Biomimetic Red Blood Cell Membrane-Mediated Nanodrugs Loading Ursolic Acid for Targeting NSCLC Therapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Animals and Cell Lines
2.3. Preparation of UA Nanoparticles (UaNPs)
2.4. Preparation of RBCM Vesicles (RVs)
2.5. Preparation of UMNPs
2.6. Determination of Nanoparticle Size and Stability Analysis
2.7. Loading and Release of UA
2.8. Membrane Protein Verification of UMNPs
2.9. Cellular Uptake Assay
2.10. In Vitro Cytotoxicity Assay
2.11. Clone Formation Assay
2.12. Cell Migration Assay
2.13. Invasion Assay
2.14. Apoptosis Assay
2.15. Assessment of Reactive Oxygen Species (ROS) Production
2.16. Western Blot
2.17. In Vivo Therapeutic Effect
2.18. Immunohistochemical (IHC) Analysis
2.19. Statistical Analysis
3. Results
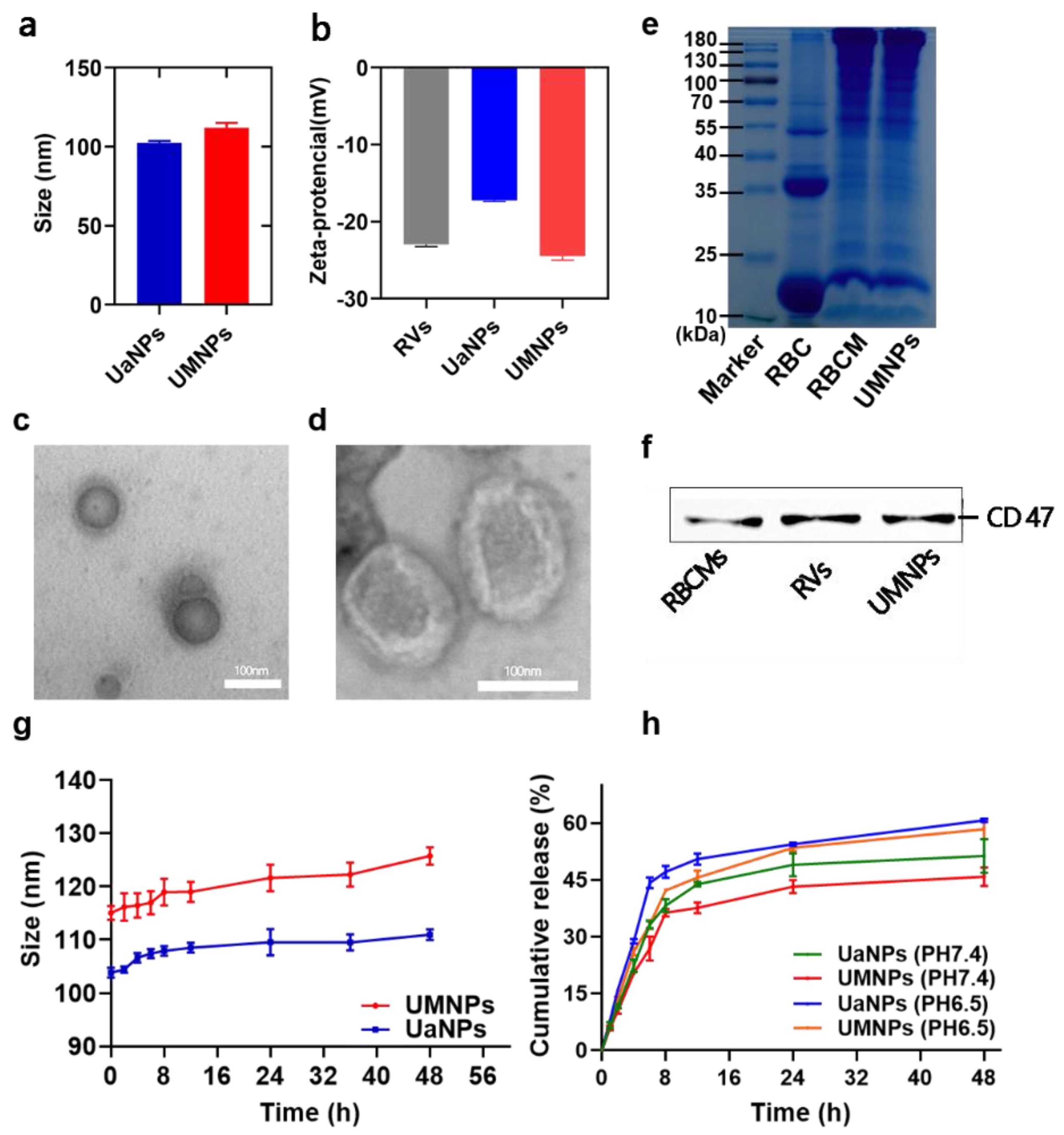
3.1. Synthesis and Characterization of UaNPs and UMNPs
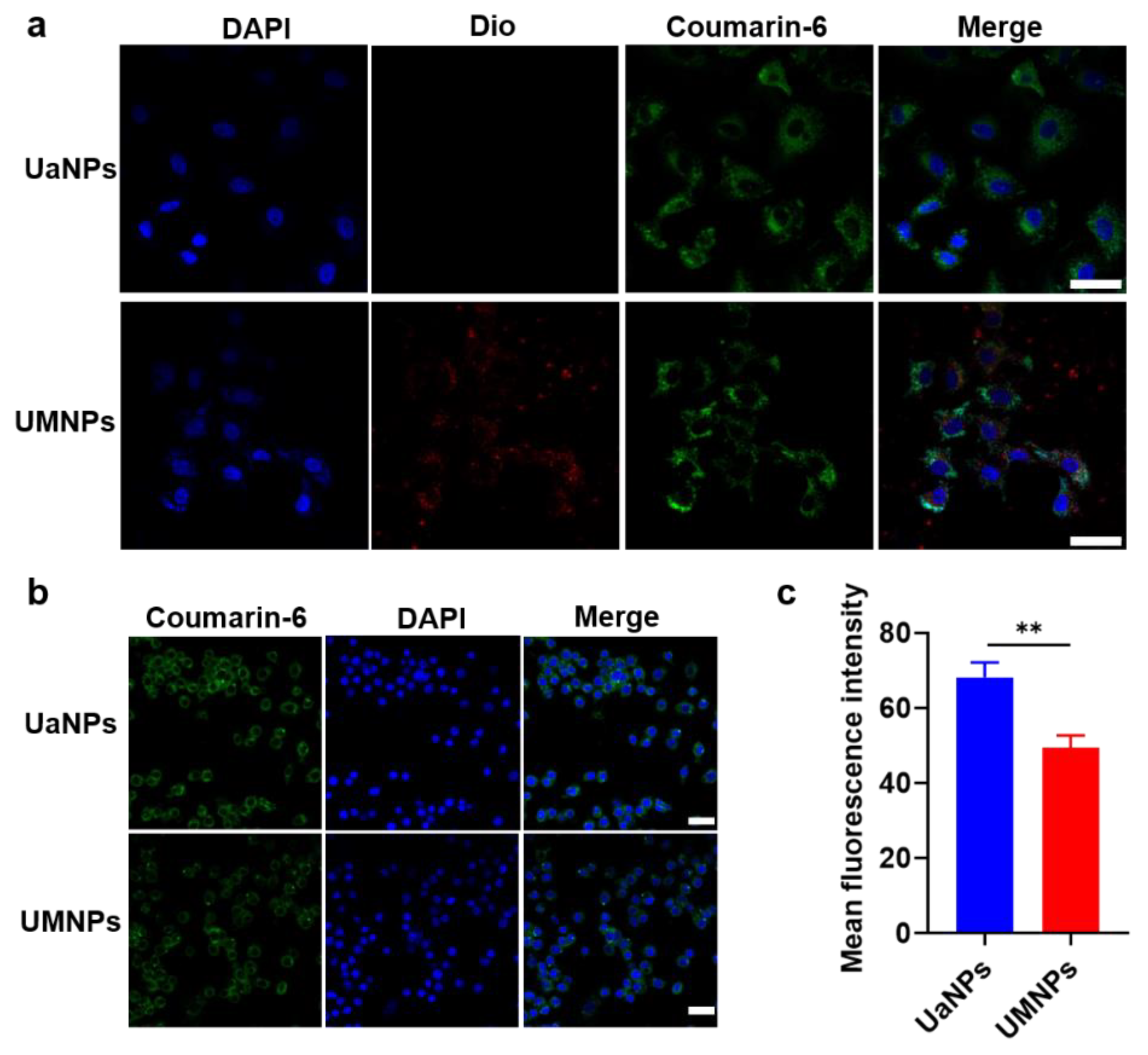
3.2. Cellular Uptake
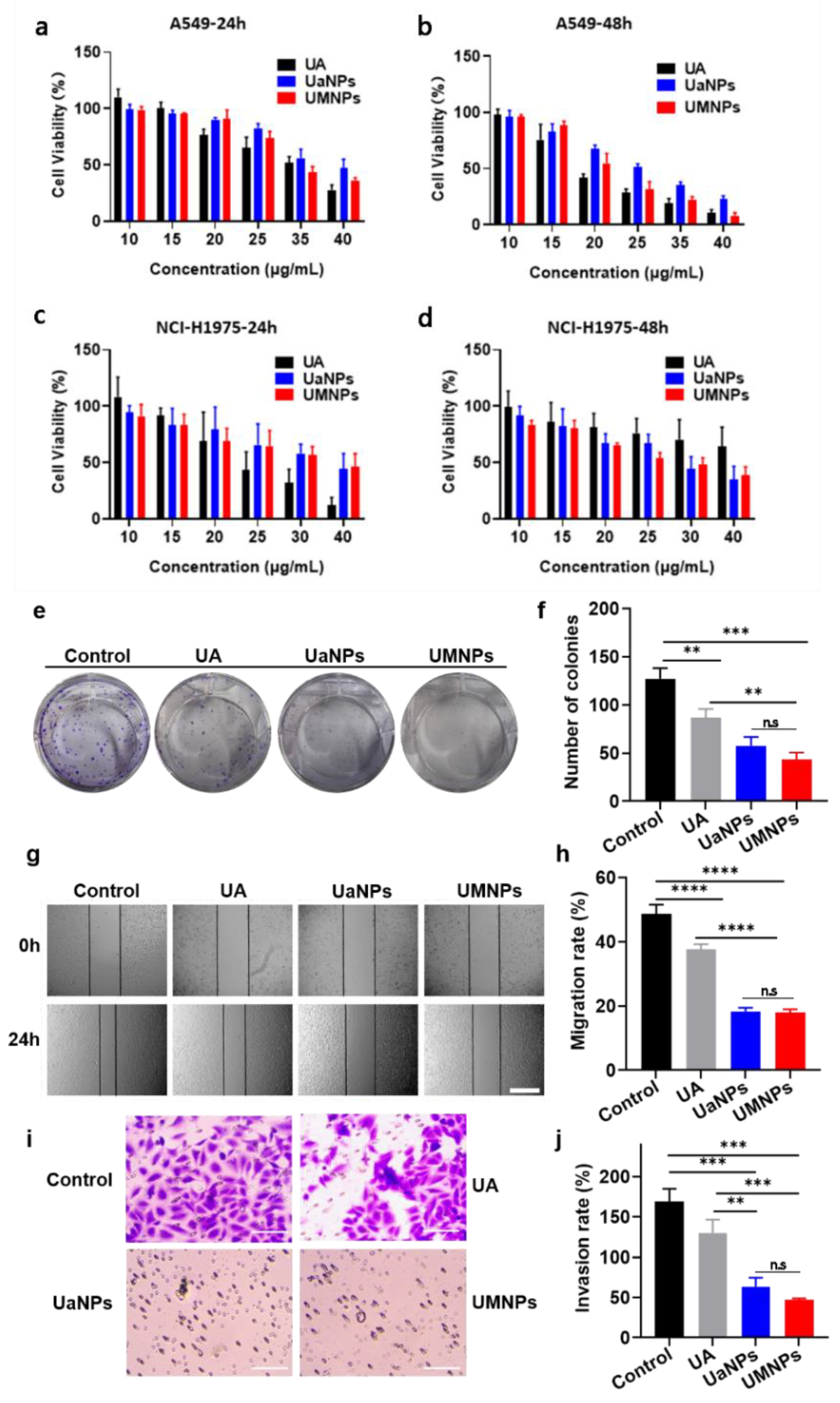
3.3. In Vitro Cytotoxicity, Migration, and Invasion Assays
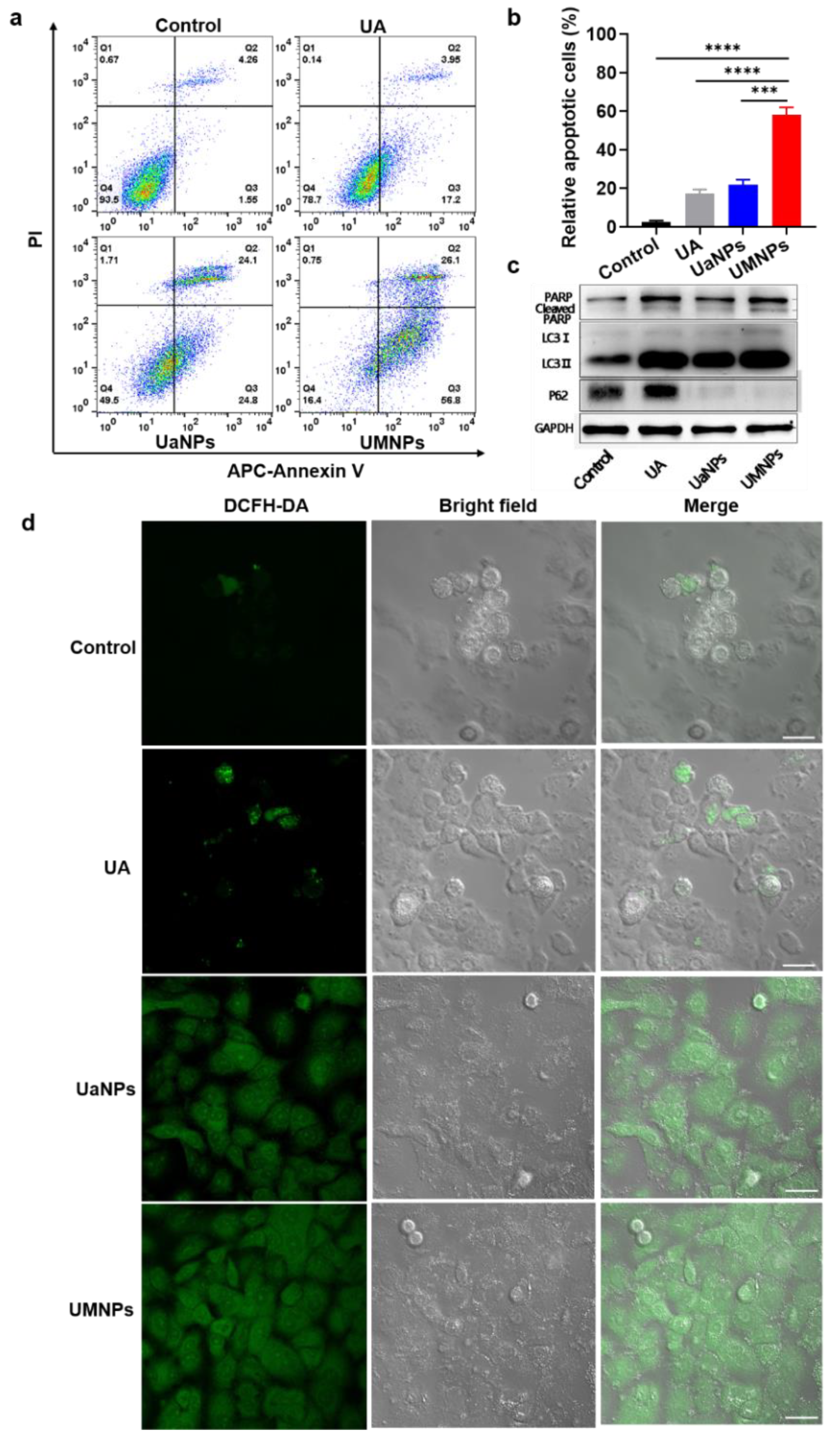
3.4. UMNPs Induce Apoptotic and Autophagic Cell Death in NSCLC Cells
3.5. In Vivo Antitumor Efficacy and Biosafety
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: Globocan Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Thai, A.A.; Solomon, B.J.; Sequist, L.V.; Gainor, J.F.; Heist, R.S. Lung Cancer. Lancet 2021, 398, 535–554. [Google Scholar] [CrossRef]
- Wu, F.; Wang, L.; Zhou, C. Lung Cancer in China: Current and Prospect. Curr. Opin. Oncol. 2021, 33, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Chen, H.-D.; Yu, Y.-W.; Li, N.; Chen, W.Q. Changing Profiles of Cancer Burden Worldwide and in China: A Secondary Analysis of the Global Cancer Statistics 2020. Chin. Med. J. 2021, 134, 783–789. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chan, J.W.Y.; Lau, R.W.H.; Cheung, W.W.; Wong, A.M.; Wong, A.M.; Wong, N.; Ng, C.S.H. Organoids in Lung Cancer Management. Front. Surg. 2021, 8, 753801. [Google Scholar] [CrossRef] [PubMed]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The Biology and Management of Non-Small Cell Lung Cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef] [PubMed]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef] [PubMed]
- Osmani, L.; Askin, F.; Gabrielson, E.; Li, Q.K. Current Who Guidelines and the Critical Role of Immunohistochemical Markers in the Subclassification of Non-Small Cell Lung Carcinoma (NSCLC): Moving from Targeted Therapy to Immunotherapy. Semin. Cancer Biol. 2018, 52, 103–109. [Google Scholar] [CrossRef]
- Imyanitov, E.N.; Iyevleva, A.G.; Levchenko, E.V. Molecular Testing and Targeted Therapy for Non-Small Cell Lung Cancer: Current Status and Perspectives. Crit. Rev. Oncol. Hematol. 2021, 157, 103194. [Google Scholar] [CrossRef]
- Wang, M.; Herbst, R.S.; Boshoff, C. Toward Personalized Treatment Approaches for Non-Small-Cell Lung Cancer. Nat. Med. 2021, 27, 1345–1356. [Google Scholar] [CrossRef]
- Genova, C.; Dellepiane, C.; Carrega, P.; Sommariva, S.; Ferlazzo, G.; Pronzato, P.; Gangemi, R.; Filaci, G.; Coco, S.; Croce, M. Therapeutic Implications of Tumor Microenvironment in Lung Cancer: Focus on Immune Checkpoint Blockade. Front. Immunol. 2022, 12, 799455. [Google Scholar] [PubMed]
- Wang, Y.; Zhang, Q.; Chen, Y.; Liang, C.L.; Liu, H.; Qiu, F.; Dai, Z. Antitumor Effects of Immunity-Enhancing Traditional Chinese Medicine. Biomed. Pharmacother. 2020, 121, 109570. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Su, L.; Zhang, F.; Zhu, X.; Zhu, Y.; Wei, L.; Jiao, X.; Hou, Y.; Chen, X.; Wang, W.; et al. Thevebioside, the Active Ingredient of Traditional Chinese Medicine, Promotes Ubiquitin-Mediated SRC-3 Degradation to Induce NSCLC Cells Apoptosis. Cancer Lett. 2020, 493, 167–177. [Google Scholar] [CrossRef] [PubMed]
- Mlala, S.; Oyedeji, A.O.; Gondwe, M.; Oyedeji, O.O. Ursolic Acid and Its Derivatives as Bioactive Agents. Molecules 2019, 24, 2751. [Google Scholar] [CrossRef]
- Kashyap, D.; Tuli, H.S.; Sharma, A.K. Ursolic Acid (UA): A Metabolite with Promising Therapeutic Potential. Life Sci. 2016, 146, 201–213. [Google Scholar] [CrossRef]
- Ramos-Hryb, A.B.; Pazini, F.L.; Kaster, M.P.; Rodrigues, A.L.S. Therapeutic Potential of Ursolic Acid to Manage Neurodegenerative and Psychiatric Diseases. CNS Drugs 2017, 31, 1029–1041. [Google Scholar]
- Yin, R.; Li, T.; Tian, J.X.; Xi, P.; Liu, R.H. Ursolic Acid, a Potential Anticancer Compound for Breast Cancer Therapy. Crit. Rev. Food Sci. Nutr. 2018, 58, 568–574. [Google Scholar] [CrossRef]
- Erdmann, J.; Kujaciński, M.; Wiciński, M. Beneficial Effects of Ursolic Acid and Its Derivatives—Focus on Potential Biochemical Mechanisms in Cardiovascular Conditions. Nutrients 2021, 13, 3900. [Google Scholar] [CrossRef]
- Tohmé, M.J.; Giménez, M.C.; Peralta, A.; Colombo, M.I.; Delgui, L.R. Ursolic Acid: A Novel Antiviral Compound Inhibiting Rotavirus Infection In Vitro. Int. J. Antimicrob. Agents 2019, 54, 601–609. [Google Scholar] [CrossRef]
- Zhang, Y.; Huang, L.; Shi, H.; Chen, H.; Tao, J.; Shen, R.; Wang, T. Ursolic Acid Enhances the Therapeutic Effects of Oxaliplatin in Colorectal Cancer by Inhibition of Drug Resistance. Cancer Sci. 2018, 109, 94–102. [Google Scholar] [CrossRef]
- Kim, K.; Shin, E.A.; Jung, J.H.; Park, J.E.; Kim, D.S.; Shim, B.S.; Kim, S.H. Ursolic Acid Induces Apoptosis in Colorectal Cancer Cells Partially via Upregulation of MicroRNA-4500 and Inhibition of JAK2/STAT3 Phosphorylation. Int. J. Mol. Sci. 2018, 20, 114. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yu, H.; Wu, R.; Chen, Z.Y.; Hu, Q.; Zhang, Y.F.; Gao, S.H.; Zhou, G.B. Autophagy Inhibition Enhances the Inhibitory Effects of Ursolic Acid on Lung Cancer Cells. Int. J. Mol. Med. 2020, 46, 1816–1826. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.; Kan, Z.; Shan, F.; Zang, J.; Zhou, J. Triple Strategies to Improve Oral Bioavailability by Fabricating Coamorphous Forms of Ursolic Acid with Piperine: Enhancing Water-Solubility, Permeability, and Inhibiting Cytochrome P450 Isozymes. Mol. Pharm. 2020, 17, 4443–4462. [Google Scholar] [CrossRef] [PubMed]
- Schito, A.M.; Caviglia, D.; Piatti, G.; Zorzoli, A.; Marimpietri, D.; Zuccari, G.; Schito, G.C.; Alfei, S. Efficacy of Ursolic Acid-Enriched Water-Soluble and Not Cytotoxic Nanoparticles against Enterococci. Pharmaceutics 2021, 13, 1976. [Google Scholar] [CrossRef]
- Wang, L.; Yin, Q.; Liu, C.; Tang, Y.; Sun, C.; Zhuang, J. Nanoformulations of Ursolic Acid: A Modern Natural Anticancer Molecule. Front. Pharmacol. 2021, 12, 706121. [Google Scholar]
- Miatmoko, A.; Mianing, E.A.; Sari, R.; Hendradi, E. Nanoparticles Use for Delivering Ursolic Acid in Cancer Therapy: A Scoping Review. Front. Pharmacol. 2021, 12, 787226. [Google Scholar]
- Moyers-Montoya, E.D.; Escobedo-González, R.G.; Vargas-Requena, C.L.; Garcia-Casillas, P.E.; Martínez-Pérez, C.A. Epithelial Growth Factor-Anchored on Polycaprolactone/6-deoxy-6-amino-β-cyclodextrin Nanofibers: In Vitro and In Vivo Evaluation. Polymers 2021, 13, 1303. [Google Scholar] [CrossRef]
- Danhier, F.; Ansorena, E.; Silva, J.M.; Coco, R.; Le Breton, A.; Préat, V. PLGA-Based Nanoparticles: An Overview of Biomedical Applications. J. Control. Release 2012, 161, 505–522. [Google Scholar]
- Kozma, G.T.; Shimizu, T.; Ishida, T.; Szebeni, J. Anti-PEG Antibodies: Properties, Formation, Testing and Role in Adverse Immune Reactions to PEGylated Nano-Biopharmaceuticals. Adv. Drug Deliv. Rev. 2020, 154, 163–175. [Google Scholar]
- Shimizu, T.; Abu Lila, A.S.; Awata, M.; Kubo, Y.; Mima, Y.; Hashimoto, Y.; Ando, H.; Okuhira, K.; Ishima, Y.; Ishida, T. A Cell Assay for Detecting Anti-PEG Immune Response against PEG-Modified Therapeutics. Pharm. Res. 2018, 35, 223. [Google Scholar] [CrossRef]
- Fang, R.H.; Kroll, A.V.; Gao, W.; Zhang, L. Cell Membrane Coating Nanotechnology. Adv. Mater. 2018, 30, 1706759. [Google Scholar] [CrossRef] [PubMed]
- Xia, Q.; Zhang, Y.; Li, Z.; Hou, X.; Feng, N. Red Blood Cell Membrane-Camouflaged Nanoparticles: A Novel Drug Delivery System for Antitumor Application. Acta Pharm. Sin. B 2019, 9, 675–689. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Wang, C.; Liu, Z. Red Blood Cells as Smart Delivery Systems. Bioconjug. Chem. 2018, 29, 852–860. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Fillmore, C.M.; Hammerman, P.S.; Kim, C.F.; Wong, K.K. Non-Small-Cell Lung Cancers: A Heterogeneous Set of Diseases. Nat. Rev. Cancer 2014, 14, 535–546. [Google Scholar]
- Xiang, Y.; Guo, Z.; Zhu, P.; Chen, J.; Huang, Y. Traditional Chinese Medicine as a Cancer Treatment: Modern Perspectives of Ancient but Advanced Science. Cancer Med. 2019, 8, 1958–1975. [Google Scholar] [CrossRef]
- Ma, Z.; Wang, N.; He, H.; Tang, X. Pharmaceutical Strategies of Improving Oral Systemic Bioavailability of Curcumin for Clinical Application. J. Control. Release 2019, 316, 359–380. [Google Scholar]
- Alves, R.C.; Fernandes, R.P.; Eloy, J.O.; Salgado, H.R.N.; Chorilli, M. Characteristics, Properties and Analytical Methods of Paclitaxel: A Review. Crit. Rev. Anal. Chem. 2018, 48, 110–118. [Google Scholar] [CrossRef]
- Xu, H.E.; Wang, T.T.; Yang, C.B.; Li, X.; Liu, G.; Yang, Z.; Singh, P.K.; Krishnan, S.; Ding, D. Supramolecular Nanofibers of Curcumin for Highly Amplified Radiosensitization of Colorectal Cancers to Ionizing Radiation. Adv. Funct. Mater. 2018, 28, 1707140. [Google Scholar] [CrossRef]
- Li, X.; Yu, N.; Li, J.; Bai, J.; Ding, D.; Tang, Q.; Xu, H. Novel “Carrier-Free” Nanofiber Codelivery Systems with the Synergistic Antitumor Effect of Paclitaxel and Tetrandrine through the Enhancement of Mitochondrial Apoptosis. ACS Appl. Mater. Interfaces 2020, 12, 10096–10106. [Google Scholar] [CrossRef]
- He, M.; Zhu, J.; Yu, N.; Kong, H.; Zeng, X.; Xie, W.; Xu, H. The Superior Antitumor Effect of Self-Assembled Paclitaxel Nanofilaments for Lung Cancer Cells. Curr. Drug Deliv. 2019, 16, 171–178. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, T.; Yan, D.; Hou, W.; Jiang, H.; Wu, M.; Wang, Y.; Chen, G.; Tang, C.; Wang, Y.; Xu, H. Biomimetic Red Blood Cell Membrane-Mediated Nanodrugs Loading Ursolic Acid for Targeting NSCLC Therapy. Cancers 2022, 14, 4520. https://doi.org/10.3390/cancers14184520
Wu T, Yan D, Hou W, Jiang H, Wu M, Wang Y, Chen G, Tang C, Wang Y, Xu H. Biomimetic Red Blood Cell Membrane-Mediated Nanodrugs Loading Ursolic Acid for Targeting NSCLC Therapy. Cancers. 2022; 14(18):4520. https://doi.org/10.3390/cancers14184520
Chicago/Turabian StyleWu, Ting, Dan Yan, Wenjun Hou, Hui Jiang, Min Wu, Yanling Wang, Gang Chen, Chunming Tang, Yijun Wang, and Huae Xu. 2022. "Biomimetic Red Blood Cell Membrane-Mediated Nanodrugs Loading Ursolic Acid for Targeting NSCLC Therapy" Cancers 14, no. 18: 4520. https://doi.org/10.3390/cancers14184520
APA StyleWu, T., Yan, D., Hou, W., Jiang, H., Wu, M., Wang, Y., Chen, G., Tang, C., Wang, Y., & Xu, H. (2022). Biomimetic Red Blood Cell Membrane-Mediated Nanodrugs Loading Ursolic Acid for Targeting NSCLC Therapy. Cancers, 14(18), 4520. https://doi.org/10.3390/cancers14184520





