Simple Summary
Arsenic is a chemical element that is toxic, and long-term exposure to it causes cancers such as lung, skin, liver, and bladder cancers. Over 150 million people around the world are affected by arsenic exposure. However, the molecular mechanism of how arsenic induces carcinogenesis is still not clear. As a carcinogen, arsenic has been demonstrated not to cause point mutations. Hence, the understanding of the dysregulation of epigenetic mechanisms caused by arsenic may help to unravel the mechanisms by which arsenic induces cancers.
Abstract
Arsenic is a crucial environmental metalloid whose high toxicity levels negatively impact human health. It poses significant health concerns to millions of people in developed and developing countries such as the USA, Canada, Bangladesh, India, China, and Mexico by enhancing sensitivity to various types of diseases, including cancers. However, how arsenic causes changes in gene expression that results in heinous conditions remains elusive. One of the proposed essential mechanisms that still has seen limited research with regard to causing disease upon arsenic exposure is the dysregulation of epigenetic components. In this review, we have extensively summarized current discoveries in arsenic-induced epigenetic modifications in carcinogenesis and angiogenesis. Importantly, we highlight the possible mechanisms underlying epigenetic reprogramming through arsenic exposure that cause changes in cell signaling and dysfunctions of different epigenetic elements.
1. Arsenic and Mechanisms of Arsenic-Induced Carcinogenesis
Arsenic (As), a chemical element, is classified as a toxic metalloid and is associated with various human cancers [1], and its toxicity depends on the molecular form and oxidation state. International Agency for Research on Cancer (IARC) and US Environmental Protection Agency (USEPA) designated arsenic as Group 1 and Group A human carcinogens, respectively [2,3]. Furthermore, it is graded as first on the substance priority list in the Agency for Toxic Substances and Disease Registry (ASTDR), USA (https://www.atsdr.cdc.gov/spl/index.html, accessed on 9 February 2022) [4]. Chronic exposure to dietary arsenic is linked to skin, bladder, liver, and lung cancer [4,5,6,7]. Drinking water contaminated with arsenic has been linked with increased mortality of both noncancerous diseases and cancers in Bangladesh [8]. Both chronic and acute exposure to arsenic is harmful to different tissues and organs in the body, such as alteration in skin pigmentation and hyperkeratosis, peripheral neuropathy, development, cognitive impairments, and cardiovascular diseases.
The contamination of arsenic increases with the finding of newer places [9]. The familiar sources of arsenic exposure include drinking water, food, and inhalation in an industrial work setting. Over 150 million people on the earth are exposed to carcinogenic (10 μg/L) levels of arsenic [9,10], and the majority of these people are affected by drinking water from aquifers contaminated with arsenic. Countries with arsenic concentrations exceeding this carcinogenic level (10 μg/L) in the drinking water include Bangladesh, India, China, Argentina, Mexico, Canada, the USA, and Chile [11]. Arsenic exposure to foods usually occurs by growing crops in the soil contaminated with arsenic and/or irrigating water contaminated with arsenic [12]. Furthermore, NIOSH estimates that approximately 1.5 million workers have been affected by arsenic or arsenic compounds [13].
Arsenic has several states; the most common valence states of arsenic are inorganic AsIII (arsenite) and AsV (arsenate). Inorganic arsenic is very toxic to humans, whereas organic arsenic has low toxicity. AsIII is the highest toxic form because it is more soluble in water than arsenic compounds. It contains a lone electron pair that can engage in chemical bonds [14,15]. Depending on the types of food, arsenic can be found in both inorganic (when combined with oxygen, chlorine, and sulfur, among other elements) and organic forms (when linked with carbon and hydrogen). Inorganic arsenic is typically found in the inorganic form in drinking water, soil, and some terrestrial foods such as rice, as either AsIII or AsV. Inorganic pentavalent arsenic AsV is absorbed by the body through drinking water and uses membrane transporters such as aquaporin and inorganic phosphate transporters (PiT) to enter the cells [16,17]. In the cell, arsenic AsV is converted to the more toxic form arsenite in a glutathione-dependent reaction (GSH), with subsequent methylation to mono-methylated (MMA) and di-methylated arsenicals (DMA), respectively [18,19]. Methylated arsenicals, especially MMAIII, are considered more toxic than inorganic AsIII both in vivo (in hamsters) [20] and in vitro (human cell lines) [21].
The mechanisms by which arsenic induces carcinogenesis are still a point of debate. However, it has been known that arsenical compounds contribute to carcinogenesis by disrupting the signaling cascade, changing gene expression, elevating levels of oxidative stress and inflammation, increasing genotoxic and DNA damage, decreasing DNA repair, inducing cell cycle arrest and apoptosis [19,22,23,24], acting as co-carcinogenesis with other environmental toxicants [25], and alterations of epigenetic regulation. There is also doubt about whether arsenic is genotoxic or not because arsenic does not cause point mutations in standard mutagenicity assays; hence it is considered to be nongenotoxic [26,27]. Although arsenic is viewed as a carcinogen, its non-mutagenic characteristics violet its function in causing genetic alteration. However, a less studied mechanism, but one that is crucial for understanding arsenic-induced carcinogenesis, is the dysregulation of epigenetic modifications. Studies investigating epigenetic regulation changes upon arsenic exposure during the last decade are increasing. The researchers have attempted to explore the role of DNA methylation, histone modification, miRNAs and lncRNAs alteration, mRNA modification, and alternative splicing in arsenic toxicity and carcinogenesis. The present review will comprehensively discuss the epigenetic regulations involved in gene expression, and their dysregulation is pivotal in arsenic-induced transformation, tumor growth, and angiogenesis. The general scheme of the mechanism of arsenic-induced carcinogenesis is shown as follows (Figure 1).
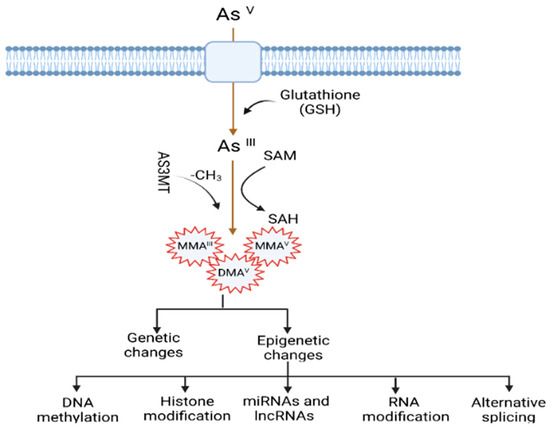
Figure 1.
Mechanisms of arsenic-induced carcinogenesis. Arsenic exposure induces carcinogenesis via its biotransformation process, which causes effects on both genetic and epigenetic levels. The biotransformation of arsenic happens via a series of reactions such as reduction, oxidation, and methylation. Pentavalent arsenic (AsV) is reduced to trivalent (AsIII) and then methylated into organic arsenic species with higher carcinogenic potential. Here, S-adenosylmethionine (SAM) acts as a methyl donor, and Glutathione (GSH) and other thiols serve as reducing agents. Epigenetic alterations induced by arsenic exposure include abnormal changes in DNA methylation, histone modification, miRNAs and lncRNAs expression, RNA modification, and alternative splicing.
2. Arsenic-Induced Changes in DNA Methylation
DNA methylation is the inclusion of the methyl group (-CH3) in the 5-carbon on the cytosine residues (5 mC) in CpG (Cytosine-Phosphate-Guanine) and non-CpG (CpA, CpT, and CpC) dinucleotides. The methyl group comes from a methyl donor, generally from S-adenyl methionine (SAM), and this process is mediated by DNA methyl transferases (DNMTs) [28]. CpG dinucleotides are concentrated in CpG islands (short CpG-rich DNA stretches) and regions of repetitive sequences such as centromeric repeats, retrotransposon elements, rDNA, etc. [29,30,31]. In cancers, the changes of methylation status mainly occur within CpG islands, which occupy ~70% of all mammalian promotors. In addition, these islands play an important role in the regulation of transcription, and their general changes have been found during malignant transformation [32,33]. The functional effect of the dysregulation of DNA methylation is context- and spatial-dependent, dynamic, tissue-specific, and trans-generationally heritable [34,35,36]. Generally, gene silencing involves promotor methylation, and constitutive gene expression is associated with gene body methylation [32]. However, the methylation of the gene body may also be found to inactivate repetitive DNA elements within the gene body [35,37] and show dramatic alteration intron-exon boundaries [38]. These complex methylation patterns underline the necessity of DNA methylation profiling to answer biological questions.
Although it is evident that the dysregulation of DNA methylation has been demonstrated in different cancers, our knowledge of the impact of inorganic arsenic (iAs) on DNA methylation is still growing. Methyl transferase (MTs) catalyze the methyl group transfer in the 5-carbon on the cytosine residues (5 mC) in CpG dinucleotides and use SAM, a coenzyme, as a methyl group donor. Long-term exposure to arsenic causes depletion of the SAM by MTs such as AS3MT [arsenic (III) methyl transferase]. Furthermore, arsenic can also control DNMTs and decrease their activities. For example, studies have found that arsenic exposure causes a reduction of mRNA levels and activity of DNMTs [39,40,41].
iAs exposure has been shown to change global DNA methylation in vitro, in animal studies as well as in population studies (Table 1). For instance, a chronic low-dose of iAs exposure induces DNA hypo-methylation in cells [42]. In addition, fish, mice, and rats exposed to iAs exhibit hepatic global DNA hypomethylation [42,43,44]. However, limited studies are available for the human population compared to in vitro and animal studies. A recent study assessed the association between arsenic exposure and global DNA methylation (∼850,000 CpGs) through drinking water among 396 Bangladeshi people who joined the Health Effects of Arsenic Longitudinal Study (HEALS). The study identified 34 CpGs associated with arsenic concentration in the urinary tract and found a positive relationship between higher arsenic concentration and DNA hypomethylation in those CpGs. Among the arsenic-associated CpGs, most of the genes were annotated to the reactive oxygen species (ROS) pathway, tumor necrosis factor-α (TNF-α) signaling, and inflammatory response via nuclear factor kappa B (NF-κB). These are essential hallmarks of cancer and aging [45]. The results are consistent with earlier studies indicating that epigenetic alterations potentially regulate arsenic toxicity [45]. Pilsner et al. showed that iAs exposure led to global hypomethylation of leukocytes in human skin. The authors observed that people with hypo methylation in the peripheral blood lymphocytes (PBL) DNA were prone to skin lesions two years later when they adjusted for age, urinary As, and other factors [46]. A whole-genome microarray-based study showed that the status of DNA methylation changed over time in people who were affected by arsenic-induced skin lesions compared to control in Bangladesh. The study found the top 20 differentially methylated CpG sites. Among these top CpG sites, the methylation percentages increased in 13 CpGs, and decreased in 7 CpGs between baseline and follow-up [47]. Bandyopadhyay et al. evaluated the association of cytogenetic damage by measuring lymphocyte micronucleus (MN) frequency and long interspersed nuclear element-1 (LINE-1) methylation status among children who were exposed to arsenic in the areas of West Bengal. They observed that a high reduction of LINE-1 methylation was associated with MN frequency in exposed children compared to unexposed children, suggesting that LINE-1 methylation is a potential epigenetic marker for arsenic toxicity in individuals [48].
Besides the changes in global DNA methylation status, iAs exposure also causes changes in DNA methylation in specific regions of targeted genes in different cancers [49]. For example, the association between arsenic exposure and hypomethylated or hypermethylated promotors of some genes was found in human skin cancer [50] and bladder cancer [51,52]. The carcinogenesis can occur due to the silence of tumor suppressor genes via hypermethylation [40]. Some studies have found that iAs exposure leads to increased methylation of the promotor for tumor suppressor genes such as p15, p16, p53, and death-associated protein kinase (DAPK) in vitro and in vivo [40,50,51], DNA repair-related genes such as ERCC2, RPA1 in human hepatocytes [53], MLH1 in whole human blood [54], and genes associated with the Wnt pathway like MYC and WNT2B [53]. However, another study involving a human population chronically exposed to arsenic demonstrated hypomethylation at the promoter of the DNA repair gene ERCC2 [55]. Smeester et al. comprehensively studied the status of DNA methylation within CpG islands for more than 14,000 genes among arsenic-exposed individuals with skin lesions and without skin lesions [56]. They identified 183 genes with differentially methylated CpG islands, of which 182 were hyper-methylated in individuals with signs of arsenicosis. Gene enrichment analysis showed that most genes involved cancer-linked pathways via genes such as p53. They also identified an arsenic-methylated tumor suppressorome, a complex of 17 known or putative tumor suppressors silenced in human cancers, which includes hypermethylated genes such as chromosome 11 open reading frame 70 (C11orf70), centromere protein E (CENPE), forkhead box F1 (FOXF1), homeobox B5 (HOXB5), homeobox B9 (HOXB9), hsa-mir-126, SWI/SNF related, matrix associated, actin dependent regulator of chromatin subfamily d member 2 (SMARCD2), T-box brain 1 (TBR1), etc. Chanda et al. showed the hypermethylation of GMDS gene fragments in the peripheral blood leukocyte DNA of individuals exposed to arsenic and with skin cancer. They indicated it as a biomarker for arsenic-induced cancer [57]. The AS3MT gene plays an essential role in the metabolism of arsenic and its toxicological mechanism. Gribble et al. found decreased methylation in the promotor region of AS3MT in an arsenic-exposed area in Arizona [58]. However, no further studies have been performed to investigate the association between skin lesion status and AS3MT promoter methylation to date. On the other hand, carcinogenesis can also occur due to the activation of oncogene genes via hypomethylation. For instance, mice treated with iAs showed hypomethylation of the promoter region of oncogene Hras1 and increased mRNA levels of Hras1 [59], which was consistent with another study showing hypomethylation and increased mRNA levels of Hras1 and c-myc in vitro [60,61]. Arsenic exposure also led to Esr1 gene overexpression via hypomethylation of its promoter region, which is closely related to arsenic-induced hepatocarcinogenesis [44]. However, a recent study by Janasik et al. found hypermethylation of genes promoter of Nuclear factor-erythroid factor 2-related factor 2 (NRF2) and Kelch-like ECH-associated protein 1 (KEAP1) among occupationally arsenic-exposed copper mill workers from Poland [62].
DNA methylation inhibition occurs in a site-specific manner by proteins known as the ten-eleven translocation (TET) enzymes [63,64]. These TET enzymes oxidize 5 mc to 5 hyrdoxymethylcytosine (5 hmc). Disruption of this group of proteins has been shown in different types of cancer. Wang et al. showed that As inhibited the TET-mediated DNA demethylation and subsequently induced the hypermethylation in the promotor region to suppress the antioxidant genes 8-oxoguanine DNA glycosylase (OGG1) and glutathione S-transferase Pi 1 (GSTP1), thus increasing oxidative stress in human bronchial epithelial (HBE) cells in vitro [65]. In another recent study, Domingo-Relloso et al. conducted an epigenome-wide association study (EWAS) to compare the association of different As exposure levels and human blood 5 mc and 5 hmc markers in two diverse populations from the Aragon Workers Health Study (AWHS, Spain) and the Folic Acid and Creatinine Trial (FACT, Bangladesh) [66]. The effect of As on site-specific 5 mC and 5 hmC was measured using the Illumina methylation EPIC array on more than 850,000 CpG sites. They indicated different epigenetic effects for low As exposure in the AWHS population and high As exposure in the FACT population. The differentially methylated (DMP) and hydroxymethylated (DHP) positions were primarily found in distinct genomic sites. For example, they found three DMPs annotated to CLEC12A, a gene that plays a role in inflammation and immune response, which was consistent with previous studies [67]. In addition, they also found one DHP annotated to NPLOC4, a gene that has protein processing function in the endoplasmic reticulum (ER) in the FACT population exposed to a high dose of As. This is invariable to a study that reported a role of As in ER stress-associated protein misfolding and apoptosis [68], for which mechanisms are known to be associated with cardiometabolic diseases and cancer.
In addition, arsenic exposure also causes transgenerational genotoxicity and the alteration of global DNA methylation patterns in the animal model. Parental chronic arsenic exposure led to genotoxic damage (F0–F3), different methylation patterns, changes in physical and reproductive parameters, abnormal morphology in the ovaries (F0 and F1) and testicles (F1–F3), and a decline in the quality of sperm (F0–F3, except F2), suggesting that an individual’s early life disruptions can negatively impact later generations’ health [36]. An association was found between low or high-dose exposure to arsenic during gestation with umbilical cord blood DNA methylation. There was increased DNA methylation in CpG sites of LINE-1 and, to a lesser extent, within the promotor region of p16 [69]. Studies also showed the sex-dependent association between arsenic exposure and cord blood DNA methylation status, and the impact was even more prominent in the boys than in the girls [70].
Arsenic doesn’t induce point mutations but causes deletion mutations and chromosomal instability [40]. One possible mechanism by which arsenical compounds contribute to carcinogenesis is the disruption of normal epigenetic marks at specific loci, which may cause changes in gene expression and carcinogenesis [71,72]. Although arsenic exposure was found to alter methylation levels in global DNA and promoters of some genes, current research is hard to understand due to the complexity and insubstantial information provided in the current studies. Further investigations are necessary to systematically explore DNA methylation on a genome-wide level in cell lines exposed to arsenic and target tissues from well-characterized arsenic-exposed populations or tumor tissues from arsenic-associated cancers. Such studies will assist in elucidating the possible biological effects of arsenic exposure on DNA methylation and carcinogenesis. Arsenic-induced alterations of DNA methylation status and carcinogenesis are summarized in Table 1.

Table 1.
Arsenic-induced alterations of DNA methylation status and carcinogenesis.
Table 1.
Arsenic-induced alterations of DNA methylation status and carcinogenesis.
| Tissue/Cells | Source of Arsenic | DNA Methylation | References | ||
|---|---|---|---|---|---|
| Global | Gene-Specific | ||||
| Hyper | Hypo | ||||
| Prostate epithelial cell line RWPE-1 | AsIII | Hypo | [73,74] | ||
| HaCaT keratinocytes | AsIII | Hypo | [39] | ||
| TRL 1215 rat liver epithelial cells | AsIII | Hypo | [42] | ||
| Goldfish | AsIII | Hypo | [75] | ||
| Fisher 344 rat | AsIII | Hypo | [43] | ||
| 129/SvJ mice | AsIII | Hypo | [44] | ||
| Blood samples | Drinking water | Hypo | [45] | ||
| Blood samples from skin lesion patients and control | 13 Hyper and 7 hypo methylation of CpG islands | [47] | |||
| Human | Hyper | [76] | |||
| Hypo (in skin lesion patients) | [46] | ||||
| Peripheral blood lymphocyte DNA from skin lesions and non-skin lesions | Drinking water (urine samples) | 182 genes out of 183 hypermethylated; Identified a silenced tumor suppressorome consists of 17 genes | [56] | ||
| MMAIII | ZHCAN12 and C1QTNF6 | ||||
| Uroepithelial SV-HUC-1 cells | AsIII | DAPK | [77] | ||
| Hamster embryo cells | AsIII | c-myc and Ha-ras | [61] | ||
| TRL 1215 rat liver epithelial cells | AsIII | c-myc | [51] | ||
| C57BL/6J mice | AsIII | c-Ha-ras | [59] | ||
| A/J mice | AsV | p16, RASSF1 | [78] | ||
| C3H mice | AsIII | ERα | [79] | ||
| Blood samples from the people of West Bengal, India | Drinking water | p53 and p21 in skin cancer patients | [50] | ||
| Tissues from arsenic-induced skin lesions (cases) and with no skin lesions (controls) | Drinking water | DAPK and p16 | [80] | ||
| Blood samples from copper mill workers and Non-occupationally exposed healthy controls in Poland | Copper mill (urine) | NRF2 and KEAP1 | [62] | ||
| Blood samples from arsenic-exposed individuals (with and without skin lesions) | Drinking water (water, urine) | MLH1 and MSH2 | [81] | ||
| Samples from bladder tumor | Drinking water (toenail) | RASSF1A and PRSS3 | [52] | ||
| Cord blood lymphocytes | Drinking water (cord blood, nails, and hair) | p53 | [82] | ||
| Blood samples from the West Bengal population and HEK293 cell lines | Drinking water(water, urine), sodium arsenite, AsIII | Increased ERCC2 expression | [55] | ||
| Blood samples from arsenic-exposed individuals (with and without skin lesions) | Drinking water (water, urine) | Increased Tfam and PGC1α expression | [83] | ||
3. Arsenic Alters Histone Post-Translational Modification (PTM)
Genomic DNA is vast, around 2 m in length, and fits into nuclei of approximately 6 µm diameter by packaging it into chromatin [84]. The nucleosome is the central unit of chromatin, which consists of around 146 DNA base pairs wrapped 1.65 times around an octamer of histone protein, comprising two copies of each histone: H2A, H2B, H3, and H4 [85] and linked by the fifth histone (H1), which helps stabilize the nucleosome and facilitates the folding of nucleosomes into chromatin compression [86]. Post-translational modifications (PTMs) of these histone proteins alter chromatin’s structure and function, leading to a change in gene expression. Besides chromatin structures and gene expressions, histone modifications also affect various biological processes such as replication, repair mechanisms, and the recombination of DNA [87]. Histone modification includes acetylation, methylation, phosphorylation, and others such as ubiquitylation, biotinylation, glycosylation, carbonylation, ADP-ribosylation, crotonylation, propionylation, N-formylation, sumoylation, citrullination, etc. [88]. In this review, we will discuss the most studied histone modifications due to arsenic exposure.
3.1. Histone Acetylation
Histone acetylation or deacetylation is a dynamic and reversible event [89]. More than 50 years ago, Alfrey et al. demonstrated that histone acetylation and transcriptional activity were positively correlated [90]. Since then, histone acetylation has been found to be an important event for gene regulation by increasing the ability to regulate and activate transcription through chromatin modification [91]. Histone acetyltransferases (HATs) and histone deacetylases (HDACs) are the two types of antagonistic enzymes that regulate histone acetylation and deacetylation, respectively. HATs catalyzed the addition of an acetyl group to the ε-amino group of specific lysine side chains within the histone’s basic N-terminal tail region by using acetyl co-A as a donor. This event neutralizes the lysine’s positive charge and weakens the interactions between histones and DNA, resulting in a relaxation of chromatin, which favors higher transcription. The acetylation at particular lysine sites can also recruit the SWItch/Sucrose Non-Fermentable (SWI/SNF) complexes which are bromodomain-containing proteins that help change the structure of chromatin to a more open state to enable it for active transcription [92]. In contrast, HDACs remove the acetyl group from the lysine residues and favor compact chromatin [93].
Increasing evidence has indicated that arsenic can change the pattern of acetylation in the histone proteins at different parts of the chromatin. In the 1980s, it was reported that arsenic exposure significantly decreased histone acetylation in Drosophila [94]. More recently, alteration in both H3 and H4 histone elements has been associated with global or dose-dependent arsenic exposure [95,96,97]. For example, both AsIII and MMAIII exposure has been shown to induce malignant transformation of human urothelial cells in vitro and to alter histone H3 acetylation patterns [98]. In addition, the same study found DNA hypermethylation in a number of promoters that are already hypoacetylated. This result leads us to believe that the genes may be targeted in a coordinated manner by arsenic via the alteration of various epigenetic mechanisms to promote malignant transformation [98]. In addition, H3K27 and H3K9 acetylation has been associated with occupational arsenic exposure [95,99]. During embryo development, arsenic also increases global H3K9 hypoacetylation [96]. Jo et al. showed that in human bladder epithelial cells, the H4K16 acetylation global level was decreased in a dose- and time-dependent manner after exposure to both AsIII and MMAIII treatment [100]. Moreover, silencing the gene MYST1, which is needed for H4K16 acetylation, caused higher cytotoxicity from arsenical exposure, suggesting that H4K16 acetylation may be crucial for resistance to arsenic-induced toxicity. Several other researchers have investigated the influence of arsenic on specific histone acetylation and noticed dissimilarities from lysine residue. Arsenic did not change H4K5ac [99,101] or H4K8ac [101]. AsIII did not change H3K14ac in the APL cell lines [102]. However, an epidemiological study observed a positive association between urinary arsenic and H3K14ac in lymphocytes [103]. Interestingly, AsIII exposure was found to upregulate the genes required in apoptosis or for the response to cell stress by inducing histone acetylation via HDACs [102,104] and by hindering HDAC genes that associate with higher global histone acetylation [105]. It has been reported that the HDAC inhibitor can restore arsenic-induced endothelial dysfunction and dementia, and inhibit malignant transformation induced by iAs [101,106].
3.2. Histone Methylation
Histone methylation takes place mainly on the lysine and arginine side chain and does not change the charge of the histone protein, as opposed to histone acetylation and phosphorylation. Furthermore, this modification adds another level of complexity. Lysine can be mono-, di-, or tri-methylated, while arginine can be mono- and di-methylated [107,108,109]. Histone methylation is commonly observed on the histones H3 and H4. However, H2A and H2B methylations are also noted. Both transcriptional activation and repression are observed with histone methylation. For example, H3K4 and H3K36 methylation was correlated with transcriptional activation, but H3K9, H3K27, and H4K20 methylation had been shown to induce transcriptional repression [110,111]. It was also believed that histone methylation was an irreversible and permanent epigenetic change [112]. However, recently, enzymes such as histone lysine demethylase and arginine deiminase were shown to directly remove a methyl group from lysine residue and antagonize histone arginine methylation, respectively.
The abnormal loss or gain of histone methylation levels has been demonstrated in tumorigenesis [113]. For example, researchers observed that H2B total methylation levels were increased by treating embryonic cells from Drosophila melanogaster with 50 μM trivalent arsenic, whereas H3 and H4 histone methylation were abolished [94,114]. However, mammalian cell responses to arsenic exposure are not simple: the varied effects of AsIII on the methylation of H3 lysine residues were observed, including higher H3K9 dimethylation (H3K9me2) and H3K4 tri-methylation (H3K4me3) and lower H3K27 tri-methylation (H3K27me3) [115]. A study on human lung carcinoma A549 cells confirmed the increased H3K4me3 after 24-h or seven-day exposures, which is consistent with a previous study that utilized RWPE1 cells initially derived from a human prostate [116,117]. Methylation at H3K4me3 has been shown to correlate with active gene transcription and is generally found in the transcription start sites [118]. However, H3K9me2 is a repression mark. Increased H3K9me2, which is catalyzed by higher levels of G9a protein, a nuclear lysine methyl transferase [115], is associated with reversible modification correlated with transcriptional repression [88], and has been demonstrated to be involved in the silencing of tumor suppressor genes in the cancer cell lines [119,120]. H3K27me3 is also a repressive epigenetic mark that is crucial for regulating genes and the inactivation X chromosome [121]. Although multiple studies have investigated arsenic effects on H3K27me3, the discoveries were inconsistent. A study did not find any alteration of H3K27me3 after treating HepG2 cells derived from a male human liver tumor with 7.5 μM AsIII [105]. However, Zhou et al. demonstrated that 2.5 and 5 μM μg/L of AsIII decreased H3K27me3 in A549 cells [115]. This result is consistent with a population-based study among Bangladeshi men in which an inverse association between As exposure and H3K27me3 in peripheral blood mononuclear cells (PBMCs) was observed [122]. In contrast, there was a positive correlation between As exposure and H3K27me3 among women [122], similar to research demonstrating that 0.5 μM AsIII enhanced this post-transcription modification using embryonic fibroblasts derived from a female mouse [123]. Arsenic also induced epigenetic modification via the generation of oxidative stress [124]. Ma et al. found a positive relationship between arsenic levels in hair and urine, and altered total H3K9me2 and H3K36me3 amounts. The alteration of H3K36me3 was found to be higher in the promoter regions of oxidative stress response (OSR) genes in HaCaT and HEK cells [103].
3.3. Histone Phosphorylation
The phosphorylation of histone is crucial for chromatin condensation and transcriptional activation during mitosis and meiosis [125]. Phosphorylation can modify all four histone core proteins (H2A, H2B, H3, and H4) and linker protein H1. It mainly occurs on serine, threonine, and tyrosine residues. Phosphatases and kinases control this modification. For example, H2A and H2B phosphorylation is catalyzed by several kinases, such as ataxia telangiectasia mutated (ATM) for H2AX [126]. In contrast, Cyclin-dependent kinases (CDKs) are responsible for H1 phosphorylation [127]. H3 phosphorylation is found during cell cycle progression and regulation of gene expression [128]. Similarly, histone H4 (serine 1) phosphorylation is increased during the cell cycle and is regulated by casein kinase 2 [129]. The function of histone phosphorylation is to loosen the chromatin by acting against the positively charged histone protein [130].
Histone phosphorylation plays an important role in arsenic-induced carcinogenesis. Exposure to a high dose AsIII (10 μM) has been shown to reduce the total H1 and H3 phosphorylation levels in Chinese hamster ovary cells [131]. However, several studies using various cell lines have demonstrated consistently that different doses and durations of both AsIII and DMAIII can globally induce H3 phosphorylation on a serine residue (H3S10ph) [102,132,133,134,135], which is essential for the regulation of chromosome segregation during mitosis [136]. Studies have also indicated that H3 phosphorylation induced by arsenic exposure may be necessary for the upregulation of the oncogenes c-fos and c-jun [104] and the induction of caspase 10, a proapoptotic factor [102]. Interestingly, nickel, another metal, has also been demonstrated to increase H3S10 (serine 10) through the activation of the c-jun N-terminal kinase/stress-activated protein kinase (JNK/SAPK) pathway [137]. Arsenic exposure activates JNK and p38/Mpk2 kinase [138], and histone H3 phosphorylation through the JNK/SAPK pathway, which may be a common mechanism of metal-induced histone modification.
Overall, these studies prove that the dysregulation of PTM occurs due to arsenic exposure. However, the findings have been inconsistent in some cases because of many factors, including dose and time of exposure differences, duration and type of arsenic compound, and compounding factors such as sex contribution and measurement error. Hence, further work is necessary to completely unravel the relationship between altered histone modification and arsenic exposure, and to elucidate the total amount of altered PTM of histone on arsenic-induced carcinogenesis and angiogenesis.
4. Abnormal Changes of MicroRNAs and lncRNAs upon Arsenic Exposure
4.1. MicroRNAs
MicroRNAs (miRNAs) are small non-coding RNAs that participate in different biological regulatory events such as RNA silencing and post-transcriptional regulation of genes. Ambros and colleagues discovered lin-4, the first miRNA in Caenorhabditis elegans, as a small non-coding RNA that affected development via regulating the expression of the protein lin-14 [139]. Since then, miRNAs have been found to be present in both invertebrates and vertebrates, and some of them are highly conserved across the species, leading us to believe that miRNA-mediated post-transcriptional regulation is a general regulatory function across species [140,141,142]. Each miRNA is believed to target several hundred mRNAs, and each mRNA may be suppressed by several different types of miRNAs [143]. MicroRNAs regulate mRNA through sequence-specific RNA–RNA interactions in the 3′ untranslated region (3′-UTR) of targeted mRNA, destabilizing the mRNA and deactivating gene expression [144,145]. Currently there are 38,589 hairpin precursors and 48,860 mature miRNAs from 271 organisms recorded in miRBase catalogs, though the roles of many miRNAs are still unknown [146]. Approximately 30% of mammalian genes are regulated by miRNAs [147]. In addition, increasing evidence has established the dysregulation of miRNA expression in cell differentiation, proliferation, and angiogenesis that can lead to carcinogenesis via different mechanisms. These mechanisms include amplification or deletion, a transcriptional control of miRNAs, dysregulated epigenetic changes, and defects in the miRNA biogenesis machinery [145].
It has been demonstrated that miRNAs are heavily dysregulated in cancers [19,148]. Because miRNAs are negative regulators of gene expression, dysregulation of these miRNAs can be tumorigenic if targeted mRNAs are either tumor suppressors or oncogenes. For instance, the let-7 miRNA family directly targets the RAS oncogene to suppress its expression, and the reduction of let-7 miRNA family members leads to the overexpression of RAS oncoprotein. Conversely, perturbation of the miR-34 family leads to the dysregulation of the p53 tumor suppressor pathway [40,149].
Although notable progress has been made regarding the biogenesis and mechanisms of miRNAs in different types of cancer, our knowledge is limited about the dysregulation of miRNAs in As-induced carcinogenesis. The very first study of arsenic-induced miRNA dysregulation in cell transformation was done by Wang et al. in 2011. They found that exposure to a low concentration of arsenic for 16 weeks led to malignant transformation and reduced miR-200b/c expression in immortalized human bronchial epithelial cells (HBECs) with p53 knockdown. The inhibition of malignant transformation occurred when the same cells were forcefully expressed with miR-200b [150]. Since then, there has been growing evidence of miRNA dysregulation in As-mediated carcinogenesis (Table 2).

Table 2.
miRNA alteration and carcinogenesis due to arsenic exposure.
Kong et al. found an association between the reduced level of miR-21 and an increase in urinary arsenic levels in Hong Kong children aged 12–19 [170]. In contrast, there was an increase of miR-21 and miR-222 in the peripheral blood of steelworkers [171]. When non-malignant human keratinocytes (HaCaT) were treated with arsenic, 30 miRNAs were differentially expressed in arsenic-exposed cells compared to control cells. This study confirmed the upregulation of previously found miRNAs involved in carcinogeneses, such as miR-21, miR-200a, and miR-141, which were indicated as potential biomarkers for the epithelial phenotype of cancer cells [172]. In addition, exposure to As led to the upregulation of miR-151 and miR-183 in liver tissues of rats [173] and miR-155 in cultured 16-HBE cells [158]. Zeng et al. evaluated the relationship between the expression of miRNAs and multiorgan damage in control and arsenic-exposed populations in China [155]. The study found associations between miR-155 and arsenic-induced skin damage between miR-21, miR-145, and liver damage, and between miR-191 and kidney damage, indicating that these miRNAs act as potential biomarkers for As-induced multiorgan injury. In a recent study, Al-Eryani et al. analyzed the miRNA expression profile in non-malignant hyperkeratosis (HK) and malignant skin lesion tissues, squamous cell carcinoma (SCC), and basal cell carcinoma (BCC) from West Bengal (India) people chronically exposed to high levels of arsenic, and found the differential expression of 35 miRNAs among the three skin lesions. They found that miR-425-5p and miR-433 were upregulated in both BCC and SCC compared to HK and were potentially associated with malignancy. However, miR-184 and miR-576-3p were upregulated in SCC alone compared to both BCC and HK. MiR-29c, miR-381, miR-452, miR-487b, miR-494, and miR-590-5p were selectively decreased in BCC compared to both SCC and HK. They summarized both phenotype- and stage-related differential miRNA expression profiles that may serve as possible biomarkers for arsenic-induced internal tumors [9,152].
Several potential mechanisms are associated with miRNA dysregulation in As-mediated cancerous outcomes. For example, As exposure led to the generation of reactive oxygen species (ROS) and conceivably changed the miRNA expression [41,163,164]. A study found that miR-21 was upregulated in the malignant transformed human embryo lung fibroblast (HELF) cells after As exposure, which was due to the activation of the ERK/NF-κB pathway by ROS [174]. We also showed that chronic As exposure led to ROS generation in human bronchial epithelial BEAS-2B cells, which induced cyclooxygenase-2 (COX2) and hypoxia-inducible factor (HIF)1-α expression through miR199a-5p suppression, thus promoting tumor growth and angiogenesis. The forced expression of miR-199a-5p suppressed COX2 and HIF1-α expression and impaired arsenic-induced angiogenesis and tumor growth [153]. Another mechanism study showed that arsenic promoted epithelial-mesenchymal transition (EMT) by inducing pro-inflammatory cytokine interleukin-6 (IL-6) secretion, mediating the signal transducer and activator of transcription 3 (STAT3) signaling, and increasing miR-21 expression in an autocrine manner [175]. In addition, miR-301a was also found to be increased in human lung epithelial BEAS-2B cells exposed to As, and miR-301 was an oncogenic miRNA that directly antagonized SMAD4 in the IL6/STAT3/miR-301a/SMAD4 signaling pathway during As-induced carcinogenesis [151]. Liu et al. found that increased miR-21 expression inhibited tumor suppressor programmed cell death protein 4 (PDCD4) and activated the ERK signaling pathway through the decreased expression of tumor suppressor PTEN [176]. Similarly, the induction of miR-222 expression by As exposure inhibited PTEN expression and was responsible for inducing cell transformation and tumor growth [164].
4.2. Long Noncoding RNAs (ln cRNAs)
Long noncoding RNA (lncRNA) is a type of RNA that is greater than 200 nucleotides and is not translated into a protein. Like mRNAs, lncRNAs are also generally transcribed by RNA polymerase II and processed with a 5′-cap structure and 3′-end poly-A, followed by RNA splicing and editing to create isoform transcripts. Because of their low expressions, lncRNAs were primarily thought to be a transcription noise. However, with the advancement of technology and better understanding, lncRNAs were observed to be involved in the transcription and post-transcription regulation via interaction with RNA, DNA, or proteins [177]. The lncRNAs can be found in the genomic loci, which are putatively intronic, intergenic, or intersected with protein-coding regions in either sense or antisense orientation, which can control the target gene expression in the downstream via cis- or trans-regulatory mechanism. In addition, lncRNAs also regulate mRNA splicing and act as predecessors to noncoding RNAs (ncRNAs), such as miRNAs [178,179,180]. They can function as tumor suppressors or oncogenes and play roles in various signaling pathways [181]. Several lncRNAs have been recognized as independent or additional biomarkers in the diagnosis and prognosis of cancer [182].
A recent study found that programmed cell death 1 ligand (PD-L1) and STAT3 were upregulated in arsenic-transformed BEAS-2B cells, and knockdown of STAT3 inhibited arsenic-induced PD-L1 upregulation. Lnc-DC, an lncRNA, was an upstream regulator to mediate arsenic-induced STAT3 activation, suggesting that Lnc-DC/STAT3 cascade may mediate PD-L1 upregulation during arsenic-induced transformation [154]. In lung cancer cells, STAT3 is directly bound to the PD-L1 promotor and is necessary for PD-L1 expression [183,184]. Ji et al. found that the expression levels of metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), one of the well-known lncRNAs, highly correlated with the tumor stage and metastasis of non-small cell lung cancer (NSCLC) [185]. In the area of arsenic exposure, the levels of MALAT1 expression were increased in hepatocellular carcinoma (HCC) patients [186]. Increased hypoxia-inducible factor 2α (HIF-2α) and MALAT1 expression levels were also found in HCC tissues and arsenic-induced transformed human hepatic epithelial (L-02) cells. Functionally, the upregulation of MALAT1 and HIF-2α enhanced the invasive capability of arsenic-transformed L-02 cells and HCC-LM3 cells. Mechanistically, As induced MALAT1 and separated the von Hippel-Lindau (VHL) protein from HIF-2α to reduce the ubiquitination of VHL-mediated HIF-2α, resulting in HIF-2α accumulation. In L-02 cells, arsenite exposure enhanced glycolysis [187]. In addition to HIF-2α upregulation as above, arsenic exposure also increased the expression of HIF-1α through the lncRNA MALAT1. Furthermore, arsenic exposure enhanced glycolysis by HIF-1α stabilization via MALAT1, but not by HIF-2α [187]. Currently, it is well established that enhanced glycolysis plays an essential role in cancer initiation and progression [188,189,190]. These discoveries give additional proof that supports a critical role of the MALAT1 upregulation in arsenic-mediated carcinogenesis. Animals exposed to arsenic treatment also showed the upregulation of MALAT1 during the progression of mouse liver fibrosis. Together, these studies show that arsenic exposure upregulates MALAT1 expression in both cultured cells and mice, suggesting a critical role of lncRNA MALAT1 in arsenic carcinogenicity and toxicity [191]. However, the role and mechanisms of these upregulated lncRNAs in arsenic-induced carcinogenesis are currently not clear and remain to be elucidated.
Overall, arsenic exposure changes the expression profiles of miRNAs and lncRNAs, which may serve as potential biological markers and provide therapeutic values for arsenic-induced carcinogenesis. However, the research is limited to lncRNAs, and more studies are necessary to unravel their function during this process. Similarly, studies are also required to investigate miRNAs in As-induced cancer in different stages, especially mechanisms related to direct vs. indirect effects of arsenic-targeted miRNAs on the population and how these miRNAs control different signaling pathways to cause cancer and other diseases.
5. Arsenic Causes Abnormal RNA Modification
RNA methylation is a reversible post-transcriptional alteration to RNA that epigenetically regulates different biological processes and is widely present in both eukaryotes and prokaryotes [192]. In this process, a methyl group is transferred from an active methyl compound to a different compound. It occurs not only in messenger RNA (mRNA), but also in other RNA species, including transfer RNA (tRNA), ribosomal RNA (rRNA), transfer-messenger RNA (tmRNA), small nucleolar (snoRNA), microRNA, viral RNA, and so on [193,194]. RNA methylation modulates RNA splicing [195], stability [196], translation [197,198], DNA damage repair [199], nuclear export [200], miRNA biogenesis initiation [201], immunogenicity [202], and the occurrence and development of cancer [203,204]. Among more than 170 types of modification that have been observed in all kinds of RNAs, methylation accounts for more than 50% of them. When methylation is found at the sixth N of the adenylate of RNA, it is called m6A methylation. Studies also found other forms of RNA methylation besides m6A methylation, such as m6Am, m7G, m1A, and m5c. However, m6A alteration has been considered the most abundant methylation alteration in the eukaryote mRNA [205] that affects every process in the life cycle of RNA [206].
M6A methyl transferases such as methyltransferase-like enzyme 3/14 (METTL3/14), Wilms tumor 1-associated protein (WTAP), RBM15/15B, and KIAA1429 catalyze m6A modification [207]. The binding proteins, called ‘readers’, which can recognize and bind to the methylated RNA, decode m6A methylation, and generate a functional signal. Readers include eukaryotic initiation factor (eIF) 3, YT521-B homology (YTH) domain-containing protein [208], heterogeneous nuclear ribonucleoprotein (HNRNP) protein family [209], and the IGF2 mRNA binding proteins (IGF2BP) family [210]. On the other hand, demethylases such as fat mass and obesity-associated protein (FTO) and alkB homolog 5 (ALKBH5) are called “erasers” because they remove the methyl group from the target mRNAs. Growing evidence has suggested that the affluence of m6A and expressions of its regulators, including writers, erasers, and readers, are often dysregulated in different types of cancers and are essential for cancer initiation, progression, metastasis, as well as drug resistance and cancer relapse [211,212,213,214,215]. For instance, METTL3 can recruit translation initiation factors directly and increase RNA translation. It promoted cell growth, survival, and invasion by upregulating EGFR and TAZ in lung adenocarcinoma [216]. Choe et al. demonstrated that METTL3 promoted the translation and transformation of oncogenes and formed dense polyribosomes by interacting with EIF3H in primary lung cancer, which might be a potential therapeutic target [217]. The role of METTL14 in lung cancer is controversial; a recent study showed that METTL14 knockdown suppressed the malignant progression of non-small cell lung cancer (NSCLC) by reducing Twist expression [218]. However, other studies showed that METTL14 was downregulated in lung adenocarcinoma (LUAD) and mediated lncRNA HCG11 [219] or miR-30c-1-3p to inhibit tumor growth [220]. The overexpression of FTO decreased m6A levels in MZF1 mRNA transcripts, increased mRNA stability, and promoted MZF1 expression, leading to the proliferation and invasion of lung squamous cell carcinoma cells [221]. Although m6A RNA modification dysregulation is associated with various cancers, the underlying mechanisms of m6A in cancer have not yet been fully understood.
Gao et al. found that As treatment reduced m6A modification near the stop codon of an endogenous inducer of somatic mutation gene APOBEC3B (A3B) in the human alveolar basal epithelial cells from adenocarcinoma. FTO was responsible for reducing m6A alteration in A3B, which led to increased A3B expression and higher DNA mutation rates of the m6A reader YTHDF2. They confirmed that A3B was a downstream target of FTO in lung tissues from As-exposed mice. FTO protein expression was positively correlated with A3B protein expression in tumor samples from human NSCLC patients [222]. Another study also confirmed that As treatment increased FTO expression, decreased m6A RNA methylation, and consequently induced malignant transformation and tumorigenesis in keratinocytes. FTO deletion inhibited arsenic-induced tumorigenesis in both in vitro and in vivo experiments. Arsenic stabilization of the FTO protein occurred via impeding p62-mediated autophagy, which led to a positive feedback loop to keep up FTO accumulation [223]. Unfortunately, few studies have been performed on As-induced dysregulation of m6A methylation; hence, further studies are necessary to understand the potential molecular mechanisms of m6A in As-induced tumorigenesis and cancer progression.
6. Arsenic Exposure and Alternative Splicing
The removal of intronic sequences and splicing together of adjacent exons are necessary for RNA maturation and translation into protein. The elimination of introns is followed by attaching exons in their DNA-corresponding order, known as consecutive splicing, which occurs at every intron-exon boundary [224]. Alternative splicing diverges from this process via mechanisms that reorder the pattern of exons into alternative coding sequences that translate to different proteins. This mechanism is an evolutionarily conserved process that significantly increases transcriptome and proteome diversity from a limited genome. Alternative splicing is necessary to maintain cellular homeostasis and is essential in regulating cell differentiation and development [225,226]. Alternative splicing is tightly controlled by other significant processes in the cell, and perturbation of this process is known to occur commonly in human cancers [63,227,228]. Alternative splicing is associated with carcinogenesis [229], angiogenesis [230], and EMT [227,228]. Growing evidence has demonstrated that the decision of alternative splicing or consecutive splicing occurs while the mRNA is still tied to the DNA and takes place transcriptionally [231]. Several studies have shown the factors that control the structure of chromatin, such as histone PTMs and DNA methylation, dictate the selection of exon candidates for splicing [232,233,234].
As discussed, exposure to iAs significantly changes DNA methylation and histone post-translational modifications. Hence, it is reasonable to believe that it may play a part in alternative splicing by changing chromatin organization. Cardoso et al. found that when human immortalized human keratinocytes (HaCaT) were treated with sodium arsenite at 100 nM for 28 weeks, a minimum of 600 different alternative splicing events were found at each time point tested. They found that chronic arsenic exposure induced the canonical isoforms of the splice regulators DDX42, RMB25, and SRRM2 [235]. Alternating splicing might occur via DNA binding inhibition by alternative splicing modifiers such as CCCTC-binding factor (CTCF), TET1/2, and poly (ADP) ribose polymerase (PARP1) [64]. Several other studies also showed that PARP-1 inhibition occurred in arsenite-exposed cells [236,237], and PARP-1 was a direct molecular target of arsenite, which selectively interacted with zinc finger domains [236,237,238]. Notably, many splicing factors are regulated by PARylation [239,240,241], and inhibition of PARP1 binding to DNA upon arsenic exposure affects the structural properties of chromatin and the PARylation activities, which indirectly controls splicing decisions. iAs also inhibits the methylcytosine dioxygenases (TET1/2), the DNA-binding proteins with zinc finger motifs. TET1/2 are necessary to oxidize 5-methylcytosine to 5-hydroxymethylcytosine and 5-carboxylcytosine [242]. Deactivation of TET1/2 allows 5-methylcytosine to assemble at the CTCF target sites and stops CTCF from attaching to its target sites, consequently leading to exon exclusion [243]. Thus, iAs may participate in splicing decisions by blocking the binding of PARP1 or CTCF to DNA.
iAs also changes alternate splicing by upregulating p52 via non-canonical NF-kB pathway activation [244]. p52 modulates the splicing factor SRSF1 by co-localizing and interacting with it [245]. SRSF1 overexpression is induced by MYC [246], which is also dysregulated in iAs exposure [53,247]. MYC increases core pre-mRNA machinery in the process of carcinogenesis and maintains the suitable splicing of alternative exons [248]. More studies are needed to further elucidate how iAs dictates alternating splicing.
7. Conclusions and Future Direction
Arsenic alone is not efficient to cause point mutation or initiate and promote tumor development in animal models. However, growing evidence has shown that arsenic causes the dysregulation of epigenetic changes, including DNA methylation, histone modification, miRNAs and lncRNAs, RNA modification, and alternative splicing, which consequently changes the gene expressions followed by severe pathologies, including cancers. Our understanding of the underlying epigenetic mechanisms is still limited, especially for RNA methylation, lncRNAs, and alternative splicing. More studies are necessary to elucidate their roles and mechanisms in arsenic-induced carcinogenesis. In addition, most studies used different cell lines and animals to characterize epigenetic changes induced by arsenic exposure. More population studies using human cohorts exposed to varying arsenic levels are necessary to unveil how individual variability, genetic background, and other confounding variables such as diet, gender, and age may influence the epigenetic responses. Studies are also required to systematically investigate epigenetic profiles to identify and validate the markers of epigenetic changes in targeted disease-relevant tissues such as the skin, bladder, kidney, and lung.
In summary, a comprehensive epigenomic approach is necessary to understand the mechanisms of arsenic-induced carcinogenesis and angiogenesis. These mechanistic comprehensions of epigenetic changes can provide potential biomarkers of arsenic exposure and develop potential therapeutic targets for mitigating the global burden of arsenic-induced diseases, including cancers.
Author Contributions
Conceptualization, L.-Z.L. and R.I.; writing—original draft preparation, R.I. and L.Z.; writing—review and editing, R.I., Y.W., G.L.-Y. and L.-Z.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Institutes of Health grants (no. R01CA232587, R01ES033197, R01CA263506 and K02ES029119 to L.-Z.L.), American Cancer Society Research Scholar (no. NEC-129306 to L.-Z.L.), Commonwealth University Research Enhancement Program grant with the Pennsylvania Department of Health (SAP#4100088563 to L.-Z.L.), and the Cancer Center grant (NIH 5P30CA056036) at Thomas Jefferson University.
Institutional Review Board Statement
Ethical review and approval were waived for this study, due to the use of already available published data.
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study by the investigator of each published study included in the present review.
Data Availability Statement
The data presented in this study are openly available in Medline and Embase.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- IARC Working Group on the Evaluation of Carcinogenic Risks to Humans; World Health Organization; International Agency for Research on Cancer. Some Drinking-Water Disinfectants and Contaminants, Including Arsenic; IARC: Lyon, France, 2004; Volume 84, pp. 1–477.
- Bardach, A.E.; Ciapponi, A.; Soto, N.; Chaparro, M.R.; Calderon, M.; Briatore, A.; Cadoppi, N.; Tassara, R.; Litter, M.I. Epidemiology of chronic disease related to arsenic in Argentina: A systematic review. Sci. Total Environ. 2015, 538, 802–816. [Google Scholar] [CrossRef] [PubMed]
- Straif, K.; Benbrahim-Tallaa, L.; Baan, R.; Grosse, Y.; Secretan, B.; El Ghissassi, F.; Bouvard, V.; Guha, N.; Freeman, C.; Galichet, L.; et al. A review of human carcinogens--Part C: Metals, arsenic, dusts, and fibres. Lancet Oncol. 2009, 10, 453–454. [Google Scholar] [CrossRef]
- Chen, C.J.; Kuo, T.L.; Wu, M.M. Arsenic and cancers. Lancet 1988, 1, 414–415. [Google Scholar] [CrossRef]
- Marshall, G.; Ferreccio, C.; Yuan, Y.; Bates, M.N.; Steinmaus, C.; Selvin, S.; Liaw, J.; Smith, A.H. Fifty-year study of lung and bladder cancer mortality in Chile related to arsenic in drinking water. J. Natl. Cancer Inst. 2007, 99, 920–928. [Google Scholar] [CrossRef]
- Smith, A.H.; Hopenhayn-Rich, C.; Bates, M.N.; Goeden, H.M.; Hertz-Picciotto, I.; Duggan, H.M.; Wood, R.; Kosnett, M.J.; Smith, M.T. Cancer risks from arsenic in drinking water. Environ. Health Perspect 1992, 97, 259–267. [Google Scholar] [CrossRef]
- Hopenhayn-Rich, C.; Biggs, M.L.; Fuchs, A.; Bergoglio, R.; Tello, E.E.; Nicolli, H.; Smith, A.H. Bladder cancer mortality associated with arsenic in drinking water in Argentina. Epidemiology 1996, 7, 117–124. [Google Scholar] [CrossRef]
- Sohel, N.; Persson, L.A.; Rahman, M.; Streatfield, P.K.; Yunus, M.; Ekström, E.C.; Vahter, M. Arsenic in drinking water and adult mortality: A population-based cohort study in rural Bangladesh. Epidemiology 2009, 20, 824–830. [Google Scholar] [CrossRef]
- Sanyal, T.; Bhattacharjee, P.; Paul, S.; Bhattacharjee, P. Recent Advances in Arsenic Research: Significance of Differential Susceptibility and Sustainable Strategies for Mitigation. Front. Public Health 2020, 8, 464. [Google Scholar] [CrossRef]
- Ozturk, M.; Metin, M.; Altay, V.; Bhat, R.A.; Ejaz, M.; Gul, A.; Unal, B.T.; Hasanuzzaman, M.; Nibir, L.; Nahar, K.; et al. Arsenic and Human Health: Genotoxicity, Epigenomic Effects, and Cancer Signaling. Biol. Trace Elem. Res. 2022, 200, 988–1001. [Google Scholar] [CrossRef]
- Chakraborti, D.; Rahman, M.M.; Paul, K.; Chowdhury, U.K.; Sengupta, M.K.; Lodh, D.; Chanda, C.R.; Saha, K.C.; Mukherjee, S.C. Arsenic calamity in the Indian subcontinent What lessons have been learned? Talanta 2002, 58, 3–22. [Google Scholar] [CrossRef]
- Chung, J.Y.; Yu, S.D.; Hong, Y.S. Environmental source of arsenic exposure. J. Prev. Med. Public Health 2014, 47, 253–257. [Google Scholar] [CrossRef] [PubMed]
- Stöhrer, G. Arsenic: Opportunity for risk assessment. Arch. Toxicol 1991, 65, 525–531. [Google Scholar] [CrossRef]
- O’Day, P.A. Chemistry and Mineralogy of Arsenic. Elements 2006, 2, 77–83. [Google Scholar] [CrossRef]
- Zampella, G.; Neupane, K.P.; De Gioia, L.; Pecoraro, V.L. The importance of stereochemically active lone pairs for influencing Pb(II) and As(III) protein binding. Chemistry 2012, 18, 2040–2050. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fang, J.; Leonard, S.S.; Rao, K.M. Cadmium inhibits the electron transfer chain and induces reactive oxygen species. Free Radic. Biol. Med. 2004, 36, 1434–1443. [Google Scholar] [CrossRef]
- Hubaux, R.; Becker-Santos, D.D.; Enfield, K.S.; Rowbotham, D.; Lam, S.; Lam, W.L.; Martinez, V.D. Molecular features in arsenic-induced lung tumors. Mol. Cancer 2013, 12, 20. [Google Scholar] [CrossRef]
- Drobna, Z.; Styblo, M.; Thomas, D.J. An Overview of Arsenic Metabolism and Toxicity. Curr Protoc Toxicol 2009, 42, 4–31. [Google Scholar] [CrossRef]
- Davey, J.C.; Nomikos, A.P.; Wungjiranirun, M.; Sherman, J.R.; Ingram, L.; Batki, C.; Lariviere, J.P.; Hamilton, J.W. Arsenic as an endocrine disruptor: Arsenic disrupts retinoic acid receptor-and thyroid hormone receptor-mediated gene regulation and thyroid hormone-mediated amphibian tail metamorphosis. Environ. Health Perspect 2008, 116, 165–172. [Google Scholar] [CrossRef]
- Petrick, J.S.; Jagadish, B.; Mash, E.A.; Aposhian, H.V. Monomethylarsonous acid (MMA(III)) and arsenite: LD(50) in hamsters and in vitro inhibition of pyruvate dehydrogenase. Chem. Res. Toxicol. 2001, 14, 651–656. [Google Scholar] [CrossRef]
- Styblo, M.; Del Razo, L.M.; Vega, L.; Germolec, D.R.; LeCluyse, E.L.; Hamilton, G.A.; Reed, W.; Wang, C.; Cullen, W.R.; Thomas, D.J. Comparative toxicity of trivalent and pentavalent inorganic and methylated arsenicals in rat and human cells. Arch. Toxicol. 2000, 74, 289–299. [Google Scholar] [CrossRef]
- Chatterjee, A.; Chatterji, U. Arsenic abrogates the estrogen-signaling pathway in the rat uterus. Reprod. Biol. Endocrinol. 2010, 8, 80. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.M.; Beck, B.D.; Rhomberg, L.R. Historical perspective on the role of cell proliferation in carcinogenesis for DNA-reactive and non-DNA-reactive carcinogens: Arsenic as an example. Toxicology 2021, 456, 152783. [Google Scholar] [CrossRef] [PubMed]
- Hei, T.K.; Filipic, M. Role of oxidative damage in the genotoxicity of arsenic. Free Radic Biol. Med. 2004, 37, 574–581. [Google Scholar] [CrossRef] [PubMed]
- Rossman, T.G.; Uddin, A.N.; Burns, F.J. Evidence that arsenite acts as a cocarcinogen in skin cancer. Toxicol. Appl. Pharmacol. 2004, 198, 394–404. [Google Scholar] [CrossRef] [PubMed]
- Rossman, T.G. Mechanism of arsenic carcinogenesis: An integrated approach. Mutat. Res. 2003, 533, 37–65. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.B.; Leszczynska, J.; Hickey, C.; Rossman, T.G. Further evidence against a direct genotoxic mode of action for arsenic-induced cancer. Toxicol. Appl. Pharmacol. 2007, 222, 289–297. [Google Scholar] [CrossRef] [PubMed]
- Razin, A.; Riggs, A.D. DNA methylation and gene function. Science 1980, 210, 604–610. [Google Scholar] [CrossRef]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar] [CrossRef]
- Takai, D.; Jones, P.A. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc. Natl. Acad. Sci. USA 2002, 99, 3740–3745. [Google Scholar] [CrossRef]
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27. [Google Scholar] [CrossRef]
- Baylin, S.B.; Jones, P.A. A decade of exploring the cancer epigenome—Biological and translational implications. Nat. Rev. Cancer 2011, 11, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef] [PubMed]
- Moore, L.D.; Le, T.; Fan, G. DNA methylation and its basic function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed]
- Yong, W.S.; Hsu, F.M.; Chen, P.Y. Profiling genome-wide DNA methylation. Epigenet. Chromatin 2016, 9, 26. [Google Scholar] [CrossRef]
- Nava-Rivera, L.E.; Betancourt-Martínez, N.D.; Lozoya-Martínez, R.; Carranza-Rosales, P.; Guzmán-Delgado, N.E.; Carranza-Torres, I.E.; Delgado-Aguirre, H.; Zambrano-Ortíz, J.O.; Morán-Martínez, J. Transgenerational effects in DNA methylation, genotoxicity and reproductive phenotype by chronic arsenic exposure. Sci. Rep. 2021, 11, 8276. [Google Scholar] [CrossRef]
- Yoder, J.A.; Walsh, C.P.; Bestor, T.H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997, 13, 335–340. [Google Scholar] [CrossRef]
- Laurent, L.; Wong, E.; Li, G.; Huynh, T.; Tsirigos, A.; Ong, C.T.; Low, H.M.; Kin Sung, K.W.; Rigoutsos, I.; Loring, J.; et al. Dynamic changes in the human methylome during differentiation. Genome Res. 2010, 20, 320–331. [Google Scholar] [CrossRef]
- Reichard, J.F.; Schnekenburger, M.; Puga, A. Long term low-dose arsenic exposure induces loss of DNA methylation. Biochem. Biophys. Res. Commun. 2007, 352, 188–192. [Google Scholar] [CrossRef]
- Ren, X.; McHale, C.M.; Skibola, C.F.; Smith, A.H.; Smith, M.T.; Zhang, L. An emerging role for epigenetic dysregulation in arsenic toxicity and carcinogenesis. Environ. Health Perspect 2011, 119, 11–19. [Google Scholar] [CrossRef]
- Chakraborty, A.; Ghosh, S.; Biswas, B.; Pramanik, S.; Nriagu, J.; Bhowmick, S. Epigenetic modifications from arsenic exposure: A comprehensive review. Sci. Total Environ. 2022, 810, 151218. [Google Scholar] [CrossRef]
- Zhao, C.Q.; Young, M.R.; Diwan, B.A.; Coogan, T.P.; Waalkes, M.P. Association of arsenic-induced malignant transformation with DNA hypomethylation and aberrant gene expression. Proc. Natl. Acad. Sci. USA 1997, 94, 10907–10912. [Google Scholar] [CrossRef] [PubMed]
- Uthus, E.O.; Davis, C. Dietary arsenic affects dimethylhydrazine-induced aberrant crypt formation and hepatic global DNA methylation and DNA methyltransferase activity in rats. Biol. Trace Elem. Res. 2005, 103, 133–145. [Google Scholar] [CrossRef]
- Chen, H.; Li, S.; Liu, J.; Diwan, B.A.; Barrett, J.C.; Waalkes, M.P. Chronic inorganic arsenic exposure induces hepatic global and individual gene hypomethylation: Implications for arsenic hepatocarcinogenesis. Carcinogenesis 2004, 25, 1779–1786. [Google Scholar] [CrossRef] [PubMed]
- Demanelis, K.; Argos, M.; Tong, L.; Shinkle, J.; Sabarinathan, M.; Rakibuz-Zaman, M.; Sarwar, G.; Shahriar, H.; Islam, T.; Rahman, M.; et al. Association of Arsenic Exposure with Whole Blood DNA Methylation: An Epigenome-Wide Study of Bangladeshi Adults. Environ. Health Perspect 2019, 127, 57011. [Google Scholar] [CrossRef]
- Pilsner, J.R.; Liu, X.; Ahsan, H.; Ilievski, V.; Slavkovich, V.; Levy, D.; Factor-Litvak, P.; Graziano, J.H.; Gamble, M.V. Folate deficiency, hyperhomocysteinemia, low urinary creatinine, and hypomethylation of leukocyte DNA are risk factors for arsenic-induced skin lesions. Environ. Health Perspect 2009, 117, 254–260. [Google Scholar] [CrossRef]
- Seow, W.J.; Kile, M.L.; Baccarelli, A.A.; Pan, W.C.; Byun, H.M.; Mostofa, G.; Quamruzzaman, Q.; Rahman, M.; Lin, X.; Christiani, D.C. Epigenome-wide DNA methylation changes with development of arsenic-induced skin lesions in Bangladesh: A case-control follow-up study. Environ. Mol. Mutagen 2014, 55, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Bandyopadhyay, A.K.; Paul, S.; Adak, S.; Giri, A.K. Reduced LINE-1 methylation is associated with arsenic-induced genotoxic stress in children. Biometals 2016, 29, 731–741. [Google Scholar] [CrossRef]
- Jones, P.A.; Baylin, S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef]
- Chanda, S.; Dasgupta, U.B.; Guhamazumder, D.; Gupta, M.; Chaudhuri, U.; Lahiri, S.; Das, S.; Ghosh, N.; Chatterjee, D. DNA hypermethylation of promoter of gene p53 and p16 in arsenic-exposed people with and without malignancy. Toxicol. Sci. 2006, 89, 431–437. [Google Scholar] [CrossRef]
- Chen, W.T.; Hung, W.C.; Kang, W.Y.; Huang, Y.C.; Chai, C.Y. Urothelial carcinomas arising in arsenic-contaminated areas are associated with hypermethylation of the gene promoter of the death-associated protein kinase. Histopathology 2007, 51, 785–792. [Google Scholar] [CrossRef]
- Marsit, C.J.; Karagas, M.R.; Danaee, H.; Liu, M.; Andrew, A.; Schned, A.; Nelson, H.H.; Kelsey, K.T. Carcinogen exposure and gene promoter hypermethylation in bladder cancer. Carcinogenesis 2006, 27, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Wu, L.; Lu, M.; Meng, X.; Gao, B.; Qiao, X.; Zhang, W.; Xue, D. Analysis of the transcriptional regulation of cancer-related genes by aberrant DNA methylation of the cis-regulation sites in the promoter region during hepatocyte carcinogenesis caused by arsenic. Oncotarget 2015, 6, 21493–21506. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.B.; Vahter, M.; Concha, G.; Broberg, K. Environmental arsenic exposure and DNA methylation of the tumor suppressor gene p16 and the DNA repair gene MLH1: Effect of arsenic metabolism and genotype. Metallomics 2012, 4, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Banerjee, N.; Chatterjee, A.; Sau, T.J.; Das, J.K.; Mishra, P.K.; Chakrabarti, P.; Bandyopadhyay, A.; Giri, A.K. Arsenic-induced promoter hypomethylation and over-expression of ERCC2 reduces DNA repair capacity in humans by non-disjunction of the ERCC2-Cdk7 complex. Metallomics 2014, 6, 864–873. [Google Scholar] [CrossRef] [PubMed]
- Smeester, L.; Rager, J.E.; Bailey, K.A.; Guan, X.; Smith, N.; García-Vargas, G.; Del Razo, L.M.; Drobná, Z.; Kelkar, H.; Stýblo, M.; et al. Epigenetic changes in individuals with arsenicosis. Chem. Res. Toxicol. 2011, 24, 165–167. [Google Scholar] [CrossRef] [PubMed]
- Chanda, S.; Dasgupta, U.B.; Mazumder, D.G.; Saha, J.; Gupta, B. Human GMDS gene fragment hypermethylation in chronic high level of arsenic exposure with and without arsenic induced cancer. Springerplus 2013, 2, 557. [Google Scholar] [CrossRef]
- Gribble, M.O.; Tang, W.Y.; Shang, Y.; Pollak, J.; Umans, J.G.; Francesconi, K.A.; Goessler, W.; Silbergeld, E.K.; Guallar, E.; Cole, S.A.; et al. Differential methylation of the arsenic (III) methyltransferase promoter according to arsenic exposure. Arch. Toxicol. 2014, 88, 275–282. [Google Scholar] [CrossRef][Green Version]
- Okoji, R.S.; Yu, R.C.; Maronpot, R.R.; Froines, J.R. Sodium arsenite administration via drinking water increases genome-wide and Ha-ras DNA hypomethylation in methyl-deficient C57BL/6J mice. Carcinogenesis 2002, 23, 777–785. [Google Scholar] [CrossRef]
- Chen, H.; Liu, J.; Zhao, C.Q.; Diwan, B.A.; Merrick, B.A.; Waalkes, M.P. Association of c-myc overexpression and hyperproliferation with arsenite-induced malignant transformation. Toxicol. Appl. Pharmacol. 2001, 175, 260–268. [Google Scholar] [CrossRef]
- Takahashi, M.; Barrett, J.C.; Tsutsui, T. Transformation by inorganic arsenic compounds of normal Syrian hamster embryo cells into a neoplastic state in which they become anchorage-independent and cause tumors in newborn hamsters. Int. J. Cancer 2002, 99, 629–634. [Google Scholar] [CrossRef]
- Janasik, B.; Reszka, E.; Stanislawska, M.; Jablonska, E.; Kuras, R.; Wieczorek, E.; Malachowska, B.; Fendler, W.; Wasowicz, W. Effect of Arsenic Exposure on NRF2-KEAP1 Pathway and Epigenetic Modification. Biol. Trace Elem. Res. 2018, 185, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, K.D.; Helin, K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Saintilnord, W.N.; Fondufe-Mittendorf, Y. Arsenic-induced epigenetic changes in cancer development. Semin. Cancer Biol. 2021, 76, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, W.; Zhang, A. TET-mediated DNA demethylation plays an important role in arsenic-induced HBE cells oxidative stress via regulating promoter methylation of OGG1 and GSTP1. Toxicol. In Vitro 2021, 72, 105075. [Google Scholar] [CrossRef] [PubMed]
- Domingo-Relloso, A.; Bozack, A.; Kiihl, S.; Rodriguez-Hernandez, Z.; Rentero-Garrido, P.; Casasnovas, J.A.; Leon-Latre, M.; Garcia-Barrera, T.; Gomez-Ariza, J.L.; Moreno, B.; et al. Arsenic exposure and human blood DNA methylation and hydroxymethylation profiles in two diverse populations from Bangladesh and Spain. Environ. Res. 2022, 204, 112021. [Google Scholar] [CrossRef]
- Prasad, P.; Sinha, D. Low-level arsenic causes chronic inflammation and suppresses expression of phagocytic receptors. Environ. Sci. Pollut Res. Int. 2017, 24, 11708–11721. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Yang, S.M.; Zhang, H.P.; Yang, Y.; Sun, S.B.; Chang, J.P.; Tao, X.C.; Yang, T.Y.; Liu, C.; Yang, Y.M. Endoplasmic reticulum stress mediates the arsenic trioxide-induced apoptosis in human hepatocellular carcinoma cells. Int. J. Biochem. Cell Biol 2015, 68, 158–165. [Google Scholar] [CrossRef]
- Kile, M.L.; Houseman, E.A.; Baccarelli, A.A.; Quamruzzaman, Q.; Rahman, M.; Mostofa, G.; Cardenas, A.; Wright, R.O.; Christiani, D.C. Effect of prenatal arsenic exposure on DNA methylation and leukocyte subpopulations in cord blood. Epigenetics 2014, 9, 774–782. [Google Scholar] [CrossRef]
- Pilsner, J.R.; Hall, M.N.; Liu, X.; Ilievski, V.; Slavkovich, V.; Levy, D.; Factor-Litvak, P.; Yunus, M.; Rahman, M.; Graziano, J.H.; et al. Influence of prenatal arsenic exposure and newborn sex on global methylation of cord blood DNA. PLoS ONE 2012, 7, e37147. [Google Scholar] [CrossRef]
- Andrew, A.S.; Jewell, D.A.; Mason, R.A.; Whitfield, M.L.; Moore, J.H.; Karagas, M.R. Drinking-water arsenic exposure modulates gene expression in human lymphocytes from a U.S. population. Environ. Health Perspect 2008, 116, 524–531. [Google Scholar] [CrossRef]
- Xie, Y.; Liu, J.; Benbrahim-Tallaa, L.; Ward, J.M.; Logsdon, D.; Diwan, B.A.; Waalkes, M.P. Aberrant DNA methylation and gene expression in livers of newborn mice transplacentally exposed to a hepatocarcinogenic dose of inorganic arsenic. Toxicology 2007, 236, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Coppin, J.F.; Qu, W.; Waalkes, M.P. Interplay between cellular methyl metabolism and adaptive efflux during oncogenic transformation from chronic arsenic exposure in human cells. J. Biol. Chem. 2008, 283, 19342–19350. [Google Scholar] [CrossRef]
- Benbrahim-Tallaa, L.; Waterland, R.A.; Styblo, M.; Achanzar, W.E.; Webber, M.M.; Waalkes, M.P. Molecular events associated with arsenic-induced malignant transformation of human prostatic epithelial cells: Aberrant genomic DNA methylation and K-ras oncogene activation. Toxicol. Appl. Pharmacol 2005, 206, 288–298. [Google Scholar] [CrossRef] [PubMed]
- Bagnyukova, T.V.; Luzhna, L.I.; Pogribny, I.P.; Lushchak, V.I. Oxidative stress and antioxidant defenses in goldfish liver in response to short-term exposure to arsenite. Environ. Mol. Mutagen 2007, 48, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Pilsner, J.R.; Liu, X.; Ahsan, H.; Ilievski, V.; Slavkovich, V.; Levy, D.; Factor-Litvak, P.; Graziano, J.H.; Gamble, M.V. Genomic methylation of peripheral blood leukocyte DNA: Influences of arsenic and folate in Bangladeshi adults. Am. J. Clin. Nutr. 2007, 86, 1179–1186. [Google Scholar] [CrossRef]
- Chai, C.Y.; Huang, Y.C.; Hung, W.C.; Kang, W.Y.; Chen, W.T. Arsenic salts induced autophagic cell death and hypermethylation of DAPK promoter in SV-40 immortalized human uroepithelial cells. Toxicol Lett. 2007, 173, 48–56. [Google Scholar] [CrossRef]
- Cui, X.; Wakai, T.; Shirai, Y.; Hatakeyama, K.; Hirano, S. Chronic oral exposure to inorganic arsenate interferes with methylation status of p16INK4a and RASSF1A and induces lung cancer in A/J mice. Toxicol Sci. 2006, 91, 372–381. [Google Scholar] [CrossRef]
- Waalkes, M.P.; Liu, J.; Chen, H.; Xie, Y.; Achanzar, W.E.; Zhou, Y.S.; Cheng, M.L.; Diwan, B.A. Estrogen signaling in livers of male mice with hepatocellular carcinoma induced by exposure to arsenic in utero. J. Natl. Cancer Inst. 2004, 96, 466–474. [Google Scholar] [CrossRef]
- Banerjee, N.; Paul, S.; Sau, T.J.; Das, J.K.; Bandyopadhyay, A.; Banerjee, S.; Giri, A.K. Epigenetic modifications of DAPK and p16 genes contribute to arsenic-induced skin lesions and nondermatological health effects. Toxicol. Sci. 2013, 135, 300–308. [Google Scholar] [CrossRef]
- Bhattacharjee, P.; Sanyal, T.; Bhattacharjee, S.; Bhattacharjee, P. Epigenetic alteration of mismatch repair genes in the population chronically exposed to arsenic in West Bengal, India. Environ. Res. 2018, 163, 289–296. [Google Scholar] [CrossRef]
- Intarasunanont, P.; Navasumrit, P.; Waraprasit, S.; Chaisatra, K.; Suk, W.A.; Mahidol, C.; Ruchirawat, M. Effects of arsenic exposure on DNA methylation in cord blood samples from newborn babies and in a human lymphoblast cell line. Environ. Health 2012, 11, 31. [Google Scholar] [CrossRef] [PubMed]
- Sanyal, T.; Paul, M.; Bhattacharjee, S.; Bhattacharjee, P. Epigenetic alteration of mitochondrial biogenesis regulatory genes in arsenic exposed individuals (with and without skin lesions) and in skin cancer tissues: A case control study. Chemosphere 2020, 258, 127305. [Google Scholar] [CrossRef] [PubMed]
- Brookes, E.; Shi, Y. Diverse epigenetic mechanisms of human disease. Annu. Rev. Genet. 2014, 48, 237–268. [Google Scholar] [CrossRef] [PubMed]
- Luger, K.; Mäder, A.W.; Richmond, R.K.; Sargent, D.F.; Richmond, T.J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 1997, 389, 251–260. [Google Scholar] [CrossRef]
- DeRouchey, J.; Hoover, B.; Rau, D.C. A comparison of DNA compaction by arginine and lysine peptides: A physical basis for arginine rich protamines. Biochemistry 2013, 52, 3000–3009. [Google Scholar] [CrossRef]
- Bhattacharjee, P.; Paul, S.; Bhattacharjee, P. Understanding the mechanistic insight of arsenic exposure and decoding the histone cipher. Toxicology 2020, 430, 152340. [Google Scholar] [CrossRef]
- Peterson, C.L.; Laniel, M.A. Histones and histone modifications. Curr. Biol. 2004, 14, R546–R551. [Google Scholar] [CrossRef]
- Glozak, M.A.; Seto, E. Histone deacetylases and cancer. Oncogene 2007, 26, 5420–5432. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in the Regulation of Rna Synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef]
- Kuo, M.H.; Allis, C.D. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays 1998, 20, 615–626. [Google Scholar] [CrossRef]
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100. [Google Scholar] [CrossRef] [PubMed]
- Gräff, J.; Tsai, L.H. Histone acetylation: Molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 2013, 14, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Arrigo, A.P. Acetylation and methylation patterns of core histones are modified after heat or arsenite treatment of Drosophila tissue culture cells. Nucleic Acids Res. 1983, 11, 1389–1404. [Google Scholar] [CrossRef]
- Cantone, L.; Nordio, F.; Hou, L.; Apostoli, P.; Bonzini, M.; Tarantini, L.; Angelici, L.; Bollati, V.; Zanobetti, A.; Schwartz, J.; et al. Inhalable metal-rich air particles and histone H3K4 dimethylation and H3K9 acetylation in a cross-sectional study of steel workers. Environ. Health Perspect 2011, 119, 964–969. [Google Scholar] [CrossRef]
- Cronican, A.A.; Fitz, N.F.; Carter, A.; Saleem, M.; Shiva, S.; Barchowsky, A.; Koldamova, R.; Schug, J.; Lefterov, I. Genome-wide alteration of histone H3K9 acetylation pattern in mouse offspring prenatally exposed to arsenic. PLoS ONE 2013, 8, e53478. [Google Scholar] [CrossRef]
- Ge, Y.; Zhu, J.; Wang, X.; Zheng, N.; Tu, C.; Qu, J.; Ren, X. Mapping dynamic histone modification patterns during arsenic-induced malignant transformation of human bladder cells. Toxicol. Appl. Pharmacol. 2018, 355, 164–173. [Google Scholar] [CrossRef]
- Jensen, T.J.; Novak, P.; Eblin, K.E.; Gandolfi, A.J.; Futscher, B.W. Epigenetic remodeling during arsenical-induced malignant transformation. Carcinogenesis 2008, 29, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Wu, D.; Zhao, L.; Yang, Y.; Ding, J.; Dong, L.; Hu, L.; Wang, F.; Zhao, X.; Cai, Y.; et al. Arsenic Trioxide Reduces Global Histone H4 Acetylation at Lysine 16 through Direct Binding to Histone Acetyltransferase hMOF in Human Cells. PLoS ONE 2015, 10, e0141014. [Google Scholar] [CrossRef]
- Jo, W.J.; Ren, X.; Chu, F.; Aleshin, M.; Wintz, H.; Burlingame, A.; Smith, M.T.; Vulpe, C.D.; Zhang, L. Acetylated H4K16 by MYST1 protects UROtsa cells from arsenic toxicity and is decreased following chronic arsenic exposure. Toxicol. Appl. Pharmacol. 2009, 241, 294–302. [Google Scholar] [CrossRef]
- Ge, Y.; Gong, Z.; Olson, J.R.; Xu, P.; Buck, M.J.; Ren, X. Inhibition of monomethylarsonous acid (MMA(III))-induced cell malignant transformation through restoring dysregulated histone acetylation. Toxicology 2013, 312, 30–35. [Google Scholar] [CrossRef]
- Li, J.; Chen, P.; Sinogeeva, N.; Gorospe, M.; Wersto, R.P.; Chrest, F.J.; Barnes, J.; Liu, Y. Arsenic trioxide promotes histone H3 phosphoacetylation at the chromatin of CASPASE-10 in acute promyelocytic leukemia cells. J. Biol. Chem. 2002, 277, 49504–49510. [Google Scholar] [CrossRef] [PubMed]
- Ma, L.; Li, J.; Zhan, Z.; Chen, L.; Li, D.; Bai, Q.; Gao, C.; Li, J.; Zeng, X.; He, Z.; et al. Specific histone modification responds to arsenic-induced oxidative stress. Toxicol. Appl. Pharmacol. 2016, 302, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Gorospe, M.; Barnes, J.; Liu, Y. Tumor promoter arsenite stimulates histone H3 phosphoacetylation of proto-oncogenes c-fos and c-jun chromatin in human diploid fibroblasts. J. Biol. Chem. 2003, 278, 13183–13191. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, T.; Brocher, J.; Stopper, H.; Hock, R. Sodium arsenite modulates histone acetylation, histone deacetylase activity and HMGN protein dynamics in human cells. Chromosoma 2008, 117, 147–157. [Google Scholar] [CrossRef]
- Sharma, B.; Sharma, P.M. Arsenic toxicity induced endothelial dysfunction and dementia: Pharmacological interdiction by histone deacetylase and inducible nitric oxide synthase inhibitors. Toxicol. Appl. Pharmacol. 2013, 273, 180–188. [Google Scholar] [CrossRef]
- Ng, S.S.; Yue, W.W.; Oppermann, U.; Klose, R.J. Dynamic protein methylation in chromatin biology. Cell Mol. Life Sci. 2009, 66, 407–422. [Google Scholar] [CrossRef]
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13. [Google Scholar] [CrossRef]
- Lan, F.; Shi, Y. Epigenetic regulation: Methylation of histone and non-histone proteins. Sci. China C Life Sci. 2009, 52, 311–322. [Google Scholar] [CrossRef]
- Arita, A.; Costa, M. Epigenetics in metal carcinogenesis: Nickel, arsenic, chromium and cadmium. Metallomics 2009, 1, 222–228. [Google Scholar] [CrossRef]
- Vakoc, C.R.; Sachdeva, M.M.; Wang, H.; Blobel, G.A. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell Biol. 2006, 26, 9185–9195. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Reversing histone methylation. Nature 2005, 436, 1103–1106. [Google Scholar] [CrossRef] [PubMed]
- Schneider, R.; Bannister, A.J.; Kouzarides, T. Unsafe SETs: Histone lysine methyltransferases and cancer. Trends Biochem. Sci. 2002, 27, 396–402. [Google Scholar] [CrossRef]
- Desrosiers, R.; Tanguay, R.M. Further characterization of the posttranslational modifications of core histones in response to heat and arsenite stress in Drosophila. Biochem. Cell Biol. 1986, 64, 750–757. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sun, H.; Ellen, T.P.; Chen, H.; Costa, M. Arsenite alters global histone H3 methylation. Carcinogenesis 2008, 29, 1831–1836. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, Q.; Arita, A.; Sun, H.; Costa, M. Effects of nickel, chromate, and arsenite on histone 3 lysine methylation. Toxicol. Appl. Pharmacol. 2009, 236, 78–84. [Google Scholar] [CrossRef]
- Treas, J.N.; Tyagi, T.; Singh, K.P. Effects of chronic exposure to arsenic and estrogen on epigenetic regulatory genes expression and epigenetic code in human prostate epithelial cells. PLoS ONE 2012, 7, e43880. [Google Scholar] [CrossRef]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Estève, P.O.; Chin, H.G.; Pradhan, S. Molecular mechanisms of transactivation and doxorubicin-mediated repression of survivin gene in cancer cells. J. Biol. Chem. 2007, 282, 2615–2625. [Google Scholar] [CrossRef]
- McGarvey, K.M.; Fahrner, J.A.; Greene, E.; Martens, J.; Jenuwein, T.; Baylin, S.B. Silenced tumor suppressor genes reactivated by DNA demethylation do not return to a fully euchromatic chromatin state. Cancer Res. 2006, 66, 3541–3549. [Google Scholar] [CrossRef]
- Yuan, W.; Xu, M.; Huang, C.; Liu, N.; Chen, S.; Zhu, B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011, 286, 7983–7989. [Google Scholar] [CrossRef]
- Chervona, Y.; Hall, M.N.; Arita, A.; Wu, F.; Sun, H.; Tseng, H.C.; Ali, E.; Uddin, M.N.; Liu, X.; Zoroddu, M.A.; et al. Associations between arsenic exposure and global posttranslational histone modifications among adults in Bangladesh. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 2252–2260. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.G.; Kim, D.J.; Li, S.; Lee, K.Y.; Li, X.; Bode, A.M.; Dong, Z. Polycomb (PcG) proteins, BMI1 and SUZ12, regulate arsenic-induced cell transformation. J. Biol. Chem. 2012, 287, 31920–31928. [Google Scholar] [CrossRef] [PubMed]
- Chervona, Y.; Arita, A.; Costa, M. Carcinogenic metals and the epigenome: Understanding the effect of nickel, arsenic, and chromium. Metallomics 2012, 4, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Fraga, M.F.; Ballestar, E.; Villar-Garea, A.; Boix-Chornet, M.; Espada, J.; Schotta, G.; Bonaldi, T.; Haydon, C.; Ropero, S.; Petrie, K.; et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat. Genet 2005, 37, 391–400. [Google Scholar] [CrossRef]
- Swank, R.A.; Th’ng, J.P.; Guo, X.W.; Valdez, J.; Bradbury, E.M.; Gurley, L.R. Four distinct cyclin-dependent kinases phosphorylate histone H1 at all of its growth-related phosphorylation sites. Biochemistry 1997, 36, 13761–13768. [Google Scholar] [CrossRef]
- Burma, S.; Chen, B.P.; Murphy, M.; Kurimasa, A.; Chen, D.J. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J. Biol. Chem. 2001, 276, 42462–42467. [Google Scholar] [CrossRef] [PubMed]
- Houben, A.; Demidov, D.; Caperta, A.D.; Karimi, R.; Agueci, F.; Vlasenko, L. Phosphorylation of histone H3 in plants--a dynamic affair. Biochim. Biophys. Acta 2007, 1769, 308–315. [Google Scholar] [CrossRef]
- Barber, C.M.; Turner, F.B.; Wang, Y.; Hagstrom, K.; Taverna, S.D.; Mollah, S.; Ueberheide, B.; Meyer, B.J.; Hunt, D.F.; Cheung, P.; et al. The enhancement of histone H4 and H2A serine 1 phosphorylation during mitosis and S-phase is evolutionarily conserved. Chromosoma 2004, 112, 360–371. [Google Scholar] [CrossRef]
- Rossetto, D.; Avvakumov, N.; Côté, J. Histone phosphorylation: A chromatin modification involved in diverse nuclear events. Epigenetics 2012, 7, 1098–1108. [Google Scholar] [CrossRef]
- Cobo, J.M.; Valdez, J.G.; Gurley, L.R. Inhibition of mitotic-specific histone phosphorylation by sodium arsenite. Toxicol. In Vitro 1995, 9, 459–465. [Google Scholar] [CrossRef]
- Ray, P.D.; Huang, B.W.; Tsuji, Y. Coordinated regulation of Nrf2 and histone H3 serine 10 phosphorylation in arsenite-activated transcription of the human heme oxygenase-1 gene. Biochim. Biophys. Acta 2015, 1849, 1277–1288. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Miyazaki, K.; Kita, K.; Ochi, T. Trivalent dimethylarsenic compound induces histone H3 phosphorylation and abnormal localization of Aurora B kinase in HepG2 cells. Toxicol. Appl. Pharmacol. 2009, 241, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Kannan-Thulasiraman, P.; Katsoulidis, E.; Tallman, M.S.; Arthur, J.S.; Platanias, L.C. Activation of the mitogen- and stress-activated kinase 1 by arsenic trioxide. J. Biol. Chem. 2006, 281, 22446–22452. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Kita, K.; Ochi, T. Phosphorylation of histone H3 at serine 10 has an essential role in arsenite-induced expression of FOS, EGR1 and IL8 mRNA in cultured human cell lines. J. Appl. Toxicol. 2013, 33, 746–755. [Google Scholar] [CrossRef]
- Prigent, C.; Dimitrov, S. Phosphorylation of serine 10 in histone H3, what for? J. Cell Sci. 2003, 116, 3677–3685. [Google Scholar] [CrossRef]
- Ke, Q.; Li, Q.; Ellen, T.P.; Sun, H.; Costa, M. Nickel compounds induce phosphorylation of histone H3 at serine 10 by activating JNK-MAPK pathway. Carcinogenesis 2008, 29, 1276–1281. [Google Scholar] [CrossRef]
- Cavigelli, M.; Li, W.W.; Lin, A.; Su, B.; Yoshioka, K.; Karin, M. The tumor promoter arsenite stimulates AP-1 activity by inhibiting a JNK phosphatase. EMBO J. 1996, 15, 6269–6279. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Lagos-Quintana, M.; Rauhut, R.; Lendeckel, W.; Tuschl, T. Identification of novel genes coding for small expressed RNAs. Science 2001, 294, 853–858. [Google Scholar] [CrossRef]
- Lau, N.C.; Lim, L.P.; Weinstein, E.G.; Bartel, D.P. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 2001, 294, 858–862. [Google Scholar] [CrossRef]
- Lee, R.C.; Ambros, V. An extensive class of small RNAs in Caenorhabditis elegans. Science 2001, 294, 862–864. [Google Scholar] [CrossRef] [PubMed]
- Hata, A.; Lieberman, J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal 2015, 8, re3. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Li, M.; Marin-Muller, C.; Bharadwaj, U.; Chow, K.H.; Yao, Q.; Chen, C. MicroRNAs: Control and loss of control in human physiology and disease. World J. Surg. 2009, 33, 667–684. [Google Scholar] [CrossRef]
- Calin, G.A.; Sevignani, C.; Dumitru, C.D.; Hyslop, T.; Noch, E.; Yendamuri, S.; Shimizu, M.; Rattan, S.; Bullrich, F.; Negrini, M.; et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. USA 2004, 101, 2999–3004. [Google Scholar] [CrossRef]
- He, L.; He, X.; Lowe, S.W.; Hannon, G.J. microRNAs join the p53 network--another piece in the tumour-suppression puzzle. Nat. Rev. Cancer 2007, 7, 819–822. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, Y.; Smith, E.; Goodall, G.J.; Drew, P.A.; Brabletz, T.; Yang, C. Reversal and prevention of arsenic-induced human bronchial epithelial cell malignant transformation by microRNA-200b. Toxicol. Sci. 2011, 121, 110–122. [Google Scholar] [CrossRef]
- Zhong, M.; Huang, Z.; Wang, L.; Lin, Z.; Cao, Z.; Li, X.; Zhang, F.; Wang, H.; Li, Y.; Ma, X. Malignant Transformation of Human Bronchial Epithelial Cells Induced by Arsenic through STAT3/miR-301a/SMAD4 Loop. Sci. Rep. 2018, 8, 13291. [Google Scholar] [CrossRef]
- Al-Eryani, L.; Jenkins, S.F.; States, V.A.; Pan, J.; Malone, J.C.; Rai, S.N.; Galandiuk, S.; Giri, A.K.; States, J.C. miRNA expression profiles of premalignant and malignant arsenic-induced skin lesions. PLoS ONE 2018, 13, e0202579. [Google Scholar] [CrossRef]
- He, J.; Wang, M.; Jiang, Y.; Chen, Q.; Xu, S.; Xu, Q.; Jiang, B.H.; Liu, L.Z. Chronic arsenic exposure and angiogenesis in human bronchial epithelial cells via the ROS/miR-199a-5p/HIF-1α/COX-2 pathway. Environ. Health Perspect 2014, 122, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, J.; Xu, W.; Li, C.; Wu, K.; Chen, G.; Cui, J. The mechanism underlying arsenic-induced PD-L1 upregulation in transformed BEAS-2B cells. Toxicol. Appl. Pharmacol. 2022, 435, 115845. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Q.; Zou, Z.; Wang, Q.; Sun, B.; Liu, Y.; Liang, B.; Liu, Q.; Zhang, A. Association and risk of five miRNAs with arsenic-induced multiorgan damage. Sci. Total Environ. 2019, 680, 1–9. [Google Scholar] [CrossRef]
- Banerjee, N.; Bandyopadhyay, A.K.; Dutta, S.; Das, J.K.; Roy Chowdhury, T.; Bandyopadhyay, A.; Giri, A.K. Increased microRNA 21 expression contributes to arsenic induced skin lesions, skin cancers and respiratory distress in chronically exposed individuals. Toxicology 2017, 378, 10–16. [Google Scholar] [CrossRef]
- Sun, B.; Xue, J.; Li, J.; Luo, F.; Chen, X.; Liu, Y.; Wang, Q.; Qi, C.; Zou, Z.; Zhang, A.; et al. Circulating miRNAs and their target genes associated with arsenism caused by coal-burning. Toxicol. Res. (Camb) 2017, 6, 162–172. [Google Scholar] [CrossRef]
- Chen, C.; Jiang, X.; Gu, S.; Zhang, Z. MicroRNA-155 regulates arsenite-induced malignant transformation by targeting Nrf2-mediated oxidative damage in human bronchial epithelial cells. Toxicol. Lett. 2017, 278, 38–47. [Google Scholar] [CrossRef]
- Beezhold, K.; Liu, J.; Kan, H.; Meighan, T.; Castranova, V.; Shi, X.; Chen, F. miR-190-mediated downregulation of PHLPP contributes to arsenic-induced Akt activation and carcinogenesis. Toxicol. Sci. 2011, 123, 411–420. [Google Scholar] [CrossRef]
- Xu, W.; Luo, F.; Sun, B.; Ye, H.; Li, J.; Shi, L.; Liu, Y.; Lu, X.; Wang, B.; Wang, Q.; et al. HIF-2α, acting via miR-191, is involved in angiogenesis and metastasis of arsenite-transformed HBE cells. Toxicol. Res. (Camb) 2016, 5, 66–78. [Google Scholar] [CrossRef]
- Chun-Zhi, Z.; Lei, H.; An-Ling, Z.; Yan-Chao, F.; Xiao, Y.; Guang-Xiu, W.; Zhi-Fan, J.; Pei-Yu, P.; Qing-Yu, Z.; Chun-Sheng, K. MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 2010, 10, 367. [Google Scholar] [CrossRef]
- Yang, Y.F.; Wang, F.; Xiao, J.J.; Song, Y.; Zhao, Y.Y.; Cao, Y.; Bei, Y.H.; Yang, C.Q. MiR-222 overexpression promotes proliferation of human hepatocellular carcinoma HepG2 cells by downregulating p27. Int. J. Clin. Exp. Med. 2014, 7, 893–902. [Google Scholar] [PubMed]
- Lu, Y.; Roy, S.; Nuovo, G.; Ramaswamy, B.; Miller, T.; Shapiro, C.; Jacob, S.T.; Majumder, S. Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein and modulating mitogenic signal. J. Biol. Chem. 2011, 286, 42292–42302. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Ge, X.; Zheng, J.; Li, D.; Liu, X.; Wang, L.; Jiang, C.; Shi, Z.; Qin, L.; Liu, J.; et al. Role and mechanism of miR-222 in arsenic-transformed cells for inducing tumor growth. Oncotarget 2016, 7, 17805–17814. [Google Scholar] [CrossRef]
- Banerjee, N.; Das, S.; Tripathy, S.; Bandyopadhyay, A.K.; Sarma, N.; Bandyopadhyay, A.; Giri, A.K. MicroRNAs play an important role in contributing to arsenic susceptibility in the chronically exposed individuals of West Bengal, India. Environ. Sci. Pollut Res. Int. 2019, 26, 28052–28061. [Google Scholar] [CrossRef]
- Cui, Y.; Han, Z.; Hu, Y.; Song, G.; Hao, C.; Xia, H.; Ma, X. MicroRNA-181b and microRNA-9 mediate arsenic-induced angiogenesis via NRP1. J. Cell Physiol. 2012, 227, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Bielenberg, D.R.; Klagsbrun, M. Targeting endothelial and tumor cells with semaphorins. Cancer Metastasis Rev. 2007, 26, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Sun, R.; Hu, Y.; Wang, H.; Guo, Y.; Yang, B.; Pi, J.; Xu, Y. miRNA-182-5p, via HIF2α, contributes to arsenic carcinogenesis: Evidence from human renal epithelial cells. Metallomics 2018, 10, 1607–1617. [Google Scholar] [CrossRef]
- Chen, Q.Y.; Li, J.; Sun, H.; Wu, F.; Zhu, Y.; Kluz, T.; Jordan, A.; DesMarais, T.; Zhang, X.; Murphy, A.; et al. Role of miR-31 and SATB2 in arsenic-induced malignant BEAS-2B cell transformation. Mol. Carcinog. 2018, 57, 968–977. [Google Scholar] [CrossRef]
- Kong, A.P.; Xiao, K.; Choi, K.C.; Wang, G.; Chan, M.H.; Ho, C.S.; Chan, I.; Wong, C.K.; Chan, J.C.; Szeto, C.C. Associations between microRNA (miR-21, 126, 155 and 221), albuminuria and heavy metals in Hong Kong Chinese adolescents. Clin. Chim. Acta 2012, 413, 1053–1057. [Google Scholar] [CrossRef]
- Bollati, V.; Marinelli, B.; Apostoli, P.; Bonzini, M.; Nordio, F.; Hoxha, M.; Pegoraro, V.; Motta, V.; Tarantini, L.; Cantone, L.; et al. Exposure to metal-rich particulate matter modifies the expression of candidate microRNAs in peripheral blood leukocytes. Environ. Health Perspect 2010, 118, 763–768. [Google Scholar] [CrossRef]
- Gonzalez, H.; Lema, C.; Kirken, R.A.; Maldonado, R.A.; Varela-Ramirez, A.; Aguilera, R.J. Arsenic-exposed Keratinocytes Exhibit Differential microRNAs Expression Profile; Potential Implication of miR-21, miR-200a and miR-141 in Melanoma Pathway. Clin. Cancer Drugs 2015, 2, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Gaile, D.P.; Gong, Z.; Qiu, W.; Ge, Y.; Zhang, C.; Huang, C.; Yan, H.; Olson, J.R.; Kavanagh, T.J.; et al. Arsenic responsive microRNAs in vivo and their potential involvement in arsenic-induced oxidative stress. Toxicol. Appl. Pharmacol. 2015, 283, 198–209. [Google Scholar] [CrossRef] [PubMed]
- Ling, M.; Li, Y.; Xu, Y.; Pang, Y.; Shen, L.; Jiang, R.; Zhao, Y.; Yang, X.; Zhang, J.; Zhou, J.; et al. Regulation of miRNA-21 by reactive oxygen species-activated ERK/NF-κB in arsenite-induced cell transformation. Free Radic. Biol. Med. 2012, 52, 1508–1518. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Xu, Y.; Ling, M.; Zhao, Y.; Xu, W.; Liang, X.; Jiang, R.; Wang, B.; Bian, Q.; Liu, Q. Arsenite evokes IL-6 secretion, autocrine regulation of STAT3 signaling, and miR-21 expression, processes involved in the EMT and malignant transformation of human bronchial epithelial cells. Toxicol. Appl. Pharmacol. 2013, 273, 27–34. [Google Scholar] [CrossRef]
- Liu, X.; Luo, F.; Ling, M.; Lu, L.; Shi, L.; Lu, X.; Xu, H.; Chen, C.; Yang, Q.; Xue, J.; et al. MicroRNA-21 activation of ERK signaling via PTEN is involved in arsenite-induced autophagy in human hepatic L-02 cells. Toxicol. Lett. 2016, 252, 1–10. [Google Scholar] [CrossRef]
- Quinn, J.J.; Chang, H.Y. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol 2017, 1008, 1–46. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Tsagakis, I.; Douka, K.; Birds, I.; Aspden, J.L. Long non-coding RNAs in development and disease: Conservation to mechanisms. J. Pathol. 2020, 250, 480–495. [Google Scholar] [CrossRef]
- Huarte, M. The emerging role of lncRNAs in cancer. Nat. Med. 2015, 21, 1253–1261. [Google Scholar] [CrossRef]
- Smolle, M.A.; Bauernhofer, T.; Pummer, K.; Calin, G.A.; Pichler, M. Current Insights into Long Non-Coding RNAs (LncRNAs) in Prostate Cancer. Int. J. Mol. Sci. 2017, 18, 473. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Xu, Z.; Xu, W.; Jiang, K.; Zhang, F.; Ding, Q.; Xu, Z.; Chen, Y. Inhibition of ATM reverses EMT and decreases metastatic potential of cisplatin-resistant lung cancer cells through JAK/STAT3/PD-L1 pathway. J. Exp. Clin. Cancer Res. 2019, 38, 149. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.B.; Yang, W.; Xuan, Y.; Lin, A.J. miR-526b-3p inhibits lung cancer cisplatin-resistance and metastasis by inhibiting STAT3-promoted PD-L1. Cell Death Dis. 2021, 12, 748. [Google Scholar] [CrossRef] [PubMed]
- Ji, P.; Diederichs, S.; Wang, W.; Böing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041. [Google Scholar] [CrossRef]
- Luo, F.; Sun, B.; Li, H.; Xu, Y.; Liu, Y.; Liu, X.; Lu, L.; Li, J.; Wang, Q.; Wei, S.; et al. A MALAT1/HIF-2α feedback loop contributes to arsenite carcinogenesis. Oncotarget 2016, 7, 5769–5787. [Google Scholar] [CrossRef] [PubMed]
- Luo, F.; Liu, X.; Ling, M.; Lu, L.; Shi, L.; Lu, X.; Li, J.; Zhang, A.; Liu, Q. The lncRNA MALAT1, acting through HIF-1α stabilization, enhances arsenite-induced glycolysis in human hepatic L-02 cells. Biochim. Et Biophys. Acta (BBA) Mol. Basis Dis. 2016, 1862, 1685–1695. [Google Scholar] [CrossRef]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef]
- Yu, L.; Chen, X.; Sun, X.; Wang, L.; Chen, S. The Glycolytic Switch in Tumors: How Many Players Are Involved? J. Cancer 2017, 8, 3430–3440. [Google Scholar] [CrossRef]
- Mirzaei, H.; Hamblin, M.R. Regulation of Glycolysis by Non-coding RNAs in Cancer: Switching on the Warburg Effect. Mol. Ther. Oncolytics 2020, 19, 218–239. [Google Scholar] [CrossRef]
- Dai, X.; Chen, C.; Xue, J.; Xiao, T.; Mostofa, G.; Wang, D.; Chen, X.; Xu, H.; Sun, Q.; Li, J.; et al. Exosomal MALAT1 derived from hepatic cells is involved in the activation of hepatic stellate cells via miRNA-26b in fibrosis induced by arsenite. Toxicol. Lett. 2019, 316, 73–84. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Xu, D.; Xiang, Z.; Ding, J.; Yang, X.; Li, D.; Han, X. m(6)A mRNA methylation regulates testosterone synthesis through modulating autophagy in Leydig cells. Autophagy 2021, 17, 457–475. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Song, J.; Yuan, W.; Zhang, W.; Sun, Z. Roles of RNA Methylation on Tumor Immunity and Clinical Implications. Front. Immunol. 2021, 12, 641507. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Wang, J.Q.; Tan, Y.; Yuan, R.; Chen, Z.S.; Zou, C. RNA methylation and cancer treatment. Pharmacol. Res. 2021, 174, 105937. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; He, C. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Trends Genet. 2016, 32, 320–321. [Google Scholar] [CrossRef]
- Du, H.; Zhao, Y.; He, J.; Zhang, Y.; Xi, H.; Liu, M.; Ma, J.; Wu, L. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef] [PubMed]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5′ UTR m(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, X.; Weng, Y.L.; Lu, Z.; Liu, Y.; Lu, Z.; Li, J.; Hao, P.; Zhang, Y.; Zhang, F.; et al. m(6)A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253. [Google Scholar] [CrossRef]
- Xiang, Y.; Laurent, B.; Hsu, C.H.; Nachtergaele, S.; Lu, Z.; Sheng, W.; Xu, C.; Chen, H.; Ouyang, J.; Wang, S.; et al. RNA m(6)A methylation regulates the ultraviolet-induced DNA damage response. Nature 2017, 543, 573–576. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Luo, G.Z.; Zhang, Z.; Wang, X.; Zhou, T.; Cui, Y.; Sha, J.; Huang, X.; Guerrero, L.; Xie, P.; et al. YTHDC1 mediates nuclear export of N(6)-methyladenosine methylated mRNAs. Elife 2017, 6, e31311. [Google Scholar] [CrossRef]
- Alarcón, C.R.; Lee, H.; Goodarzi, H.; Halberg, N.; Tavazoie, S.F. N6-methyladenosine marks primary microRNAs for processing. Nature 2015, 519, 482–485. [Google Scholar] [CrossRef]
- Karikó, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N(6)-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef]
- Huang, W.; Chen, T.Q.; Fang, K.; Zeng, Z.C.; Ye, H.; Chen, Y.Q. N6-methyladenosine methyltransferases: Functions, regulation, and clinical potential. J. Hematol. Oncol. 2021, 14, 117. [Google Scholar] [CrossRef]
- Meyer, K.D.; Jaffrey, S.R. Rethinking m(6)A Readers, Writers, and Erasers. Annu. Rev. Cell Dev. Biol. 2017, 33, 319–342. [Google Scholar] [CrossRef]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Haussmann, I.U.; Bodi, Z.; Sanchez-Moran, E.; Mongan, N.P.; Archer, N.; Fray, R.G.; Soller, M. m(6)A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature 2016, 540, 301–304. [Google Scholar] [CrossRef]
- Berulava, T.; Buchholz, E.; Elerdashvili, V.; Pena, T.; Islam, M.R.; Lbik, D.; Mohamed, B.A.; Renner, A.; von Lewinski, D.; Sacherer, M.; et al. Changes in m6A RNA methylation contribute to heart failure progression by modulating translation. Eur. J. Heart Fail. 2020, 22, 54–66. [Google Scholar] [CrossRef]
- Dorn, L.E.; Lasman, L.; Chen, J.; Xu, X.; Hund, T.J.; Medvedovic, M.; Hanna, J.H.; van Berlo, J.H.; Accornero, F. The N(6)-Methyladenosine mRNA Methylase METTL3 Controls Cardiac Homeostasis and Hypertrophy. Circulation 2019, 139, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Lan, Q.; Liu, P.Y.; Haase, J.; Bell, J.L.; Hüttelmaier, S.; Liu, T. The Critical Role of RNA m(6)A Methylation in Cancer. Cancer Res. 2019, 79, 1285–1292. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Weng, H.; Chen, J. m(6)A Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 2020, 37, 270–288. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Shi, X.; Dai, C.; Shen, F.; Rocco, G.; Chen, J.; Huang, Z.; Chen, C.; He, C.; Huang, T.; et al. RNA m(6)A Modification in Cancers: Molecular Mechanisms and Potential Clinical Applications. Innovation 2020, 1, 100066. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef] [PubMed]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramirez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef]
- Yang, F.; Yuan, W.Q.; Li, J.; Luo, Y.Q. Knockdown of METTL14 suppresses the malignant progression of non-small cell lung cancer by reducing Twist expression. Oncol. Lett. 2021, 22, 847. [Google Scholar] [CrossRef]
- Mao, J.; Qiu, H.; Guo, L. LncRNA HCG11 mediated by METTL14 inhibits the growth of lung adenocarcinoma via IGF2BP2/LATS1. Biochem. Biophys. Res. Commun. 2021, 580, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhao, J.; Wang, L.; Chi, Y.; Huang, X.; Liu, W. METTL14-Mediated miR-30c-1-3p Maturation Represses the Progression of Lung Cancer via Regulation of MARCKSL1 Expression. Mol. Biotechnol. 2022, 64, 199–212. [Google Scholar] [CrossRef]
- Liu, J.; Ren, D.; Du, Z.; Wang, H.; Zhang, H.; Jin, Y. m(6)A demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018, 502, 456–464. [Google Scholar] [CrossRef]
- Gao, M.; Qi, Z.; Feng, W.; Huang, H.; Xu, Z.; Dong, Z.; Xu, M.; Han, J.; Kloeber, J.A.; Huang, J.; et al. m6A demethylation of cytidine deaminase APOBEC3B mRNA orchestrates arsenic-induced mutagenesis. J. Biol. Chem. 2022, 298, 101563. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.H.; Yang, S.; Wei, J.; Shea, C.R.; Zhong, W.; Wang, F.; Shah, P.; Kibriya, M.G.; Cui, X.; Ahsan, H.; et al. Autophagy of the m(6)A mRNA demethylase FTO is impaired by low-level arsenic exposure to promote tumorigenesis. Nat. Commun. 2021, 12, 2183. [Google Scholar] [CrossRef] [PubMed]
- Faustino, N.A.; Cooper, T.A. Pre-mRNA splicing and human disease. Genes Dev. 2003, 17, 419–437. [Google Scholar] [CrossRef] [PubMed]
- Ast, G. How did alternative splicing evolve? Nat. Rev. Genet. 2004, 5, 773–782. [Google Scholar] [CrossRef] [PubMed]
- Cherry, S.; Lynch, K.W. Alternative splicing and cancer: Insights, opportunities, and challenges from an expanding view of the transcriptome. Genes Dev. 2020, 34, 1005–1016. [Google Scholar] [CrossRef]
- Brown, R.L.; Reinke, L.M.; Damerow, M.S.; Perez, D.; Chodosh, L.A.; Yang, J.; Cheng, C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Investig. 2011, 121, 1064–1074. [Google Scholar] [CrossRef]
- Xu, Y.; Gao, X.D.; Lee, J.H.; Huang, H.; Tan, H.; Ahn, J.; Reinke, L.M.; Peter, M.E.; Feng, Y.; Gius, D.; et al. Cell type-restricted activity of hnRNPM promotes breast cancer metastasis via regulating alternative splicing. Genes Dev. 2014, 28, 1191–1203. [Google Scholar] [CrossRef]
- David, C.J.; Chen, M.; Assanah, M.; Canoll, P.; Manley, J.L. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature 2010, 463, 364–368. [Google Scholar] [CrossRef]
- Nowak, D.G.; Woolard, J.; Amin, E.M.; Konopatskaya, O.; Saleem, M.A.; Churchill, A.J.; Ladomery, M.R.; Harper, S.J.; Bates, D.O. Expression of pro- and anti-angiogenic isoforms of VEGF is differentially regulated by splicing and growth factors. J. Cell Sci. 2008, 121, 3487–3495. [Google Scholar] [CrossRef]
- Pandya-Jones, A.; Black, D.L. Co-transcriptional splicing of constitutive and alternative exons. RNA 2009, 15, 1896–1908. [Google Scholar] [CrossRef]
- Lev Maor, G.; Yearim, A.; Ast, G. The alternative role of DNA methylation in splicing regulation. Trends Genet. 2015, 31, 274–280. [Google Scholar] [CrossRef] [PubMed]
- Naftelberg, S.; Schor, I.E.; Ast, G.; Kornblihtt, A.R. Regulation of alternative splicing through coupling with transcription and chromatin structure. Annu. Rev. Biochem 2015, 84, 165–198. [Google Scholar] [CrossRef] [PubMed]
- Rahhal, R.; Seto, E. Emerging roles of histone modifications and HDACs in RNA splicing. Nucleic Acids Res. 2019, 47, 4911–4926. [Google Scholar] [CrossRef] [PubMed]
- Ferragut Cardoso, A.P.; Banerjee, M.; Al-Eryani, L.; Sayed, M.; Wilkey, D.W.; Merchant, M.L.; Park, J.W.; States, J.C. Temporal Modulation of Differential Alternative Splicing in HaCaT Human Keratinocyte Cell Line Chronically Exposed to Arsenic for up to 28 Wk. Environ. Health Perspect 2022, 130, 17011. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Sun, X.; Cooper, K.L.; Wang, F.; Liu, K.J.; Hudson, L.G. Arsenite interacts selectively with zinc finger proteins containing C3H1 or C4 motifs. J. Biol. Chem. 2011, 286, 22855–22863. [Google Scholar] [CrossRef]
- Ding, W.; Liu, W.; Cooper, K.L.; Qin, X.J.; de Souza Bergo, P.L.; Hudson, L.G.; Liu, K.J. Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage. J. Biol. Chem. 2009, 284, 6809–6817. [Google Scholar] [CrossRef]
- Sun, X.; Zhou, X.; Du, L.; Liu, W.; Liu, Y.; Hudson, L.G.; Liu, K.J. Arsenite binding-induced zinc loss from PARP-1 is equivalent to zinc deficiency in reducing PARP-1 activity, leading to inhibition of DNA repair. Toxicol. Appl. Pharmacol. 2014, 274, 313–318. [Google Scholar] [CrossRef]
- Ji, Y.; Tulin, A.V. Post-transcriptional regulation by poly(ADP-ribosyl)ation of the RNA-binding proteins. Int. J. Mol. Sci 2013, 14, 16168–16183. [Google Scholar] [CrossRef]
- Stueckle, T.A.; Lu, Y.; Davis, M.E.; Wang, L.; Jiang, B.H.; Holaskova, I.; Schafer, R.; Barnett, J.B.; Rojanasakul, Y. Chronic occupational exposure to arsenic induces carcinogenic gene signaling networks and neoplastic transformation in human lung epithelial cells. Toxicol. Appl. Pharmacol. 2012, 261, 204–216. [Google Scholar] [CrossRef]
- Malanga, M.; Czubaty, A.; Girstun, A.; Staron, K.; Althaus, F.R. Poly(ADP-ribose) binds to the splicing factor ASF/SF2 and regulates its phosphorylation by DNA topoisomerase I. J. Biol. Chem. 2008, 283, 19991–19998. [Google Scholar] [CrossRef]
- Liu, S.; Jiang, J.; Li, L.; Amato, N.J.; Wang, Z.; Wang, Y. Arsenite Targets the Zinc Finger Domains of Tet Proteins and Inhibits Tet-Mediated Oxidation of 5-Methylcytosine. Environ. Sci. Technol. 2015, 49, 11923–11931. [Google Scholar] [CrossRef] [PubMed]
- Marina, R.J.; Sturgill, D.; Bailly, M.A.; Thenoz, M.; Varma, G.; Prigge, M.F.; Nanan, K.K.; Shukla, S.; Haque, N.; Oberdoerffer, S. TET-catalyzed oxidation of intragenic 5-methylcytosine regulates CTCF-dependent alternative splicing. EMBO J. 2016, 35, 335–355. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Shi, Y.; Yadav, S.; Wang, H. p52-Bcl3 complex promotes cyclin D1 expression in BEAS-2B cells in response to low concentration arsenite. Toxicology 2010, 273, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Pradeepa, M.M.; Sutherland, H.G.; Ule, J.; Grimes, G.R.; Bickmore, W.A. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012, 8, e1002717. [Google Scholar] [CrossRef]
- Das, S.; Anczuków, O.; Akerman, M.; Krainer, A.R. Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 2012, 1, 110–117. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ramos, R.; López-Carrillo, L.; Albores, A.; Hernández-Ramírez, R.U.; Cebrian, M.E. Sodium arsenite alters cell cycle and MTHFR, MT1/2, and c-Myc protein levels in MCF-7 cells. Toxicol. Appl. Pharmacol. 2009, 241, 269–274. [Google Scholar] [CrossRef]
- Koh, C.M.; Bezzi, M.; Low, D.H.; Ang, W.X.; Teo, S.X.; Gay, F.P.; Al-Haddawi, M.; Tan, S.Y.; Osato, M.; Sabò, A.; et al. MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 2015, 523, 96–100. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).