Identification and Validation of the lncRNA MYOSLID as a Regulating Factor of Necroptosis and Immune Cell Infiltration in Colorectal Cancer following Necroptosis-Related LncRNA Model Establishment
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Acquisition of Data
2.2. Differential Expression Analysis
2.3. Extraction of Necroptosis Genes
2.4. Identification of Necroptosis-Related lncRNAs
2.5. Establishment of the Co-Expression Network
2.6. Establishment of the Prognostic Model
2.7. Functional Annotation and Pathway Enrichment Analysis
2.8. Gene Set Enrichment Analysis
2.9. Assessment of Immune Cell Infiltration and Cibersort Analysis
2.10. Drug Sensitivity Prediction
2.11. Tissue Sample Collection and Colon Cancer Cell Line Culture
2.12. RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.13. In Vitro Experiments
2.14. Mouse Tumour Models
2.15. In Vivo Experiments
2.16. Statistical Analysis
3. Results
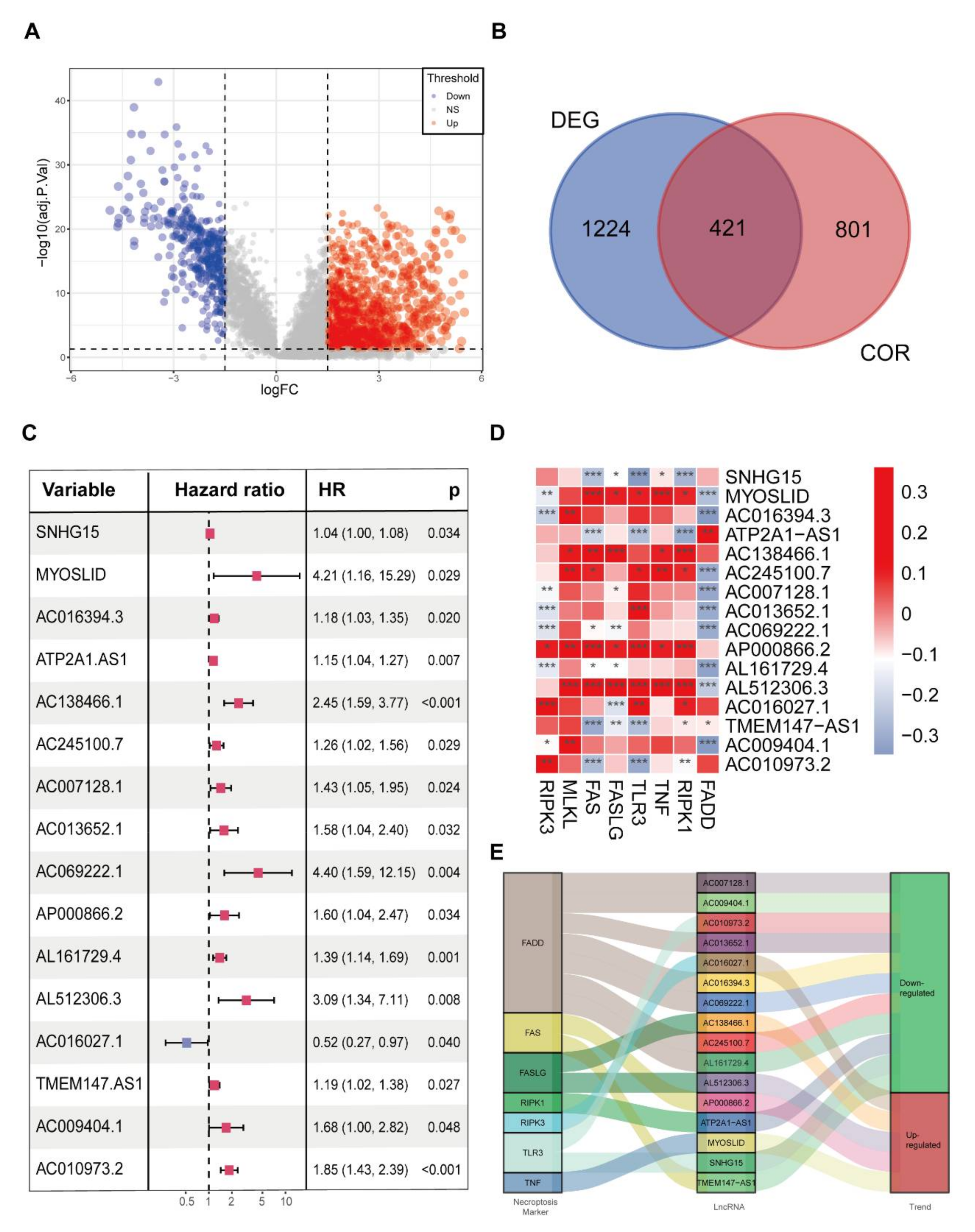
3.1. Identification of Necroptosis-Related Differentially Expressed lncRNAs in CRC
3.2. The Screening of Prognostic NRDELs in CRC Patients
3.3. Generation and Validation of an NRL Prognostic Model
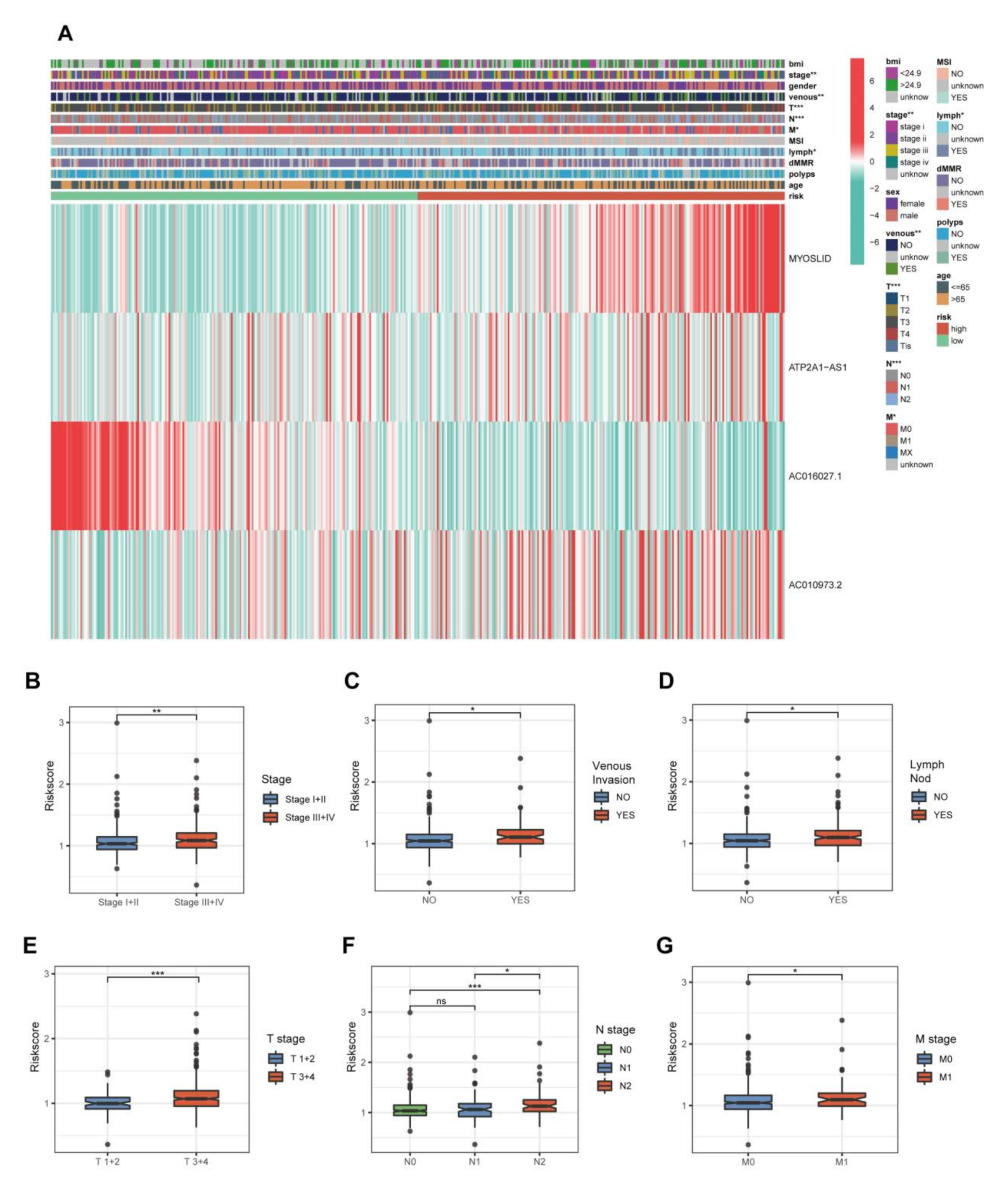
3.4. The Relationship between the 4-NRL Model and Other Clinicopathological Features
3.5. Discovery of Pathways and Functions of the New Model by GSEA and Gene Ontology Analysis
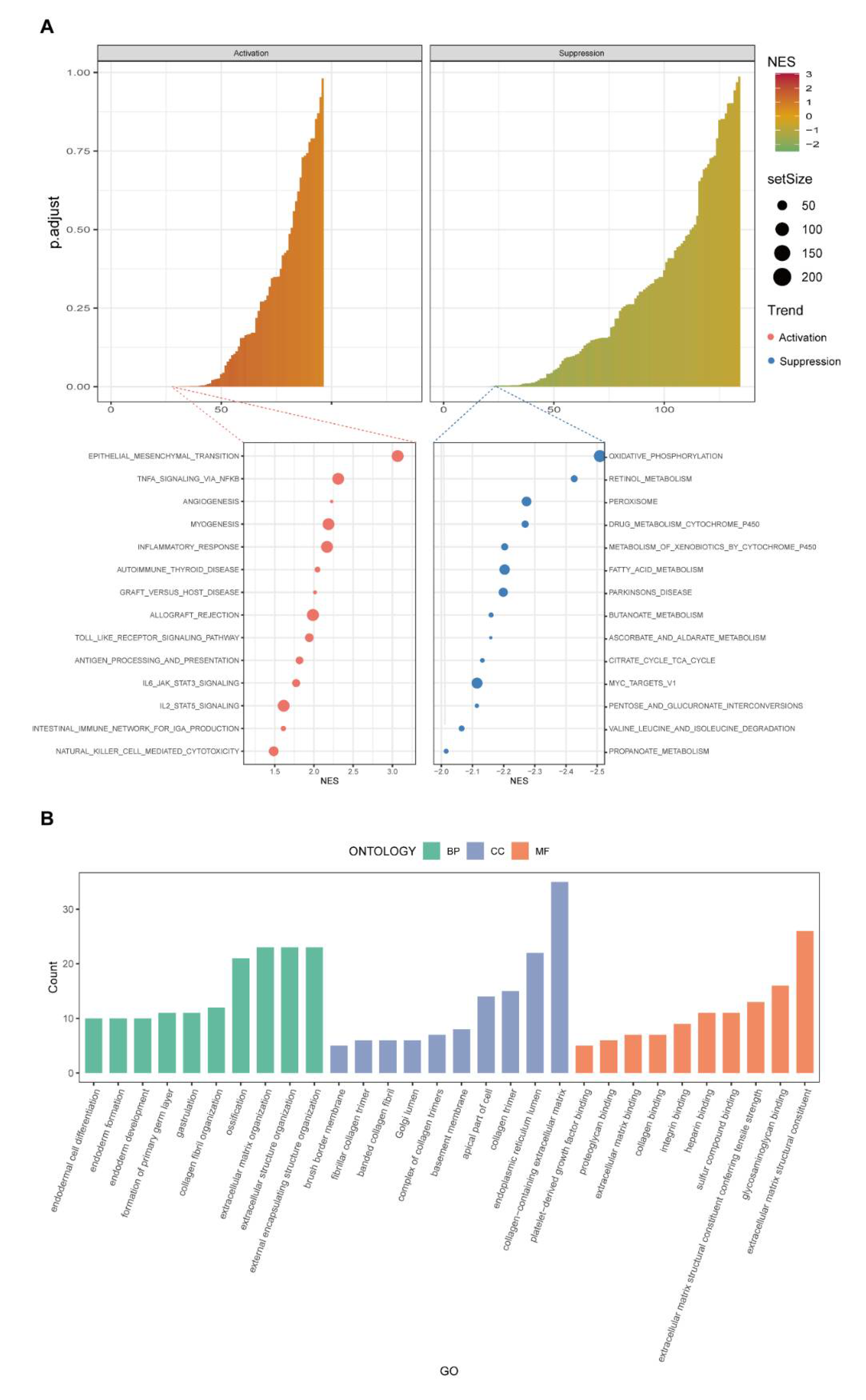
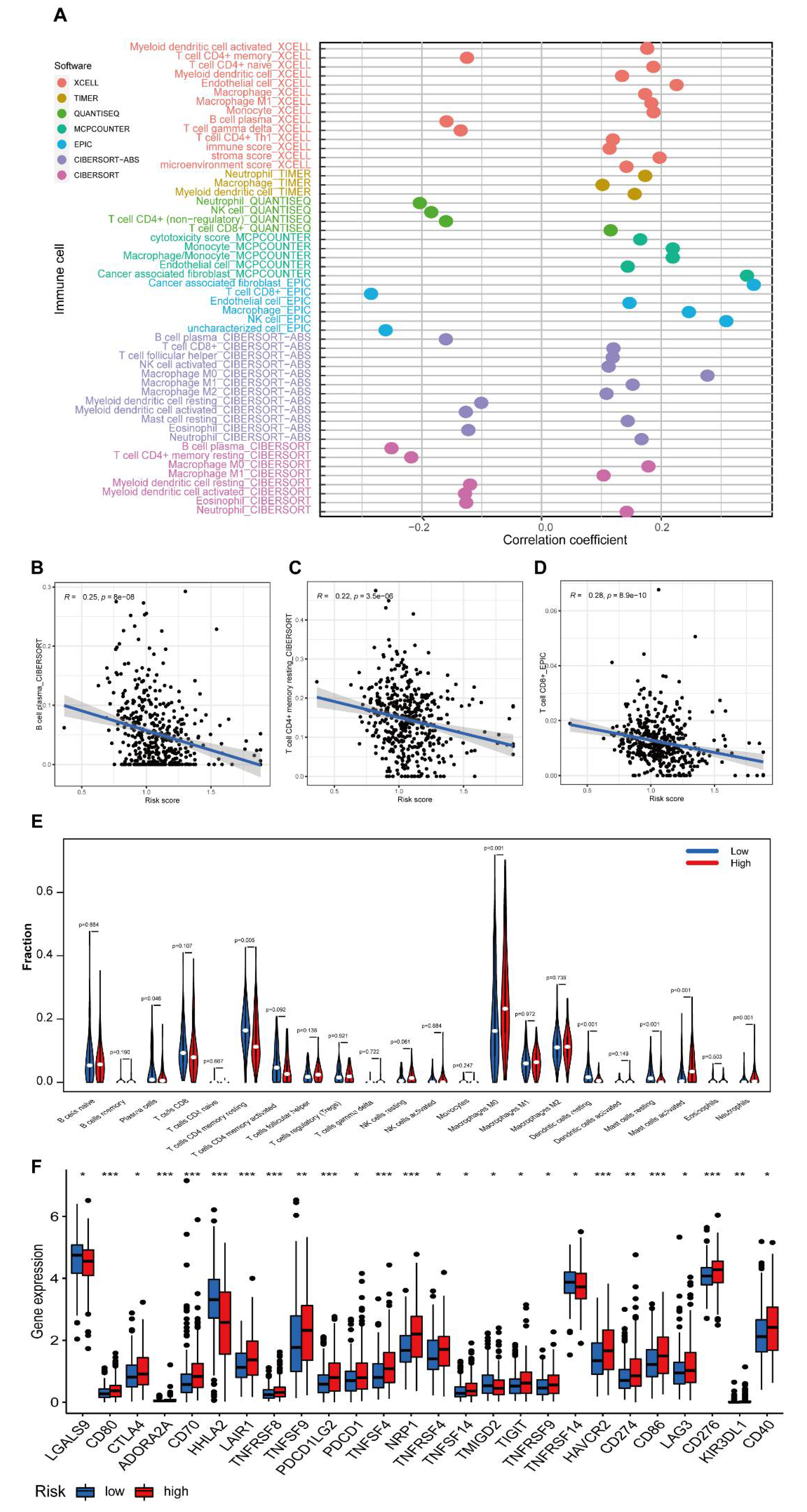
3.6. Investigation of the Immunity Landscape Using the Established Model
3.7. Exploration of Clinical Immune Treatment and Drug Sensitivity Tests in Two Risk Subgroups
3.8. The lncRNA MYOSLID Is a Regulatory Factor of Immune Infiltration in CRC
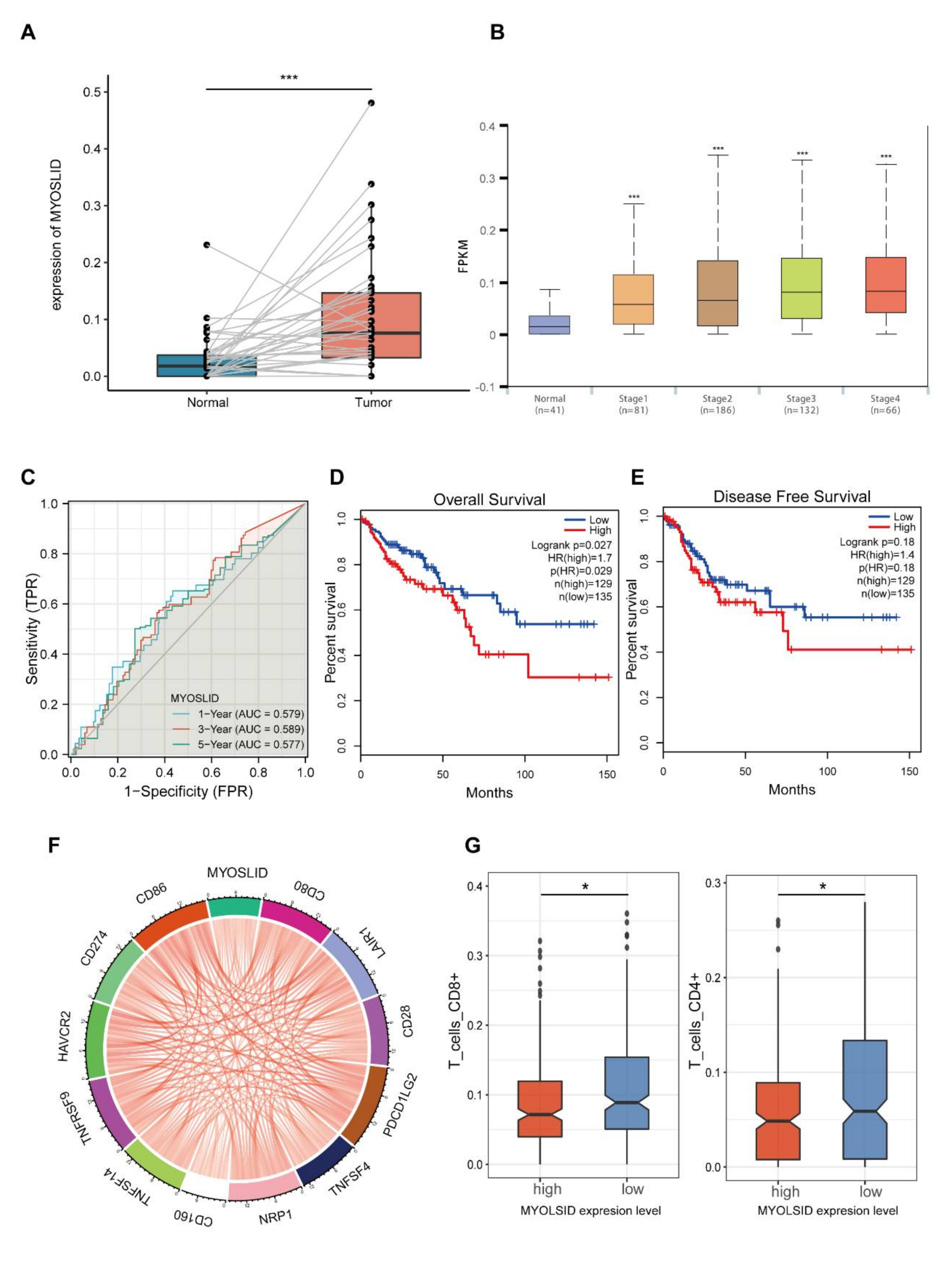
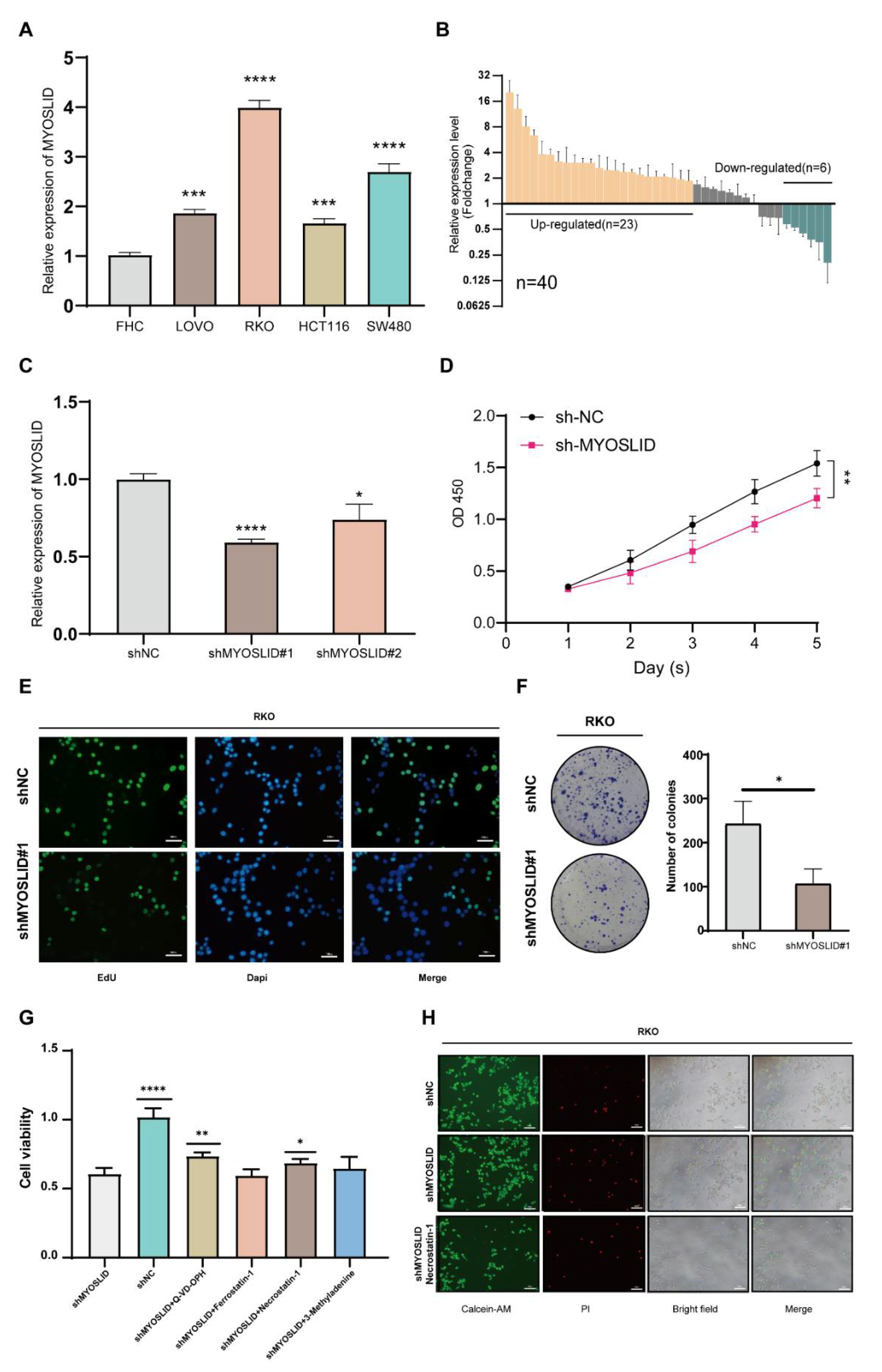
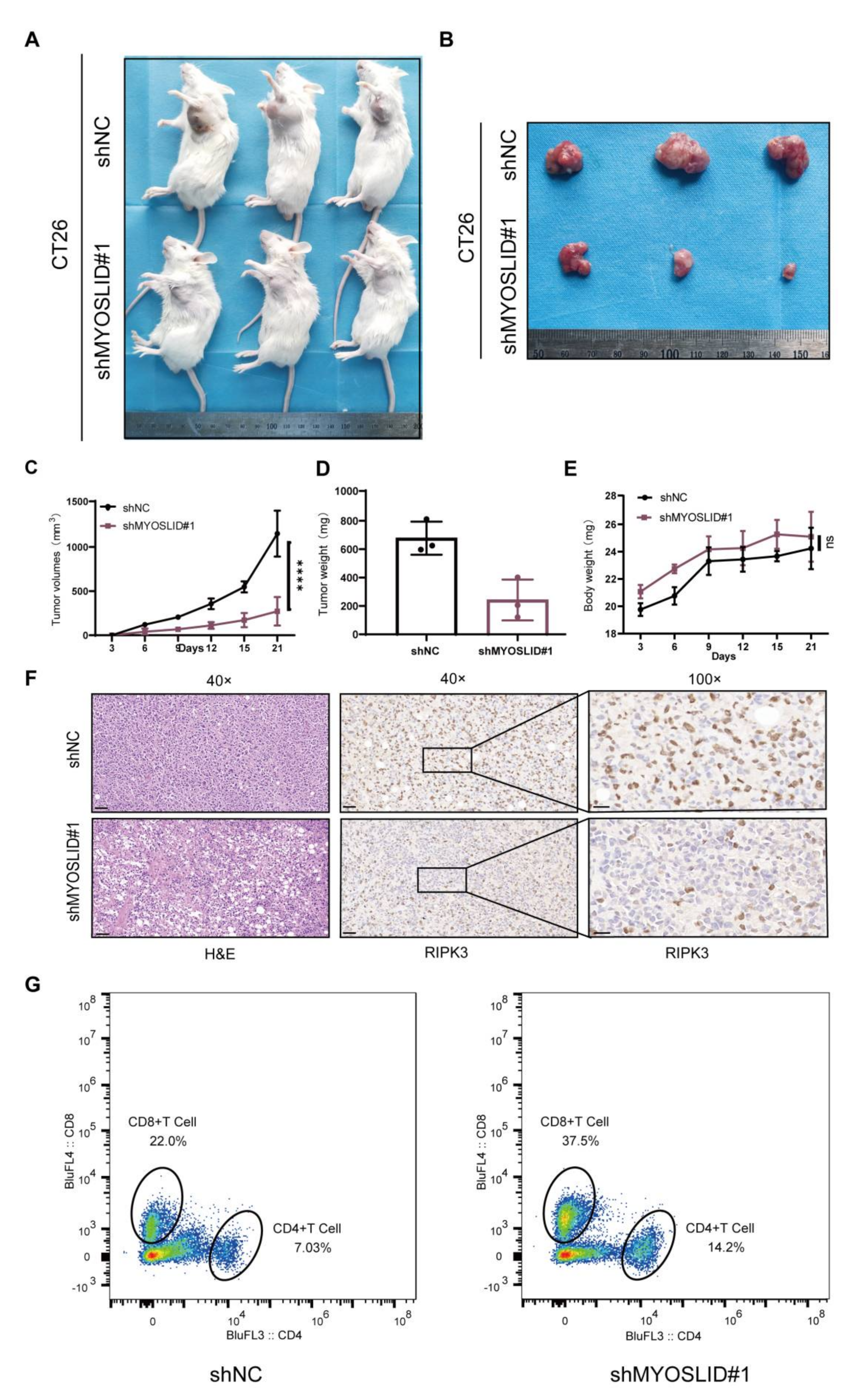
3.9. lncRNA MYOSLID Promotes the Growth of CRC by Inhibiting Necroptosis In Vitro
3.10. The LncRNA MYOSLID Promotes CRC Growth and Inhibits the Infiltration of CD4+ and CD8+ T Cells In Vivo
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2021. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Erickson, L.A. Adenocarcinoma of the Colon and Microsatellite Instability. Mayo Clin. Proc. 2018, 93, 669–670. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, J.; Prenen, H. Molecular genetics of colorectal cancer. Ann. Gastroenterol. 2014, 27, 9–14. [Google Scholar]
- Lim, K.G.; Lee, C.S.; Chin, D.H.J.; Ooi, Y.S.; Veettil, S.K.; Ching, S.M.; Burud, I.A.S.; Zakaria, J. Clinical characteristics and predictors of 5-year survival among colorectal cancer patients in a tertiary hospital in Malaysia. J. Gastrointest. Oncol. 2020, 11, 250–259. [Google Scholar] [CrossRef]
- Sammarco, G.; Gallo, G.; Vescio, G.; Picciariello, A.; De Paola, G.; Trompetto, M.; Curro, G.; Ammendola, M. Mast Cells, microRNAs and Others: The Role of Translational Research on Colorectal Cancer in the Forthcoming Era of Precision Medicine. J. Clin. Med. 2020, 9, 2852. [Google Scholar] [CrossRef]
- van den Bulk, J.; Verdegaal, E.M.; de Miranda, N.F. Cancer immunotherapy: Broadening the scope of targetable tumours. Open Biol. 2018, 8, 180037. [Google Scholar] [CrossRef]
- Piawah, S.; Venook, A.P. Targeted therapy for colorectal cancer metastases: A review of current methods of molecularly targeted therapy and the use of tumor biomarkers in the treatment of metastatic colorectal cancer. Cancer 2019, 125, 4139–4147. [Google Scholar] [CrossRef]
- Kishore, C.; Bhadra, P. Current advancements and future perspectives of immunotherapy in colorectal cancer research. Eur. J. Pharmacol. 2021, 893, 173819. [Google Scholar] [CrossRef]
- Mohammad, R.M.; Muqbil, I.; Lowe, L.; Yedjou, C.; Hsu, H.Y.; Lin, L.T.; Siegelin, M.D.; Fimognari, C.; Kumar, N.B.; Dou, Q.P.; et al. Broad targeting of resistance to apoptosis in cancer. Semin. Cancer Biol. 2015, 35, S78–S103. [Google Scholar] [CrossRef]
- Pan, H.; Pan, J.; Li, P.; Gao, J. Characterization of PANoptosis patterns predicts survival and immunotherapy response in gastric cancer. Clin. Immunol. 2022, 238, 109019. [Google Scholar] [CrossRef] [PubMed]
- Etxeberria, I.; Glez-Vaz, J.; Teijeira, Á.; Melero, I. New emerging targets in cancer immunotherapy: CD137/4-1BB costimulatory axis. ESMO Open 2020, 4, e000733. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.M.; Tan, Y.; Wang, H.; Peng, L.; Chen, H.T.; Meng, X.J.; Li, L.L.; Liu, Y.; Li, W.F.; Shan, H. The relationship between autophagy and the immune system and its applications for tumor immunotherapy. Mol. Cancer 2019, 18, 17. [Google Scholar] [CrossRef] [PubMed]
- Tang, R.; Xu, J.; Zhang, B.; Liu, J.; Liang, C.; Hua, J.; Meng, Q.; Yu, X.; Shi, S. Ferroptosis, necroptosis, and pyroptosis in anticancer immunity. J. Hematol. Oncol. 2020, 13, 110. [Google Scholar] [CrossRef]
- Chan, V.S.H.; Lam, T.P.W.; Lam, W.W.M. Nephroptosis: The wandering kidney. Kidney Res. Clin. Pract. 2018, 37, 306–307. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Lee, J.H.; Lee, S.Y.; Hong, M.J.; Choi, J.E.; Park, S.; Jeong, J.Y.; Lee, E.B.; Choi, S.H.; Lee, Y.H.; et al. Expression of key regulatory genes in necroptosis and its effect on the prognosis in non-small cell lung cancer. J. Cancer 2020, 11, 5503–5510. [Google Scholar] [CrossRef]
- Degterev, A.; Hitomi, J.; Germscheid, M.; Ch’en, I.L.; Korkina, O.; Teng, X.; Abbott, D.; Cuny, G.D.; Yuan, C.; Wagner, G.; et al. Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 2008, 4, 313–321. [Google Scholar] [CrossRef]
- Gong, Y.; Fan, Z.; Luo, G.; Yang, C.; Huang, Q.; Fan, K.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. The role of necroptosis in cancer biology and therapy. Mol. Cancer 2019, 18, 100. [Google Scholar] [CrossRef]
- Snyder, A.G.; Hubbard, N.W.; Messmer, M.N.; Kofman, S.B.; Hagan, C.E.; Orozco, S.L.; Chiang, K.; Daniels, B.P.; Baker, D.; Oberst, A. Intratumoral activation of the necroptotic pathway components RIPK1 and RIPK3 potentiates antitumor immunity. Sci. Immunol. 2019, 4, eaaw2004. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, M.; Cao, D.; Yang, C.; Jin, J.; Wu, L.; Hong, X.; Li, W.; Lu, L.; Li, J.; et al. ZBP1-MLKL necroptotic signaling potentiates radiation-induced antitumor immunity via intratumoral STING pathway activation. Sci. Adv. 2021, 7, eabf6290. [Google Scholar] [CrossRef]
- Seifert, L.; Werba, G.; Tiwari, S.; Ly, N.N.G.; Alothman, S.; Alqunaibit, D.; Avanzi, A.; Barilla, R.; Daley, D.; Greco, S.H.; et al. Author Correction: The necrosome promotes pancreatic oncogenesis via CXCL1 and Mincle-induced immune suppression. Nature 2021, 591, E28. [Google Scholar] [CrossRef] [PubMed]
- Jiang, N.; Zhang, X.; Gu, X.; Li, X.; Shang, L. Progress in understanding the role of lncRNA in programmed cell death. Cell Death Discov. 2021, 7, 30. [Google Scholar] [CrossRef] [PubMed]
- Jia, Z.; An, J.; Liu, Z.; Zhang, F. Non-Coding RNAs in Colorectal Cancer: Their Functions and Mechanisms. Front. Oncol. 2022, 12, 783079. [Google Scholar] [CrossRef] [PubMed]
- Tao, H.; Liu, X.; Liu, X.; Liu, W.; Wu, D.; Wang, R.; Lv, G. LncRNA MEG3 inhibits trophoblast invasion and trophoblast-mediated VSMC loss in uterine spiral artery remodeling. Mol. Reprod. Dev. 2019, 86, 686–695. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.R.; Xiang, S.; Song, Z.; Wu, M. The p53-inducible long noncoding RNA TRINGS protects cancer cells from necrosis under glucose starvation. Embo J. 2017, 36, 3483–3500. [Google Scholar] [CrossRef] [PubMed]
- Tran, D.D.H.; Kessler, C.; Niehus, S.E.; Mahnkopf, M.; Koch, A.; Tamura, T. Myc target gene, long intergenic noncoding RNA, Linc00176 in hepatocellular carcinoma regulates cell cycle and cell survival by titrating tumor suppressor microRNAs. Oncogene 2018, 37, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Huang, L.; Chen, W.; Zhang, G.; Li, Y.; Wu, Y.; Xiong, J.; Jie, Z. Comprehensive Analysis of Necroptosis-Related Long Noncoding RNA Immune Infiltration and Prediction of Prognosis in Patients with Colon Cancer. Front. Mol. Biosci. 2022, 9, 811269. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; He, X.; Wang, Y.; Hu, Z.; Yu, D.; Huang, H.; Zhao, S.; Wei, P.; Li, D. A Hypoxia-Related lncRNA Signature Correlates with Survival and Tumor Microenvironment in Colorectal Cancer. J. Immunol. Res. 2022, 2022, 9935705. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Wu, Z.; Lu, Z.; Li, L.; Ma, M.; Long, F.; Wu, R.; Huang, L.; Chou, J.; Yang, K.; Zhang, Y.; et al. Identification and Validation of Ferroptosis-Related LncRNA Signatures as a Novel Prognostic Model for Colon Cancer. Front. Immunol. 2021, 12, 783362. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamarudin, A.N.; Cox, T.; Kolamunnage-Dona, R. Time-dependent ROC curve analysis in medical research: Current methods and applications. BMC Med. Res. Methodol. 2017, 17, 53. [Google Scholar] [CrossRef]
- Wu, T.; Hu, E.; Xu, S.; Chen, M.; Guo, P.; Dai, Z.; Feng, T.; Zhou, L.; Tang, W.; Zhan, L.; et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2021, 2, 100141. [Google Scholar] [CrossRef] [PubMed]
- Hanzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef] [PubMed]
- Newton, K.; Manning, G. Necroptosis and Inflammation. Annu. Rev. Biochem. 2016, 85, 743–763. [Google Scholar] [CrossRef] [PubMed]
- Jiao, H.; Wachsmuth, L.; Kumari, S.; Schwarzer, R.; Lin, J.; Eren, R.O.; Fisher, A.; Lane, R.; Young, G.R.; Kassiotis, G.; et al. Z-nucleic-acid sensing triggers ZBP1-dependent necroptosis and inflammation. Nature 2020, 580, 391–395. [Google Scholar] [CrossRef]
- Zhang, Z.; Ju, F.; Chen, F.; Wu, H.; Chen, J.; Zhong, J.; Shao, L.; Zheng, S.; Wang, L.; Xue, M. GDC-0326 Enhances the Effects of 5-Fu in Colorectal Cancer Cells by Inducing Necroptotic Death. Onco Targets Ther. 2021, 14, 2519–2530. [Google Scholar] [CrossRef]
- Min, W.; Sun, L.; Li, B.; Gao, X.; Zhang, S.; Zhao, Y. lncCRLA Enhanced Chemoresistance in Lung Adenocarcinoma That Underwent EpithelialMesenchymal Transition. Oncol Res 2022, 28, 857–872. [Google Scholar] [CrossRef]
- Li, X.; Chen, M.; Shi, Q.; Zhang, H.; Xu, S. Hydrogen sulfide exposure induces apoptosis and necroptosis through lncRNA3037/miR-15a/BCL2-A20 signaling in broiler trachea. Sci. Total Environ. 2020, 699, 134296. [Google Scholar] [CrossRef]
- Mouasni, S.; Tourneur, L. FADD at the Crossroads between Cancer and Inflammation. Trends. Immunol. 2018, 39, 1036–1053. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.W.; Seo, J.; Jeong, M.; Lee, S.; Song, J. The roles of FADD in extrinsic apoptosis and necroptosis. BMB Rep. 2012, 45, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Irrinki, K.M.; Mallilankaraman, K.; Thapa, R.J.; Chandramoorthy, H.C.; Smith, F.J.; Jog, N.R.; Gandhirajan, R.K.; Kelsen, S.G.; Houser, S.R.; May, M.J.; et al. Requirement of FADD, NEMO, and BAX/BAK for aberrant mitochondrial function in tumor necrosis factor alpha-induced necrosis. Mol. Cell Biol. 2011, 31, 3745–3758. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.W.; Shao, J.; Lin, J.; Zhang, N.; Lu, B.J.; Lin, S.C.; Dong, M.Q.; Han, J. RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 2009, 325, 332–336. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Zhao, Y.; Shi, L.; Li, W.; Chen, K.; Li, M.; Chen, X.; Zhang, H.; Li, T.; Matsuzawa-Ishimoto, Y.; et al. Gut epithelial TSC1/mTOR controls RIPK3-dependent necroptosis in intestinal inflammation and cancer. J. Clin. Investig. 2020, 130, 2111–2128. [Google Scholar] [CrossRef]
- Robinson, N.; Ganesan, R.; Hegedűs, C.; Kovács, K.; Kufer, T.A.; Virág, L. Programmed necrotic cell death of macrophages: Focus on pyroptosis, necroptosis, and parthanatos. Redox Biol. 2019, 26, 101239. [Google Scholar] [CrossRef]
- Rosenbaum, S.R.; Wilski, N.A.; Aplin, A.E. Fueling the Fire: Inflammatory Forms of Cell Death and Implications for Cancer Immunotherapy. Cancer Discov. 2021, 11, 266–281. [Google Scholar] [CrossRef]
- Ostroumov, D.; Fekete-Drimusz, N.; Saborowski, M.; Kühnel, F.; Woller, N. CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell Mol. Life Sci. 2018, 75, 689–713. [Google Scholar] [CrossRef]
- Yeong, J.; Lim, J.C.T.; Lee, B.; Li, H.; Chia, N.; Ong, C.C.H.; Lye, W.K.; Putti, T.C.; Dent, R.; Lim, E.; et al. High Densities of Tumor-Associated Plasma Cells Predict Improved Prognosis in Triple Negative Breast Cancer. Front. Immunol. 2018, 9, 1209. [Google Scholar] [CrossRef]
- Aponte-López, A.; Muñoz-Cruz, S. Mast Cells in the Tumor Microenvironment. Adv. Exp. Med. Biol. 2020, 1273, 159–173. [Google Scholar] [CrossRef]
- Xia, Y.; Rao, L.; Yao, H.; Wang, Z.; Ning, P.; Chen, X. Engineering Macrophages for Cancer Immunotherapy and Drug Delivery. Adv. Mater. 2020, 32, e2002054. [Google Scholar] [CrossRef] [PubMed]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade-based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhang, W.; Lin, M.; Wu, W.; Jiang, P.; Tou, E.; Xue, M.; Richards, A.; Jourd’heuil, D.; Asif, A.; et al. MYOSLID Is a Novel Serum Response Factor-Dependent Long Noncoding RNA That Amplifies the Vascular Smooth Muscle Differentiation Program. Arter. Thromb. Vasc. Biol. 2016, 36, 2088–2099. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Wu, N.; Jiang, M.; Chu, Y.; Wang, Z.; Liu, H.; Cao, J.; Liu, H.; Xu, B.; Xie, X. Long non-coding RNA MYOSLID functions as a competing endogenous RNA to regulate MCL-1 expression by sponging miR-29c-3p in gastric cancer. Cell Prolif. 2019, 52, e12678. [Google Scholar] [CrossRef]
- Yang, S.; Chen, M.; Lin, C. A Novel lncRNA MYOSLID/miR-1286/RAB13 Axis Plays a Critical Role in Osteosarcoma Progression. Cancer Manag. Res. 2019, 11, 10345–10351. [Google Scholar] [CrossRef]
- Xiong, H.G.; Li, H.; Xiao, Y.; Yang, Q.C.; Yang, L.L.; Chen, L.; Bu, L.L.; Zhang, W.F.; Zhang, J.L.; Sun, Z.J. Long noncoding RNA MYOSLID promotes invasion and metastasis by modulating the partial epithelial-mesenchymal transition program in head and neck squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 278. [Google Scholar] [CrossRef]




| Characteristics | Training Group | Testing Group | p Value | ||
|---|---|---|---|---|---|
| No. | % | No. | % | ||
| Age | − | − | − | ||
| ≤65 | 89 | 87 | >0.05 | ||
| >65 | 123 | 125 | − | ||
| Sex | − | − | − | ||
| Female | 98 | 98 | >0.05 | ||
| Male | 114 | 114 | − | ||
| Stage | − | − | − | ||
| I | 41 | 32 | >0.05 | ||
| II | 84 | 81 | − | ||
| III | 54 | 63 | − | ||
| IV | 27 | 31 | − | ||
| Pathology T stage | − | − | − | ||
| pT1 | 4 | 6 | >0.05 | ||
| pT2 | 42 | 32 | − | ||
| pT3 | 139 | 151 | − | ||
| pT4 | 27 | 22 | − | ||
| Pathology N stage | − | − | − | ||
| pN0 | 129 | 124 | >0.05 | ||
| pN1 | 49 | 48 | − | ||
| pN2 | 34 | 40 | − | ||
| Pathology M stage | − | − | − | ||
| pM0 | 158 | 157 | >0.05 | ||
| pM1 | 27 | 31 | — |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Z.; Zhang, F.; Wang, Y.; Lu, Z.; Lin, C. Identification and Validation of the lncRNA MYOSLID as a Regulating Factor of Necroptosis and Immune Cell Infiltration in Colorectal Cancer following Necroptosis-Related LncRNA Model Establishment. Cancers 2022, 14, 4364. https://doi.org/10.3390/cancers14184364
Wu Z, Zhang F, Wang Y, Lu Z, Lin C. Identification and Validation of the lncRNA MYOSLID as a Regulating Factor of Necroptosis and Immune Cell Infiltration in Colorectal Cancer following Necroptosis-Related LncRNA Model Establishment. Cancers. 2022; 14(18):4364. https://doi.org/10.3390/cancers14184364
Chicago/Turabian StyleWu, Zhiwei, Fan Zhang, Yaohui Wang, Zhixing Lu, and Changwei Lin. 2022. "Identification and Validation of the lncRNA MYOSLID as a Regulating Factor of Necroptosis and Immune Cell Infiltration in Colorectal Cancer following Necroptosis-Related LncRNA Model Establishment" Cancers 14, no. 18: 4364. https://doi.org/10.3390/cancers14184364
APA StyleWu, Z., Zhang, F., Wang, Y., Lu, Z., & Lin, C. (2022). Identification and Validation of the lncRNA MYOSLID as a Regulating Factor of Necroptosis and Immune Cell Infiltration in Colorectal Cancer following Necroptosis-Related LncRNA Model Establishment. Cancers, 14(18), 4364. https://doi.org/10.3390/cancers14184364






