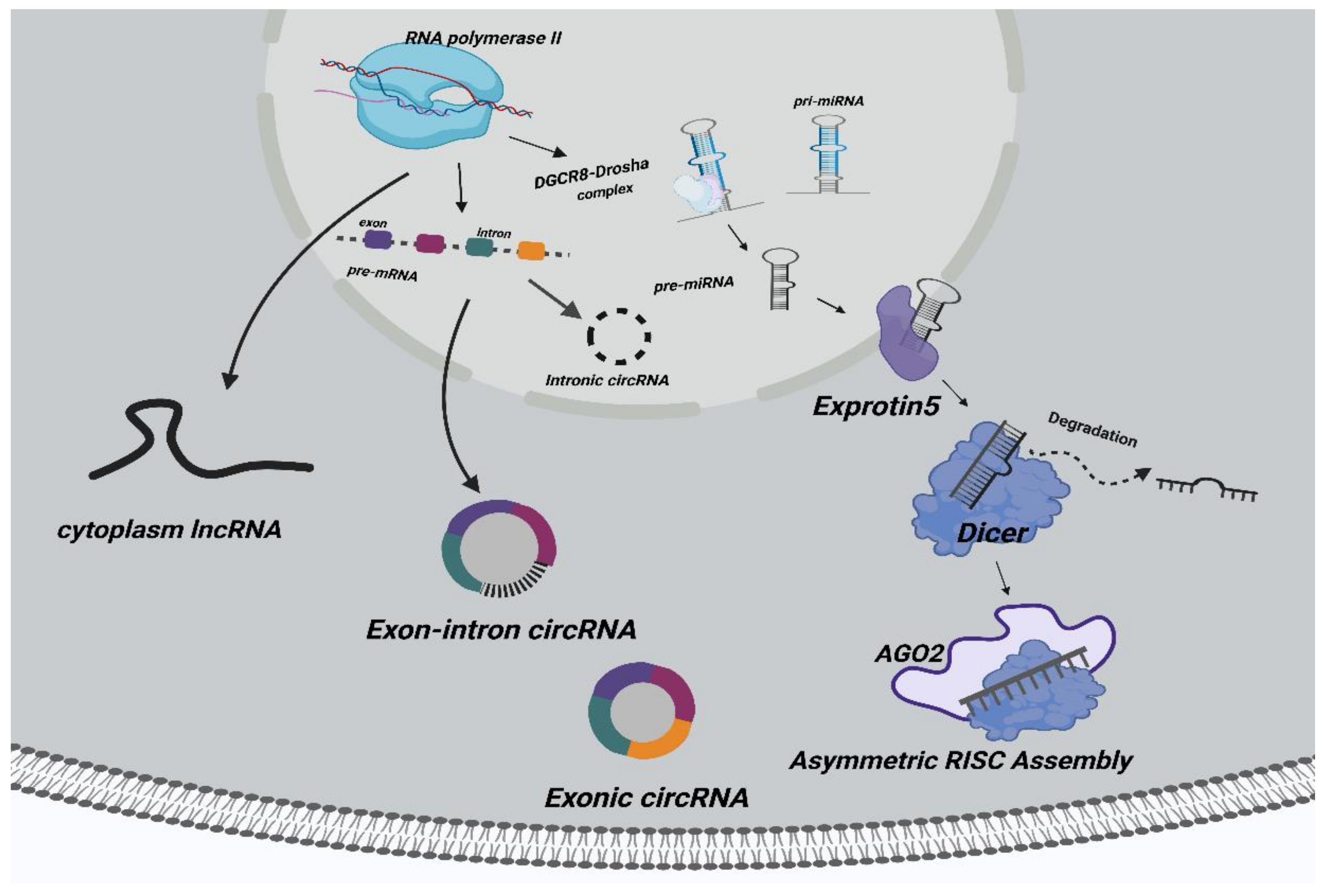
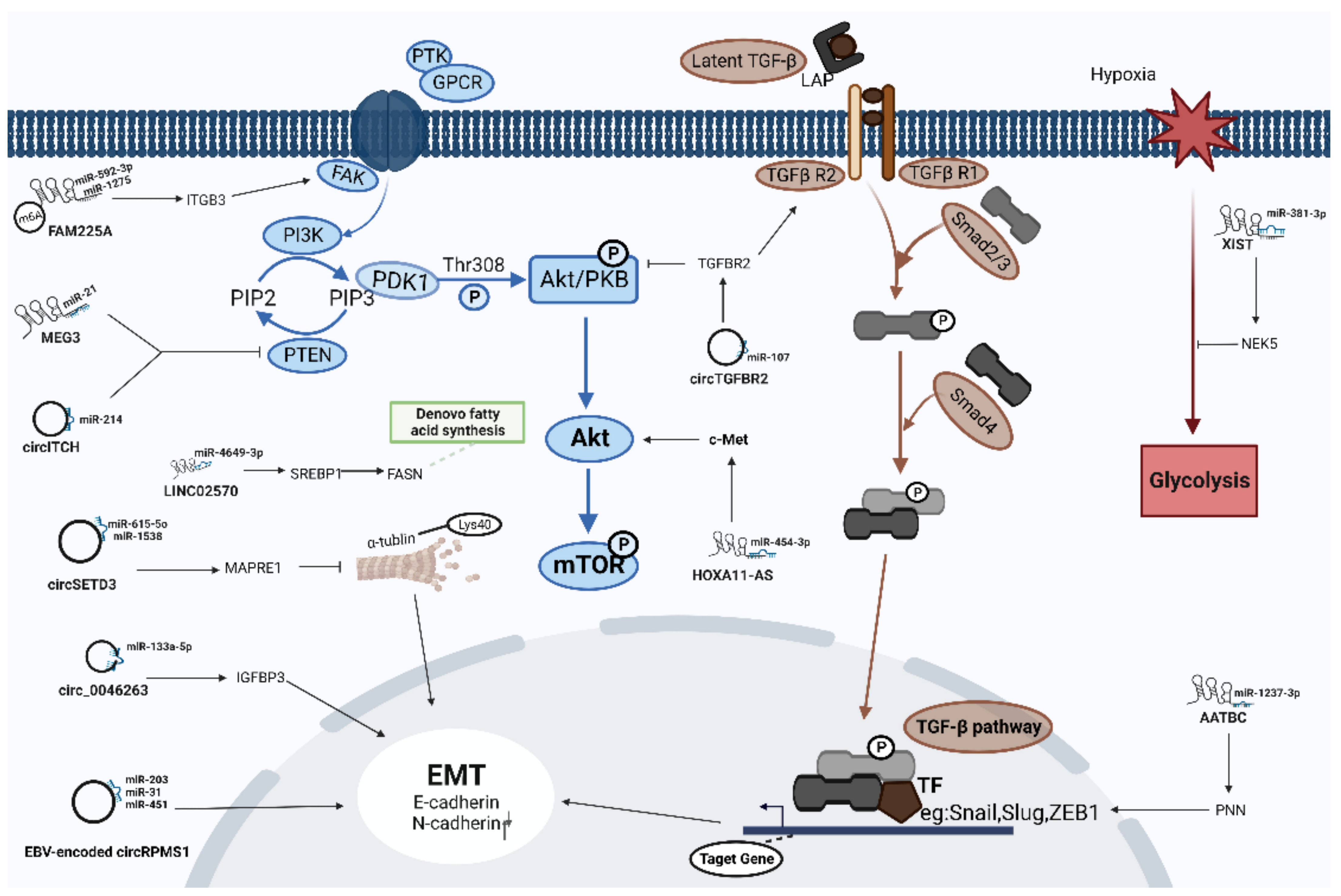
| LncRNA | miRNA | mRNA | Function | Reference |
|---|---|---|---|---|
| ZFAS1 | miR-7-5p | ENO2 | Proliferation, apoptosis, radiation resistance | [39] |
| SNHG5 | miR-1179 | HMGB3 | Proliferation, migration and invasion | [40] |
| SNHG7 | miR-514-5p | ELAVL1 | Proliferation, migration | [41] |
| DRAIC | miR-122 | SATB1 | Proliferation, migration and invasion | [42] |
| SOX2-OT | miR-146b-5p | HNRNPA2B | Proliferation, apoptosis, migration, invasion and metastasis | [43] |
| XIST | miR-148a-3p | ADAM17 | Proliferation, apoptosis, migration, invasion, EMT and metastasis | [44] |
| FAM225A | miR-590-3p miR-1275 | ITGB3 | Proliferation, migration, invasion, metastasis and FAK/PI3K/AKT pathway | [45] |
| CYTOR | miR-613 | ANXA2 | Proliferation, migration, invasion and metastasis | [46] |
| LINC02570 | miR-4649-3p | SREBP1 | Proliferation, invasion, and migration | [47] |
| HOXC13-AS | miR-383-3p | HMGA2 | Proliferation, invasion, and migration | [48] |
| SMAD5-AS1 | miR-106a-5p | SMAD5 | Proliferation, invasion, migration and EMT | [49] |
| PTPRG-AS1 | miR-194-3p | PRC1 | Proliferation, apoptosis, invasion, migration, metastasis and radiosensitivity | [50] |
| PTPRC-AS1 | miR-124-3p | LHX2 | Proliferation, apoptosis and radiosensitivity | [51] |
| FOXD3-AS1 | miR-185-3p | FOXD3 | Proliferation, invasion, migration and cell stemness | [52] |
| MEG3 | miR-21 | PTEN | Apoptosis and autophagy | [53] |
| NEAT1 | miR-129 | Bcl-2 | Apoptosis in SAHA tolerance NPC cell lines | [54] |
| CircRNA | miRNA | mRNA | Function | Reference |
|---|---|---|---|---|
| CircCTDP1 | miR-320b | HOXA10 | Proliferation, invasion, migration, apoptosis and TGFβ2 pathway | [55] |
| CircRNA_000543 | miR-9 | PDGFRB | Proliferation, apoptosis and radiosensitivity | [26] |
| CircHIPK3 | miR-4288 | ELF3 | Proliferation, invasion, and migration | [56] |
| CircTGFBR2 | miR-107 | TGFBR2 | Proliferation, invasion, migration, EMT, TGF-β and PI3K/Akt pathway | [57] |
| CircITCH | miR-214 | PTEN | Proliferation, migration and invasion | [58] |
| LncRNA/CircRNA | miRNA | mRNA | Function | Reference |
|---|---|---|---|---|
| CCAT1 | miR-181a | CPEB2 | Paclitaxel resistance | [66] |
| MAGI2-AS3 | miR-218-5p | GDPD5 SEC61A1 | Cisplatin resistance and EMT Proliferation and migration | [63] |
| CircNRIP1 | miR-515-5p | IL-25 | 5-Fu and cisplatin resistance | [27] |
| CircCRIM1 | miR-422a | FOXQ1 | Docetaxel chemosensitivity, invasion, migration, metastasis and EMT | [38] |
| XIST | miR-381-3p | NEK5 | Glycolysis, migration, invasion and metastasis under hypoxic conditions | [67] |
| HOXA11-AS | miR-454-3p | c-Met | Cisplatin resistance, C-Met/AKT/mTOR pathway | [62] |
| LncRNA/CircRNA | miRNA | mRNA | Function | Reference |
|---|---|---|---|---|
| AATBC | miR-1237-3p | PNN | Migration, invasion, EMT and metastasis | [72] |
| AFAP1-AS1 | miR-423-5p | FOSL2 RAB11B LASP1 | Invasion, migration, metastasis and Rho/Rac pathway, invasion | [73] |
| CircSETD3 | miR-615-5p miR-1538 | MAPRE1 | Invasion, migration and metastasis | [25] |
| Circ_0046263 | miR-133a-5p | IGFBP3 | Proliferation, invasion, EMT and metastasis | [74] |
| EBV-encoded CircRPMS1 | miR-203 miR-31 miR-451 | Proliferation, invasion, EMT and metastasis | [75] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, S.; Li, Y.; Xin, S.; Yang, L.; Jiang, M.; Xin, Y.; Wang, Y.; Yang, J.; Lu, J. Insight into LncRNA- and CircRNA-Mediated CeRNAs: Regulatory Network and Implications in Nasopharyngeal Carcinoma—A Narrative Literature Review. Cancers 2022, 14, 4564. https://doi.org/10.3390/cancers14194564
Zhang S, Li Y, Xin S, Yang L, Jiang M, Xin Y, Wang Y, Yang J, Lu J. Insight into LncRNA- and CircRNA-Mediated CeRNAs: Regulatory Network and Implications in Nasopharyngeal Carcinoma—A Narrative Literature Review. Cancers. 2022; 14(19):4564. https://doi.org/10.3390/cancers14194564
Chicago/Turabian StyleZhang, Senmiao, Yanling Li, Shuyu Xin, Li Yang, Mingjuan Jiang, Yujie Xin, Yiwei Wang, Jing Yang, and Jianhong Lu. 2022. "Insight into LncRNA- and CircRNA-Mediated CeRNAs: Regulatory Network and Implications in Nasopharyngeal Carcinoma—A Narrative Literature Review" Cancers 14, no. 19: 4564. https://doi.org/10.3390/cancers14194564
APA StyleZhang, S., Li, Y., Xin, S., Yang, L., Jiang, M., Xin, Y., Wang, Y., Yang, J., & Lu, J. (2022). Insight into LncRNA- and CircRNA-Mediated CeRNAs: Regulatory Network and Implications in Nasopharyngeal Carcinoma—A Narrative Literature Review. Cancers, 14(19), 4564. https://doi.org/10.3390/cancers14194564







