U-Net Based Segmentation and Characterization of Gliomas
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. Histopathological Data
2.3. Image Acquisition
2.4. U-Net Based Auto-Detection and Segmentation of Gliomas
2.5. Volume Acquisition and Texture Analysis
2.6. Statistical Analysis
3. Results
3.1. Clinical Characteristics of Patient Population
3.2. Testing Dataset
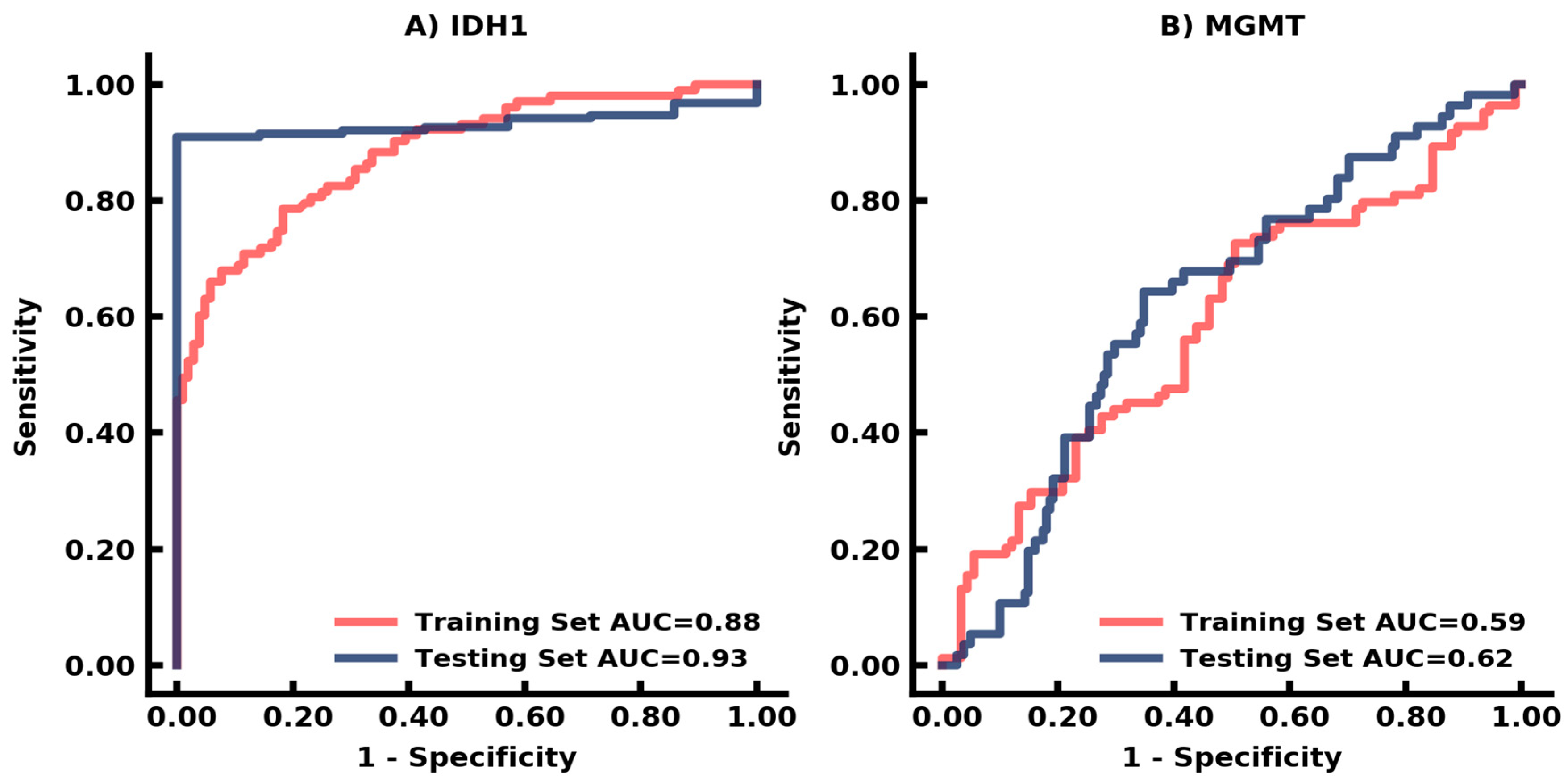
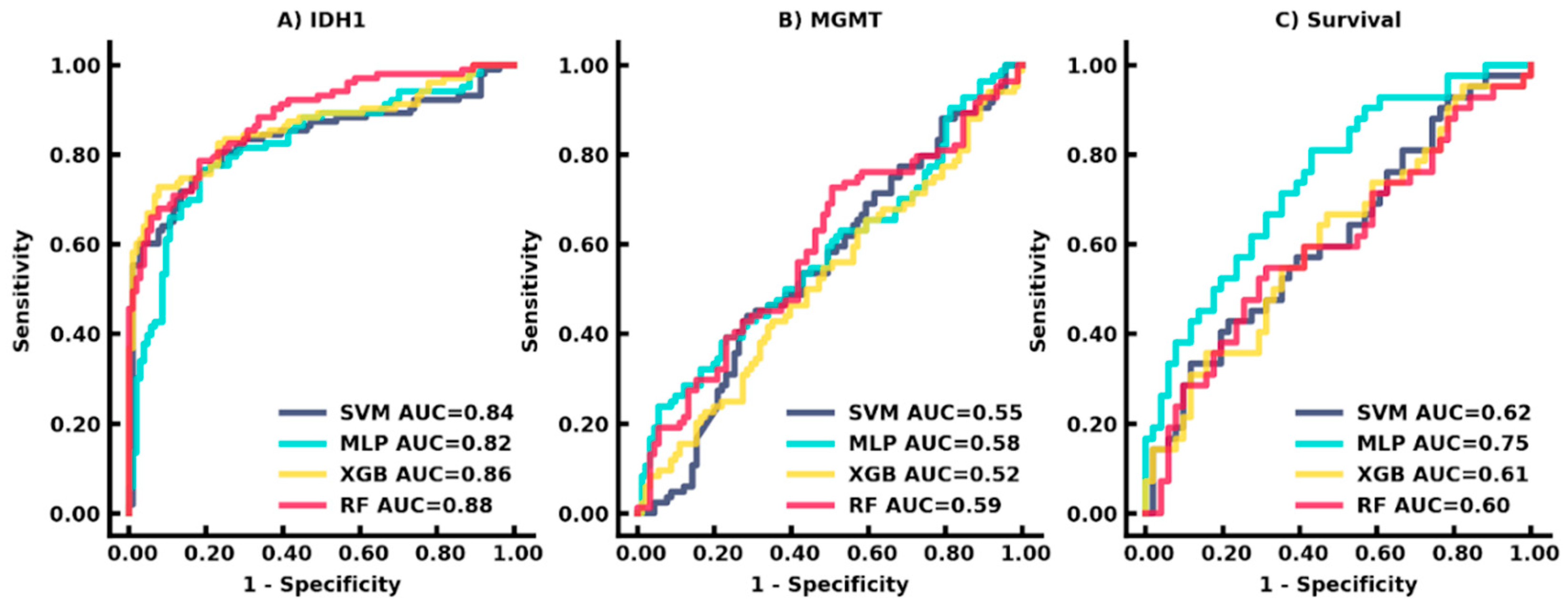
3.3. Auto-Segmentation and Prediction of Biomarkers
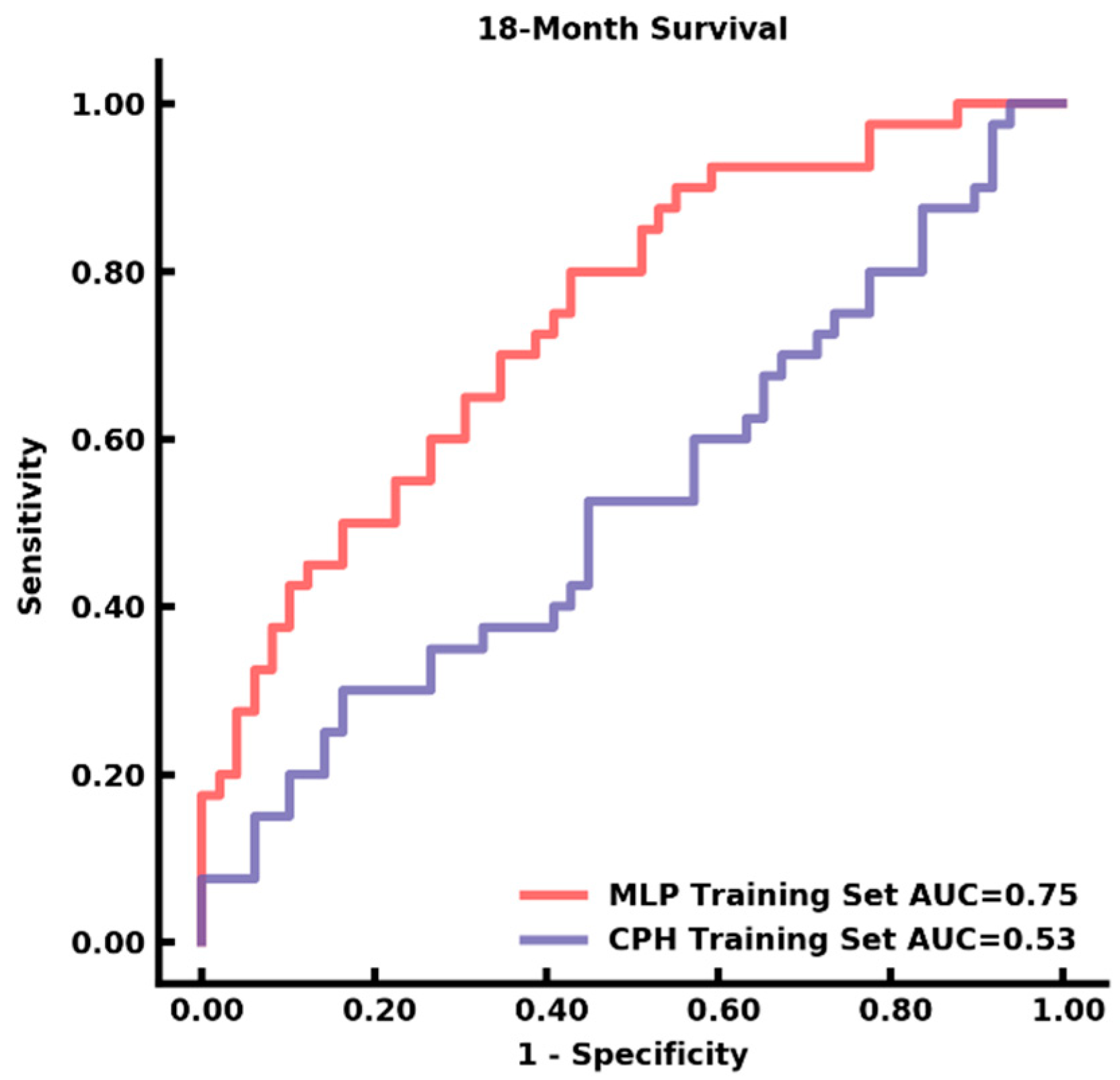
3.4. Auto-Segmentation and Prediction of Survival
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Singh, G.; Manjila, S.; Sakla, N.; True, A.; Wardeh, A.H.; Beig, N.; Vaysberg, A.; Matthews, J.; Prasanna, P.; Spektor, V. Radiomics and radiogenomics in gliomas: A contemporary update. Br. J. Cancer 2021, 125, 641–657. [Google Scholar] [CrossRef]
- Yan, H.; Parsons, D.W.; Jin, G.; McLendon, R.; Rasheed, B.A.; Yuan, W.; Kos, I.; Batinic-Haberle, I.; Jones, S.; Riggins, G.J.; et al. IDH1 and IDH2 mutations in gliomas. N. Engl. J. Med. 2009, 360, 765–773. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.-C.; Gorlia, T.; Hamou, M.-F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Nandakumar, P.; Mansouri, A.; Das, S. The Role of ATRX in Glioma Biology. Front. Oncol. 2017, 7, 236. [Google Scholar] [CrossRef]
- Hatanpaa, K.J.; Burma, S.; Zhao, D.; Habib, A.A. Epidermal Growth Factor Receptor in Glioma: Signal Transduction, Neuropathology, Imaging, and Radioresistance. Neoplasia 2010, 12, 675–684. [Google Scholar] [CrossRef]
- Wick, W.; Roth, P.; Hartmann, C.; Hau, P.; Nakamura, M.; Stockhammer, F.; Sabel, M.C.; Wick, A.; Koeppen, S.; Ketter, R.; et al. Long-term analysis of the NOA-04 randomized phase III trial of sequential radiochemotherapy of anaplastic glioma with PCV or temozolomide. Neuro-Oncol. 2016, 18, 1529–1537. [Google Scholar] [CrossRef]
- Korfiatis, P.; Kline, T.L.; Coufalova, L.; Lachance, D.H.; Parney, I.F.; Carter, R.E.; Buckner, J.C.; Erickson, B.J. MRI texture features as biomarkers to predict MGMT methylation status in glioblastomas. Med. Phys. 2016, 43, 2835–2844. [Google Scholar] [CrossRef]
- Drabycz, S.; Roldán, G.; de Robles, P.; Adler, D.; McIntyre, J.B.; Magliocco, A.M.; Cairncross, J.G.; Mitchell, J.R. An analysis of image texture, tumor location, and MGMT promoter methylation in glioblastoma using magnetic resonance imaging. Neuroimage 2010, 49, 1398–1405. [Google Scholar] [CrossRef]
- Kihira, S.; Tsankova, N.M.; Bauer, A.; Sakai, Y.; Mahmoudi, K.; Zubizarreta, N.; Houldsworth, J.; Khan, F.; Salamon, N.; Hormigo, A.; et al. Multiparametric MRI texture analysis in prediction of glioma biomarker status: Added value of MR diffusion. Neuro-Oncol. Adv. 2021, 3, vdab051. [Google Scholar] [CrossRef]
- Li, Z.-C.; Bai, H.; Sun, Q.; Zhao, Y.; Lv, Y.; Zhou, J.; Liang, C.; Chen, Y.; Liang, D.; Zheng, H. Multiregional radiomics profiling from multiparametric MRI: Identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med. 2018, 7, 5999–6009. [Google Scholar] [CrossRef]
- Lee, M.H.; Kim, J.; Kim, S.-T.; Shin, H.-M.; You, H.-J.; Choi, J.W.; Seol, H.J.; Nam, D.-H.; Lee, J.-I.; Kong, D.-S. Prediction of IDH1 Mutation Status in Glioblastoma Using Machine Learning Technique Based on Quantitative Radiomic Data. World Neurosurg. 2019, 125, e688–e696. [Google Scholar] [CrossRef]
- Ren, Y.; Zhang, X.; Rui, W.; Pang, H.; Qiu, T.; Wang, J.; Xie, Q.; Jin, T.; Zhang, H.; Chen, H.; et al. Noninvasive Prediction of IDH1 Mutation and ATRX Expression Loss in Low-Grade Gliomas Using Multiparametric MR Radiomic Features. J. Magn. Reson. Imaging 2019, 49, 808–817. [Google Scholar] [CrossRef]
- Zhang, B.; Chang, K.; Ramkissoon, S.; Tanguturi, S.; Bi, W.L.; Reardon, D.A.; Ligon, K.L.; Alexander, B.M.; Wen, P.Y.; Huang, R.Y. Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro-Oncol. 2017, 19, 109–117. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Xu, K.; Qian, Z.; Wang, K.; Fan, X.; Li, S.; Wang, Y.; Jiang, T. MRI features can predict EGFR expression in lower grade gliomas: A voxel-based radiomic analysis. Eur. Radiol. 2017, 28, 356–362. [Google Scholar] [CrossRef]
- Kickingereder, P.; Bonekamp, D.; Nowosielski, M.; Kratz, A.; Sill, M.; Burth, S.; Wick, A.; Eidel, O.; Schlemmer, H.-P.; Radbruch, A.; et al. Radiogenomics of Glioblastoma: Machine Learning–based Classification of Molecular Characteristics by Using Multiparametric and Multiregional MR Imaging Features. Radiology 2016, 281, 907–918. [Google Scholar] [CrossRef]
- Li, Y.; Liu, X.; Qian, Z.; Sun, Z.; Xu, K.; Wang, K.; Fan, X.; Zhang, Z.; Li, S.; Wang, Y.; et al. Genotype prediction of ATRX mutation in lower-grade gliomas using an MRI radiomics signature. Eur. Radiol. 2018, 28, 2960–2968. [Google Scholar] [CrossRef]
- Li, Y.; Liang, Y.; Sun, Z.; Xu, K.; Fan, X.; Li, S.; Zhang, Z.; Jiang, T.; Liu, X.; Wang, Y. Radiogenomic analysis of PTEN mutation in glioblastoma using preoperative multi-parametric magnetic resonance imaging. Neuroradiology 2019, 61, 1229–1237. [Google Scholar] [CrossRef]
- Carlson, M.R.; Pope, W.B.; Horvath, S.; Braunstein, J.G.; Nghiemphu, P.; Tso, C.-L.; Mellinghoff, I.; Lai, A.; Liau, L.M.; Mischel, P.S.; et al. Relationship between Survival and Edema in Malignant Gliomas: Role of Vascular Endothelial Growth Factor and Neuronal Pentraxin 2. Clin. Cancer Res. 2007, 13, 2592–2598. [Google Scholar] [CrossRef]
- Eliat, P.-A.; Olivié, D.; Saïkali, S.; Carsin, B.; Saint-Jalmes, H.; de Certaines, J.D. Can Dynamic Contrast-Enhanced Magnetic Resonance Imaging Combined with Texture Analysis Differentiate Malignant Glioneuronal Tumors from Other Glioblastoma? Neurol. Res. Int. 2012, 2012, 195176. [Google Scholar] [CrossRef][Green Version]
- Rao, A.; Rao, G.; Gutman, D.A.; Flanders, A.E.; Hwang, S.N.; Rubin, D.L.; Colen, R.R.; Zinn, P.O.; Jain, R.; Wintermark, M.; et al. A combinatorial radiographic phenotype may stratify patient survival and be associated with invasion and proliferation characteristics in glioblastoma. J. Neurosurg. 2016, 124, 1008–1017. [Google Scholar] [CrossRef]
- Lotan, E.; Zhang, B.; Dogra, S.; Wang, W.; Carbone, D.; Fatterpekar, G.; Oermann, E.; Lui, Y. Development and Practical Implementation of a Deep Learning–Based Pipeline for Automated Pre- and Postoperative Glioma Segmentation. Am. J. Neuroradiol. 2022, 43, 24–32. [Google Scholar] [CrossRef]
- Ghaffari, M.; Sowmya, A.; Oliver, R. Automated Brain Tumor Segmentation Using Multimodal Brain Scans: A Survey Based on Models Submitted to the BraTS 2012–2018 Challenges. IEEE Rev. Biomed. Eng. 2019, 13, 156–168. [Google Scholar] [CrossRef]
- Zhou, C.; Ding, C.; Wang, X.; Lu, Z.; Tao, D. One-Pass Multi-Task Networks with Cross-Task Guided Attention for Brain Tumor Segmentation. IEEE Trans. Image Process. 2020, 29, 4516–4529. [Google Scholar] [CrossRef]
- Chen, L.; Wu, Y.; Souza, A.M.D.; Abidin, A.Z.; Wismüller, A.; Xu, C. MRI tumor segmentation with densely connected 3D CNN. In Proceedings of the Medical Imaging 2018: Image Processing, SPIE, Houston, TX, USA, 11–13 February 2018; pp. 357–364. [Google Scholar] [CrossRef]
- Lachinov, D.; Vasiliev, E.; Turlapov, V. Glioma Segmentation with Cascaded U-Net. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries; Springer International Publishing: Cham, Switzerland, 2019; pp. 189–198. [Google Scholar]
- Paolillo, M.; Boselli, C.; Schinelli, S. Glioblastoma under Siege: An Overview of Current Therapeutic Strategies. Brain Sci. 2018, 8, 15. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely Connected Convolutional Networks. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 2261–2269. [Google Scholar]
- Mei, X.; Liu, Z.; Robson, P.M.; Marinelli, B.; Huang, M.; Doshi, A.; Jacobi, A.; Cao, C.; Link, K.E.; Yang, T.; et al. RadImageNet: An Open Radiologic Deep Learning Research Dataset for Effective Transfer Learning. Radiol. Artif. Intell. 2022, 4, e210315. [Google Scholar] [CrossRef]
- van Griethuysen, J.J.M.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.G.H.; Fillion-Robin, J.-C.; Pieper, S.; Aerts, H.J.W.L. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Amadasun, M.; King, R. Textural features corresponding to textural properties. IEEE Trans. Syst. Man. Cybern. 1989, 19, 1264–1274. [Google Scholar] [CrossRef]
- Cook, G.J.R.; Azad, G.; Owczarczyk, K.; Siddique, M.; Goh, V. Challenges and Promises of PET Radiomics. Eur. J. Nucl. Med. Mol. Imaging 2013, 40, 133–140. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural Features for Image Classification. IEEE Trans. Syst. Man. Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- Liao, X.; Cai, B.; Tian, B.; Luo, Y.; Song, W.; Li, Y. Machine-learning based radiogenomics analysis of MRI features and metagenes in glioblastoma multiforme patients with different survival time. J. Cell. Mol. Med. 2019, 23, 4375–4385. [Google Scholar] [CrossRef]
- Soni, N.; Priya, S.; Bathla, G. Texture Analysis in Cerebral Gliomas: A Review of the Literature. Am. J. Neuroradiol. 2019, 40, 928–934. [Google Scholar] [CrossRef]
- Kickingereder, P.; Burth, S.; Wick, A.; Goetz, M.; Eidel, O.; Schlemmer, H.-P.; Maier-Hein, K.H.; Wick, W.; Bendszus, M.; Radbruch, A.; et al. Radiomic Profiling of Glioblastoma: Identifying an Imaging Predictor of Patient Survival with Improved Performance over Established Clinical and Radiologic Risk Models. Radiology 2016, 280, 880–889. [Google Scholar] [CrossRef]
- Sakai, Y.; Yang, C.; Kihira, S.; Tsankova, N.; Khan, F.; Hormigo, A.; Lai, A.; Cloughesy, T.; Nael, K. MRI Radiomic Features to Predict IDH1 Mutation Status in Gliomas: A Machine Learning Approach using Gradient Tree Boosting. Int. J. Mol. Sci. 2020, 21, 8004. [Google Scholar] [CrossRef] [PubMed]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Gardner, M.W.; Dorling, S.R. Artificial neural networks (the multilayer perceptron)—A review of applications in the atmospheric sciences. Atmos. Environ. 1998, 32, 2627–2636. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kaplan, E.L.; Meier, P. Nonparametric estimation from incomplete observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Cox, D.R. Regression models and life-tables. J. R. Stat. Soc. Ser. B 1972, 34, 187–202. [Google Scholar] [CrossRef]
- Delong, E.R.; Delong, D.M.; Clarke-Pearson, D.L. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Baid, U.; Ghodasara, S.; Mohan, S.; Bilello, M.; Calabrese, E.; Colak, E.; Farahani, K.; Kalpathy-Cramer, J.; Kitamura, F.C.; Pati, S.; et al. The RSNA-ASNR-MICCAI BraTS 2021 Benchmark on Brain Tumor Segmentation and Radiogenomic Classification. arXiv 2021, arXiv:2107.02314. [Google Scholar]
- Yogananda, C.G.B.; Shah, B.R.; Vejdani-Jahromi, M.; Nalawade, S.S.; Murugesan, G.K.; Yu, F.F.; Pinho, M.C.; Wagner, B.C.; Emblem, K.E.; Bjørnerud, A.; et al. A Fully Automated Deep Learning Network for Brain Tumor Segmentation. Tomography 2020, 6, 186–193. [Google Scholar] [CrossRef] [PubMed]
- Bakas, S.; Akbari, H.; Sotiras, A.; Bilello, M.; Rozycki, M.; Kirby, J.S.; Freymann, J.B.; Farahani, K.; Davatzikos, C. Advancing the Cancer Genome Atlas glioma MRI collections with expert segmentation labels and radiomic features. Sci. Data 2017, 4, 170117. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, Y.; Chen, W.; Huang, G.; Nie, S. 3D brain glioma segmentation in MRI through integrating multiple densely connected 2D convolutional neural networks. J. Zhejiang Univ. Sci. B 2021, 22, 462–475. [Google Scholar] [CrossRef]
- Hussain, S.; Anwar, S.M.; Majid, M. Brain tumor segmentation using cascaded deep convolutional neural network. In Proceedings of the 2017 39th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Jeju, Korea, 11–15 July 2017; pp. 1998–2001. [Google Scholar]
- Wang, G.; Li, W.; Ourselin, S.; Vercauteren, T. Automatic Brain Tumor Segmentation Based on Cascaded Convolutional Neural Networks with Uncertainty Estimation. Front. Comput. Neurosci. 2019, 13, 56. [Google Scholar] [CrossRef] [PubMed]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.-F. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Martinez, R.; Esteller, M. The DNA methylome of glioblastoma multiforme. Neurobiol. Dis. 2010, 39, 40–46. [Google Scholar] [CrossRef]
- Bae, S.; Choi, Y.S.; Ahn, S.S.; Chang, J.H.; Kang, S.-G.; Kim, E.H.; Kim, S.H.; Lee, S.-K. Radiomic MRI Phenotyping of Glioblastoma: Improving Survival Prediction. Radiology 2018, 289, 797–806. [Google Scholar] [CrossRef]
- Choi, Y.; Nam, Y.; Jang, J.; Shin, N.-Y.; Lee, Y.S.; Ahn, K.-J.; Kim, B.-S.; Park, J.-S.; Jeon, S.-S.; Gil Hong, Y. Radiomics may increase the prognostic value for survival in glioblastoma patients when combined with conventional clinical and genetic prognostic models. Eur. Radiol. 2021, 31, 2084–2093. [Google Scholar] [CrossRef]
- Ma, C.; Yao, Z.; Zhang, Q.; Zou, X. Quantitative integration of radiomic and genomic data improves survival prediction of low-grade glioma patients. Math. Biosci. Eng. 2021, 18, 727–744. [Google Scholar] [CrossRef]

| Network | Pretrained Source | DSC on Validation Set | DSC on Test Set |
|---|---|---|---|
| ResNet50 | ImageNet | 0.94 | 0.83 |
| ResNet50 | RadImageNet | 0.94 | 0.89 |
| DenseNet121 | ImageNet | 0.92 | 0.83 |
| DenseNet121 | RadImageNet | 0.96 | 0.93 |
| Sensitivity | Specificity | Negative Predictive Value | Positive Predictive Value | |
|---|---|---|---|---|
| Training Set | 0.9 | 1 | 0.28 | 1 |
| Testing Set | 0.98 | 0.32 | 0.94 | 0.59 |
| Sensitivity | Specificity | Negative Predictive Value | Positive Predictive Value | |
|---|---|---|---|---|
| Training Set | 0.63 | 0.65 | 0.83 | 0.38 |
| Testing Set | 0.45 | 0.68 | 0.57 | 0.57 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kihira, S.; Mei, X.; Mahmoudi, K.; Liu, Z.; Dogra, S.; Belani, P.; Tsankova, N.; Hormigo, A.; Fayad, Z.A.; Doshi, A.; et al. U-Net Based Segmentation and Characterization of Gliomas. Cancers 2022, 14, 4457. https://doi.org/10.3390/cancers14184457
Kihira S, Mei X, Mahmoudi K, Liu Z, Dogra S, Belani P, Tsankova N, Hormigo A, Fayad ZA, Doshi A, et al. U-Net Based Segmentation and Characterization of Gliomas. Cancers. 2022; 14(18):4457. https://doi.org/10.3390/cancers14184457
Chicago/Turabian StyleKihira, Shingo, Xueyan Mei, Keon Mahmoudi, Zelong Liu, Siddhant Dogra, Puneet Belani, Nadejda Tsankova, Adilia Hormigo, Zahi A. Fayad, Amish Doshi, and et al. 2022. "U-Net Based Segmentation and Characterization of Gliomas" Cancers 14, no. 18: 4457. https://doi.org/10.3390/cancers14184457
APA StyleKihira, S., Mei, X., Mahmoudi, K., Liu, Z., Dogra, S., Belani, P., Tsankova, N., Hormigo, A., Fayad, Z. A., Doshi, A., & Nael, K. (2022). U-Net Based Segmentation and Characterization of Gliomas. Cancers, 14(18), 4457. https://doi.org/10.3390/cancers14184457







