A microRNA Prognostic Signature in Patients with Diffuse Intrinsic Pontine Gliomas through Non-Invasive Liquid Biopsy
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Standard Protocol Approvals and Patient Consents
2.2. Clinical Endpoints
2.3. Statistical Analysis
3. Results
3.1. Study Sample: Recruitment and Clinical Characteristics
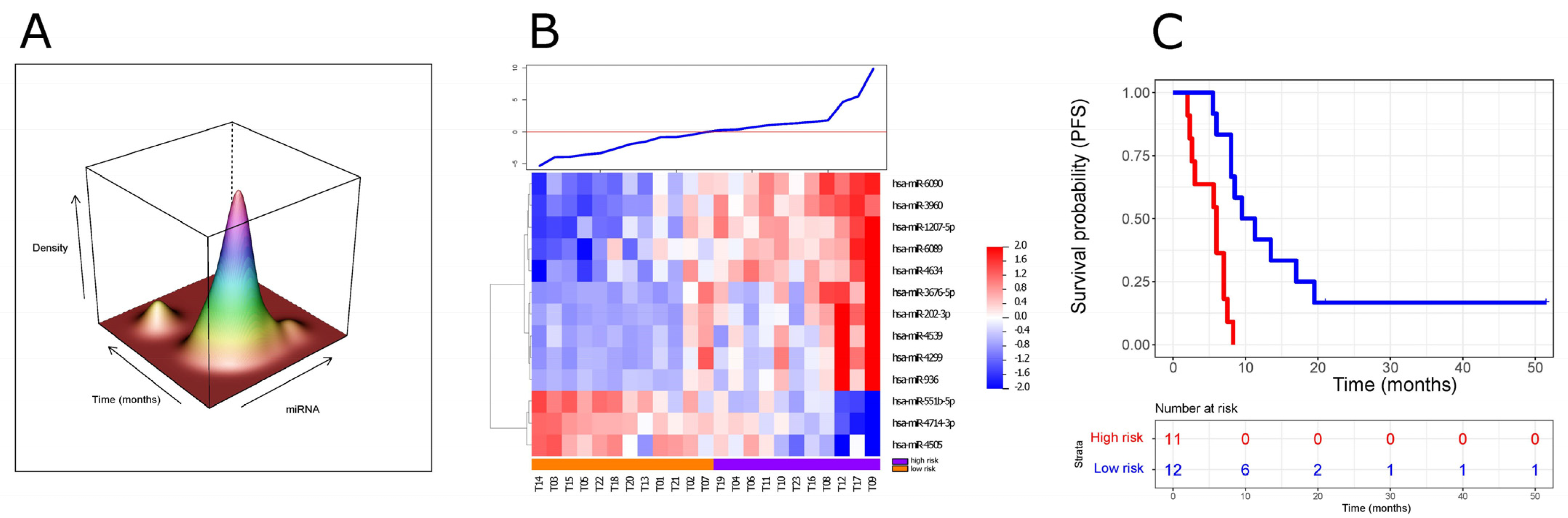
3.2. Development of a ct-miRNA Signature
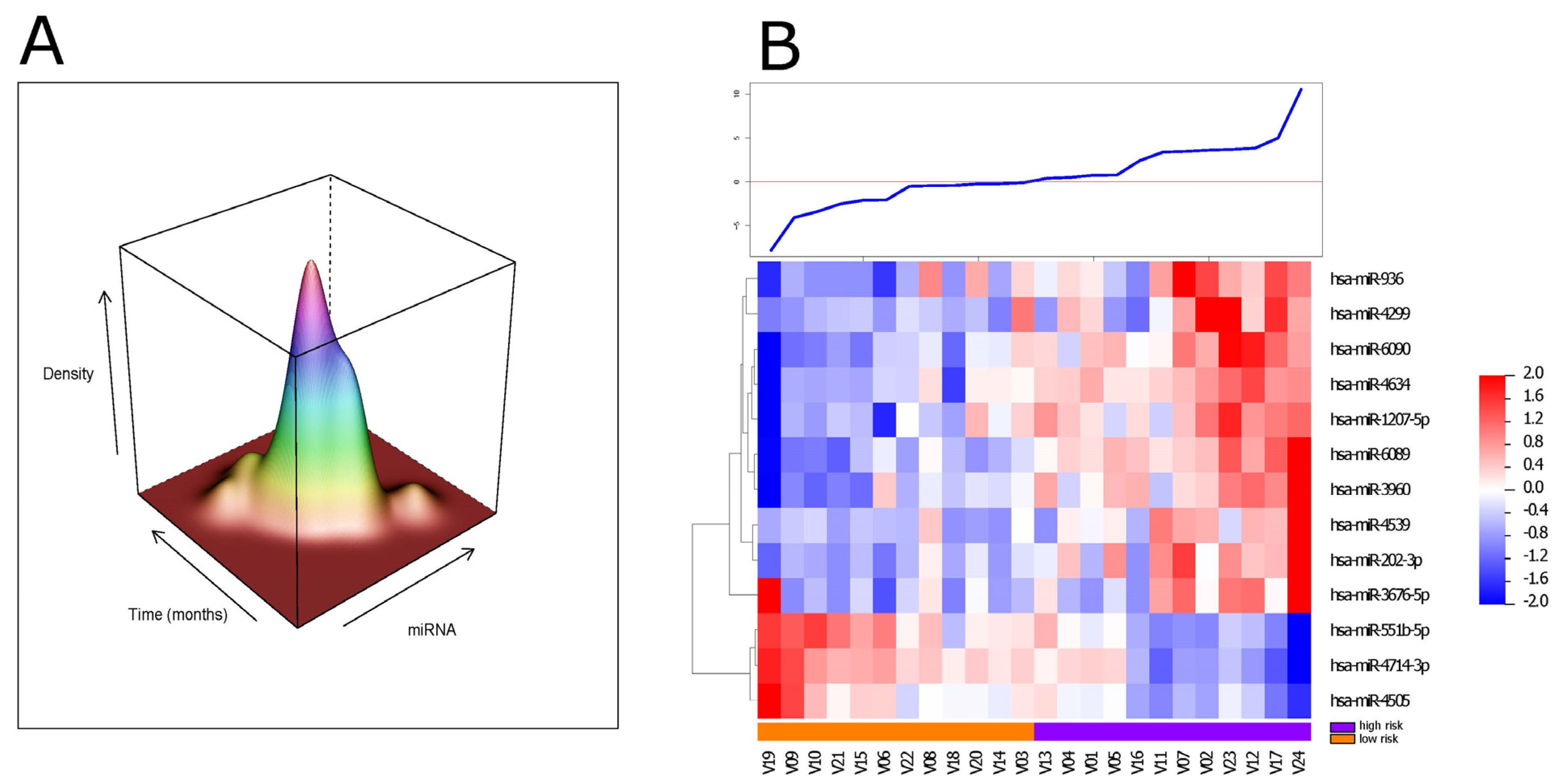
3.3. Independent Validation of Our ct-miRNA Signature
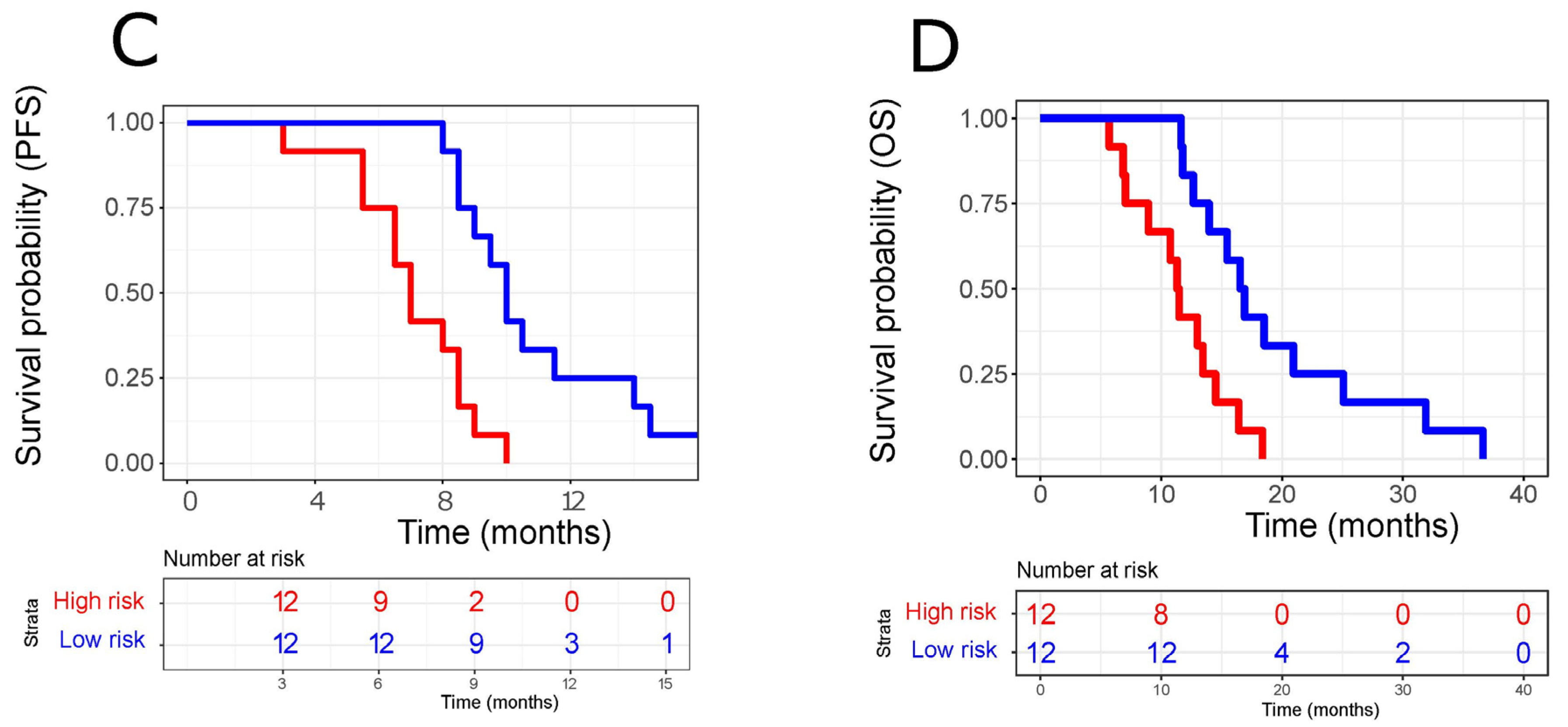
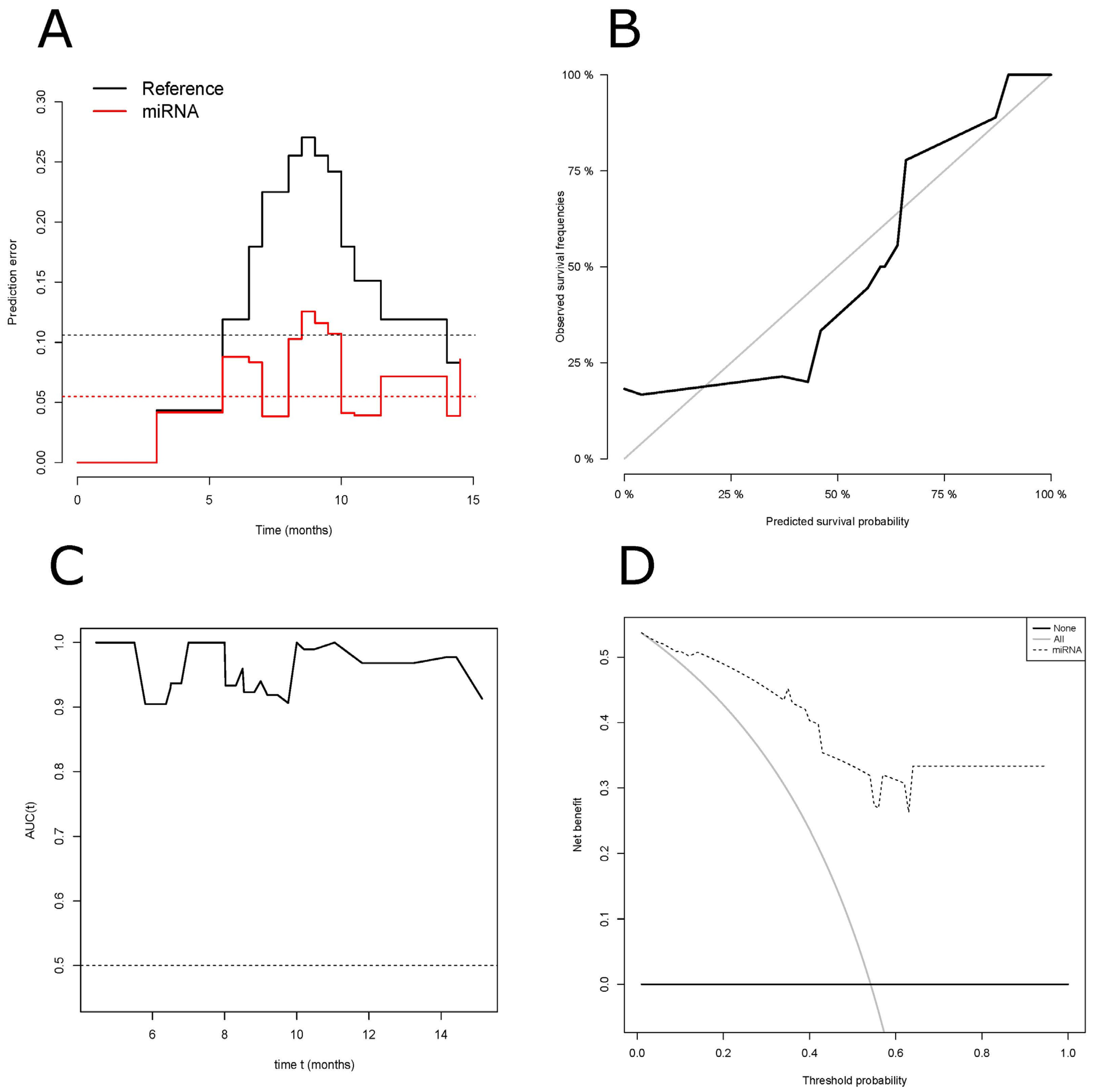
3.4. Performance of Our ct-miRNA Signature
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- DeWitt, J.C.; Mock, A.; Louis, D.N. The 2016 WHO classification of central nervous system tumors: What neurologists need to know. Curr. Opin. Neurol. 2017, 30, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.K.; Rutka, J.T. Diffuse intrinsic pontine glioma: Clinical features, molecular genetics, and novel targeted therapeutics. J. Korean Neurosurg. Soc. 2018, 61, 343–351. [Google Scholar] [CrossRef] [PubMed]
- Beuriat, P.A.; Szathmari, A.; Di Rocco, F.; Kanold, J.; Mottolese, C.; Frappaz, D. Diffuse intrinsic pontine glioma in children: Document or treat? World Neurosurg. 2016, 93, 485.e11–485.e14. [Google Scholar]
- Minh Thong, P.; Minh Duc, N. The Role of Apparent Diffusion Coefficient in the Differentiation between Cerebellar Medulloblastoma and Brainstem Glioma. Neurol. Int. 2020, 12, 34–40. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, T.J. Diffuse intrinsic pontine glioma (DIPG): Time to biopsy again? Pediatr. Blood Cancer. 2012, 58, 487–488. [Google Scholar] [CrossRef] [PubMed]
- Kieran, M.W. Time to rethink the unthinkable: Upfront biopsy of children with newly diagnosed diffuse intrinsic pontine glioma (DIPG). Pediatr. Blood Cancer. 2015, 62, 3–4. [Google Scholar] [CrossRef]
- Vitanza, N.A.; Monje, M. Diffuse intrinsic pontine glioma: From diagnosis to next-generation clinical trials. Curr. Treat Options Neurol. 2019, 21, 37. [Google Scholar] [CrossRef]
- Rechberger, J.S.; Lu, V.M.; Zhang, L.; Power, E.A.; Daniels, D.J. Clinical trials for diffuse intrinsic pontine glioma: The current state of affairs. Childs Nerv. Syst. 2020, 36, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Aquino-Parsons, C.; Hukin, J.; Green, A. Concurrent carbogen and radiation therapy in children with high-risk brainstem gliomas. Pediatr. Blood Cancer. 2008, 50, 397–399. [Google Scholar] [CrossRef]
- Bradley, K.A.; Zhou, T.; McNall-Knapp, R.Y.; Jakacki, R.I.; Levy, A.S.; Vezina, G.; Pollack, I.F. Motexafin-gadolinium and involved field radiation therapy for intrinsic pontine glioma of childhood: A Children’s Oncology Group phase 2 study. Int. J. Radiat. Oncol. Biol. Phys. 2013, 85, e55–e60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashed, W.M.; Maher, E.; Adel, M.; Saber, O.; Zaghloul, M.S. Pediatric diffuse intrinsic pontine glioma: Where do we stand? Cancer Metastasis Rev. 2019, 38, 759–770. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, L.M.; Veldhuijzen van Zanten, S.E.M.; Colditz, N.; Baugh, J.; Chaney, B.; Hoffmann, M.; Lane, A.; Fuller, C.; Miles, L.; Hawkins, C.; et al. Clinical, radiologic, pathologic, and molecular characteristics of long-term survivors of diffuse intrinsic pontine glioma (DIPG): A collaborative report from the International and European Society for Pediatric Oncology DIPG Registries. J. Clin. Oncol. 2018, 36, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Hargrave, D.; Bartels, U.; Bouffet, E. Diffuse brainstem glioma in children: Critical review of clinical trials. Lancet Oncol. 2006, 7, 241–248. [Google Scholar] [CrossRef]
- Jansen, M.H.; van Vuurden, D.G.; Vandertop, W.P.; Kaspers, G.J. Diffuse intrinsic pontine gliomas: A systematic update on clinical trials and biology. Cancer Treat Rev. 2012, 38, 27–35. [Google Scholar] [CrossRef]
- Lu, V.M.; Power, E.A.; Zhang, L.; Daniels, D.J. Liquid biopsy for diffuse intrinsic pontine glioma: An update. J. Neurosurg. Pediatr. 2019, 1–8. [Google Scholar] [CrossRef]
- Massimino, M.; Biassoni, V.; Miceli, R.; Schiavello, E.; Warmuth-Metz, M.; Modena, P.; Casanova, M.; Pecori, E.; Giangaspero, F.; Antonelli, M.; et al. Results of nimotuzumab and vinorelbine, radiation and re-irradiation for diffuse pontine glioma in childhood. J. Neurooncol. 2014, 118, 305–312. [Google Scholar] [CrossRef]
- Cooney, T.M.; Cohen, K.J.; Guimaraes, C.V.; Dhall, G.; Leach, J.; Massimino, M.; Erbetta, A.; Chiapparini, L.; Malbari, F.; Kramer, K.; et al. Response assessment in diffuse intrinsic pontine glioma: Recommendations from the Response Assessment in Pediatric Neuro-Oncology (RAPNO) working group. Lancet Oncol. 2020, 21, e330–e336. [Google Scholar] [CrossRef]
- Bair, E.; Tibshirani, R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004, 2, E108. [Google Scholar] [CrossRef]
- Bounajem, M.T.; Karsy, M.; Jensen, R.L. Liquid biopsies for the diagnosis and surveillance of primary pediatric central nervous system tumors: A review for practicing neurosurgeons. Neurosurg. Focus. 2020, 48, E8. [Google Scholar] [CrossRef] [PubMed]
- Panditharatna, E.; Kilburn, L.B.; Aboian, M.S.; Kambhampati, M.; Gordish-Dressman, H.; Magge, S.N.; Gupta, N.; Myseros, J.S.; Hwang, E.I.; Kline, C.; et al. Clinically Relevant and Minimally Invasive Tumor Surveillance of Pediatric Diffuse Midline Gliomas Using Patient-Derived Liquid Biopsy. Clin. Cancer Res. 2018, 24, 5850–5859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Toden, S.; Goel, A. Non-coding RNAs as liquid biopsy biomarkers in cancer. Br. J. Cancer. 2022, 126, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Freese, C.; Takiar, V.; Fouladi, M.; DeWire, M.; Breneman, J.; Pater, L. Radiation and subsequent reirradiation outcomes in the treatment of diffuse intrinsic pontine glioma and a systematic review of the reirradiation literature. Pract. Radiat. Oncol. 2017, 7, 86–92. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.M.; Wang, S.; Wen, X.; Han, X.R.; Wang, Y.J.; Fan, S.H.; Zhang, Z.F.; Shan, Q.; Lu, J.; Zheng, Y.L. MicroRNA-1275 promotes proliferation, invasion and migration of glioma cells via SERPINE1. J. Cell Mol. Med. 2018, 22, 4963–4974. [Google Scholar] [CrossRef]
- Wang, D.; Zhi, T.; Xu, X.; Bao, Z.; Fan, L.; Li, Z.; Ji, J.; Liu, N. MicroRNA-936 induces cell cycle arrest and inhibits glioma cell proliferation by targeting CKS1. Am. J. Cancer. Res. 2017, 7, 2131–2143. [Google Scholar] [PubMed]
- Tűzesi, Á.; Kling, T.; Wenger, A.; Lunavat, T.R.; Jang, S.C.; Rydenhag, B.; Lötvall, J.; Pollard, S.M.; Danielsson, A.; Carén, H. Pediatric brain tumor cells release exosomes with a miRNA repertoire that differs from exosomes secreted by normal cells. Oncotarget 2017, 8, 90164–90175. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.D.; Sun, X.Y.; Niu, W.; Kong, L.M.; He, M.J.; Fan, H.M.; Li, W.S.; Zhong, A.F.; Zhang, L.Y.; Lu, J. Correlation between the level of microRNA expression in peripheral blood mononuclear cells and symptomatology in patients with generalized anxiety disorder. Compr. Psychiatry. 2016, 69, 216–224. [Google Scholar] [CrossRef]
- Keller, A.; Leidinger, P.; Steinmeyer, F.; Stähler, C.; Franke, A.; Hemmrich-Stanisak, G.; Kappel, A.; Wright, I.; Dörr, J.; Paul, F.; et al. Comprehensive analysis of microRNA profiles in multiple sclerosis including next-generation sequencing. Mult. Scler. 2014, 20, 295–303. [Google Scholar] [CrossRef]
- Wu, S.; Gu, Y.; Huang, Y.; Wong, T.C.; Ding, H.; Liu, T.; Zhang, Y.; Zhang, X. Novel biomarkers for non-functioning invasive pituitary adenomas were identified by using analysis of microRNAs expression profile. Biochem. Genet. 2017, 55, 253–267. [Google Scholar] [CrossRef]
- Stylli, S.S.; Adamides, A.A.; Koldej, R.M.; Luwor, R.B.; Ritchie, D.S.; Ziogas, J.; Kaye, A.H. miRNA expression profiling of cerebrospinal fluid in patients with aneurysmal subarachnoid hemorrhage. J. Neurosurg. 2017, 126, 1131–1139. [Google Scholar] [CrossRef]
- Moskwa, P.; Zinn, P.O.; Choi, Y.E.; Shukla, S.A.; Fendler, W.; Chen, C.C.; Lu, J.; Golub, T.R.; Hjelmeland, A.; Chowdhury, D.A. A functional screen identifies miRs that induce radioresistance in glioblastomas. Mol. Cancer. Res. 2014, 12, 1767–1778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Zhang, G.; Huang, H.; Li, H.; Lin, S.; Wang, Y. Differentially expressed microRNAs in radioresistant and radiosensitive atypical meningioma: A clinical study in Chinese patients. Front. Oncol. 2020, 10, 501. [Google Scholar] [CrossRef] [PubMed]
- Teplyuk, N.M.; Mollenhauer, B.; Gabriely, G.; Giese, A.; Kim, E.; Smolsky, M.; Kim, R.Y.; Saria, M.G.; Pastorino, S.; Kesari, S.; et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro. Oncol. 2012, 14, 689–700. [Google Scholar]
- Micheel, C.M.; Nass, S.J.; Omenn, G.S. Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials; Board on Health Care Services; Board on Health Sciences Policy; Institute of Medicine. In Evolution of Translational OMICS: Lessons Learned and the Path Forward; The National Academies Press: Washington, DC, USA, 2012. [Google Scholar]
- Altman, D.G.; McShane, L.M.; Sauerbrei, W.; Taube, S.E. Reporting Recommendations for Tumor Marker Prognostic Studies (REMARK): Explanation and elaboration. PLoS Med. 2012, 9, e1001216. [Google Scholar]
- Huang, Y.; Liu, Z.; Zhong, L.; Wen, Y.; Ye, Q.; Cao, D.; Li, P.; Liu, Y. Construction of an 11-microRNA-based signature and a prognostic nomogram to predict the overall survival of head and neck squamous cell carcinoma patients. BMC Genomics. 2020, 21, 691. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Li, Y.C.; Geng, C.Y.; Wang, H.J.; Chen, W.M. Potential Relationship between Clinical Significance and Serum Exosomal miRNAs in Patients with Multiple Myeloma. Biomed. Res. Int. 2019, 2019, 1575468. [Google Scholar] [CrossRef]
- Kim, S.; Bae, W.J.; Ahn, J.M.; Heo, J.H.; Kim, K.M.; Choi, K.W.; Sung, C.O.; Lee, D. MicroRNA signatures associated with lymph node metastasis in intramucosal gastric cancer. Mod. Pathol. 2021, 34, 672–683. [Google Scholar] [CrossRef]
- Wang, Y.N.; Chen, Z.H.; Chen, W.C. Novel circulating microRNAs expression profile in colon cancer: A pilot study. Eur. J. Med. Res. 2017, 22, 51. [Google Scholar] [CrossRef]
- Jiang, X.; Jiang, M.; Xu, M.; Xu, J.; Li, Y. Identification of diagnostic utility and molecular mechanisms of circulating miR-551b-5p in gastric cancer. Pathol. Res. Pract. 2019, 215, 900–904. [Google Scholar] [CrossRef]
- Liu, L.; Ning, Y.; Yi, J.; Yuan, J.; Fang, W.; Lin, Z.; Zeng, Z. miR-6089/MYH9/β-catenin/c-Jun negative feedback loop inhibits ovarian cancer carcinogenesis and progression. Biomed. Pharmacother. 2020, 125, 109865. [Google Scholar] [CrossRef]
- Usuba, W.; Urabe, F.; Yamamoto, Y.; Matsuzaki, J.; Sasaki, H.; Ichikawa, M.; Takizawa, S.; Aoki, Y.; Niida, S.; Kato, K.; et al. Circulating miRNA panels for specific and early detection in bladder cancer. Cancer Sci. 2019, 110, 408–419. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kong, J.; Nie, Z.; Chen, D.; Qiang, J.; Gao, W.; Chen, X. Circular RNA Hsa_circ_0066755 as an Oncogene via sponging miR-651 and as a Promising Diagnostic Biomarker for Nasopharyngeal Carcinoma. Int. J. Med. Sci. 2020, 17, 1499–1507. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Lü, M.H.; Zhang, D.; Hao, N.B.; Fan, Y.H.; Wu, Y.Y.; Wang, S.M.; Xie, R.; Fang, D.C.; Zhang, H.; et al. miR-1207-5p and miR-1266 suppress gastric cancer growth and invasion by targeting telomerase reverse transcriptase. Cell Death Dis. 2014, 5, e1034. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Fan, B.; Zhao, Y.; Fang, J. MicroRNA-202 inhibits cell proliferation, migration and invasion of glioma by directly targeting metadherin. Oncol. Rep. 2017, 38, 1670–1678. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.; Li, H.; Lv, Q.; Wu, K.; Zhang, W.; Zhang, J.; Zhu, D.; Liu, Q.; Zhang, W. miR-202 inhibits the progression of human cervical cancer through inhibition of cyclin D1. Oncotarget 2016, 7, 72067–72075. [Google Scholar] [CrossRef]
- Qin, X.; Yu, S.; Zhou, L.; Shi, M.; Hu, Y.; Xu, X.; Shen, B.; Liu, S.; Yan, D.; Feng, J. Cisplatin-resistant lung cancer cell-derived exosomes increase cisplatin resistance of recipient cells in exosomal miR-100-5p-dependent manner. Int. J. Nanomedicine. 2017, 12, 3721–3733. [Google Scholar] [CrossRef]
- Liu, S.; Zang, H.; Zheng, H.; Wang, W.; Wen, Q.; Zhan, Y.; Yang, Y.; Ning, Y.; Wang, H.; Fan, S. miR-4634 augments the anti-tumor effects of RAD001 and associates well with clinical prognosis of non-small cell lung cancer. Sci. Rep. 2020, 10, 13079. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, C.D.; Ma, M.H.; Dai, D.Q. Three-microRNA signature identified by bioinformatics analysis predicts prognosis of gastric cancer patients. World J. Gastroenterol. 2018, 24, 1206–1215. [Google Scholar] [CrossRef]
- Yang, W.B.; Zhang, W.P.; Shi, J.L.; Wang, J.W. MiR-4299 suppresses non-small cell lung cancer cell proliferation, migration and invasion through modulating PTEN/AKT/PI3K pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 3408–3414. [Google Scholar] [CrossRef]
- Abu Zohair, L.M. Prediction of Student’s performance by modelling small dataset size. Int. J. Educ. Technol. High. Educ. 2019, 16. [Google Scholar] [CrossRef]
- Appierto, V.; Callari, M.; Cavadini, E.; Morelli, D.; Daidone, M.G.; Tiberio, P. A lipemia-independent NanoDrop®-based score to identify hemolysis in plasma and serum samples. Bioanalysis 2014, 6, 1215–1226. [Google Scholar] [CrossRef] [PubMed]
- Blanche, P.; Dartigues, J.-F.; Jacqmin-Gadda, H. Estimating and comparing time-dependent areas under receiver operating characteristic curves for censored event times with competing risks. Stat. Med. 2013, 32, 5381–5397. [Google Scholar] [CrossRef]
- Brier, G.W. Verification of forecasts expressed in terms of probability. Mon. Wea. Rev. 1950, 78, 1–3. [Google Scholar] [CrossRef]
- Kirschner, M.B.; Edelman, J.B.; Kao, S.C.-H.; Vallely, M.P.; Van Zandwijk, N.; Reid, G. The Impact of Hemolysis on Cell-Free microRNA Biomarkers. Front. Genet. 2013, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Kohn, M.A.; Senyak, J. Sample Size Calculators. UCSF CTSI. 20 December 2021. Available online: https://www.sample-size.net/ (accessed on 7 January 2022).
- Gerds, T.A.; Schumacher, M. Consistent Estimation of the Expected Brier Score in General Survival Models with Right-Censored Event Times. Biom. J. 2006, 48, 1029–1040. [Google Scholar] [CrossRef]
- Gerds, T.A.; Schumacher, M. Efron-Type Measures of Prediction Error for Survival Analysis. Biometrics 2007, 63, 1283–1287. [Google Scholar] [CrossRef]
- Heagerty, P.J.; Lumley, T.; Pepe, M.S. Time-Dependent ROC Curves for Censored Survival Data and a Diagnostic Marker. Biometrics 2000, 56, 337–344. [Google Scholar] [CrossRef]
- Irizarry, R.A.; Hobbs, B.; Collin, F.; Beazer-Barclay, Y.D.; Antonellis, K.J.; Scherf, U.; Speed, T. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 2003, 4, 249–264. [Google Scholar] [CrossRef]
- Johnson, W.; Li, C.; Rabinovic, A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics 2006, 8, 118–127. [Google Scholar] [CrossRef]
- López-Romero, P. Pre-processing and differential expression analysis of Agilent microRNA arrays using the AgiMicroRna Bioconductor library. BMC Genom. 2011, 12, 64. [Google Scholar] [CrossRef]
- Martínez-Camblor, P.; Pardo-Fernandez, J.C. Smooth time-dependent receiver operating characteristic curve estimators. Stat. Methods Med Res. 2017, 27, 651–674. [Google Scholar] [CrossRef] [PubMed]
- Mogensen, U.B.; Ishwaran, H.; Gerds, T.A. Evaluating Random Forests for Survival Analysis Using Prediction Error Curves. J. Stat. Softw. 2012, 50, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Oller, R.; Langohr, K. FHtest: An R Package for the Comparison of Survival Curves with Censored Data. J. Stat. Softw. 2017, 81, 1–25. [Google Scholar] [CrossRef] [Green Version]
- Porzelius, C.; Binder, H.; Schumacher, M. Parallelized prediction error estimation for evaluation of high-dimensional models. Bioinformatics 2009, 25, 827–829. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Lam, A.; Li, M.-C.; Ngan, M.; Menenzes, S.; Zhao, Y. Analysis of Gene Expression Data Using BRB-Array Tools. Cancer Informatics 2007, 3, 11–17. [Google Scholar] [CrossRef]
- Therneau, T.M. A Package for Survival Analysis in R. R Package Version 2.38. 2015. Available online: https://CRAN.R-project.org/package=survival (accessed on 1 March 2022).
- Vickers, A.J.; Elkin, E.B. Decision Curve Analysis: A Novel Method for Evaluating Prediction Models. Med Decis. Mak. 2006, 26, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Wessa, P. Free Statistics Software, Office for Research Development and Education, Version 1.2.1. 2021. Available online: https://www.wessa.net/ (accessed on 1 March 2022).
| Training Set (n = 23) | Validation Set (n = 24) | p-Value | ||
|---|---|---|---|---|
| Age (median, range) | 6.68 (2–17 y) | 7.07 (2–21 y) | 0.512 Ŧ | |
| Sex | Male | 10 | 12 | 0.654 ŦŦ |
| Female | 13 | 12 | ||
| Hydrocephalus | Yes | 4 | 8 | 0.21 ŦŦ |
| No | 19 | 16 | ||
| Pattern of progression | Local | 14 | 20 | 0.145 ŦŦ |
| Disseminated | 7 | 4 | ||
| No progression | 2 | 0 |
| Univariate Analysis (PFS) | Multivariate Analysis (PFS) | |||
|---|---|---|---|---|
| PFS | HR (95% CI) | p-value | HR (95% CI) | p-value |
| Hydrocephalus (presence vs. absence) | 0.807 (0.33–1.971) | 0.638 | 1.481 (0.517–4.246) | 0.465 |
| Age | 0.9926 (0.92–1.07) | 0.849 | 1.009 (0.935–1.09) | 0.825 |
| ct-miRNA (high vs. low risk) | 5.506 (2.034–14.9) | 0.000786 | 6.525 (2.129–20.0) | 0.00103 |
| Univariate analysis (OS) | Multivariate analysis (OS) | |||
| OS | HR (95% CI) | p-value | HR (95% CI) | p-value |
| Hydrocephalus (presence vs. absence) | 1.936 (0.787–4.759) | 0.15 | 2.8751 (1.111–7.44) | 0.0295 |
| Age | 0.998 (0.925–1.076) | 0.961 | 0.994 (0.922–1.072) | 0.8846 |
| ct-miRNA (high vs. low risk) | 4.119 (1.57–10.81) | 0.0042 | 5.351 (1.939–14.771) | 0.0012 |
| Gene Id | Weights (wi) | Circulating miRNA in Liquid Biopsy | Involment in Neurological Diseases | Suggested/Documented Functional Role in Neurological Disease | References | Reported in Other Tumors | Suggested/Documented Functional Role in Tumor Other Than Brain | References |
|---|---|---|---|---|---|---|---|---|
| hsa-miR-4714-3p | −0.889482 | Reported | blood from patients with multiple sclerosis | not investigated | Keller, 2014 [28] | Head-Neck squamous cell carcinoma | not investigated | Huang Y, 2020 [36] |
| hsa-miR-6090 | 0.401593 | Reported | cerebrospinal fluid from patients with intracerebral haemorrhage | pathological condition of brain | Stylli, 2017 [30] | downregulatd in Multiple Myeloma patients | not investigated | Zhang, 2019 [37] |
| hsa-miR-4505 | −0.402474 | Reported | nervous system | nervous system development, nerve growth factor receptor signaling | Chen, 2016 [27] | Myeloma Patients | downregulation is associated with progression of disease | Zhang, 2019 [37] |
| cerebrospinal fluid from patients with intracerebral haemorrhage | pathological condition of brain | Stylli, 2017 [30] | metastatic-intramucosal carcinoma patients | not investigated | Kim S, 2020 [38] | |||
| patients with generalized anxiety disorder | not investigated | Wu, 2018 [24] | upregulated in colon cancer pantients | not investigated | Wang, 2017 [39] | |||
| hsa-miR-551b-5p | −0.850107 | Reported | glioblastoma tissue | not investigated | Wu, 2018 [24] | downregulated in Gastric Cancer patients | regulation of ubiquitin-dependent protein catabolic process, cell division, and mRNA stability | Jiang X, 2019 [40] |
| hsa-miR-6089 | 0.54622 | Reported | cerebrospinal fluid from patients with intracerebral haemorrhage | pathological condition of brain | Stylli, 2017 [30] | Ovarian Cancer | promotes cancer cell proliferation, migration, invasion and metastasis | Liu L, 2020 [41] |
| hsa-miR-3960 | 0.431525 | Reported | cerebrospinal fluid from patients with intracerebral haemorrhage | pathological condition of brain | Stylli, 2017 [30] | downregulated in Bladder Cancer patients | not investigated | Usuba, 2018 [42] |
| hsa-miR-936 | 0.170501 | Not reported | glioma tissue | downregulation is associated to worse overall survival | Wang, 2017 [25] | nasopharyngeal carcinoma | not investigated | Wang 2020 [43] |
| hsa-miR-1207-5p | 0.466562 | Reported | cerebrospinal fluid from patients with intracerebral haemorrhage | pathological condition of brain | Stylli, 2017 [30] | gastric cancer tissues | downregulation promote proliferation, invasion and induces cell cycle arrest in gastric cancer cells in vitro and in vivo | Chen L, 2014 [44] |
| hsa-miR-202-3p | 0.345363 | Reported | glioma tissue | involvement in cell proliferation, migration and proliferation | Yang, 2017 [45] | differentially expressed in cervial cancer | not investigated | Yi, 2016 [46] |
| hsa-miR-3676-5p | 0.151234 | Reported | pituitary adenoma | regulation of tumor suppressor genes involved in invasion | Wu S, 2017 [29] | lung cancer | not investigated | Qin, 2017 [47] |
| hsa-miR-4634 | 0.46722 | Reported | cerebrospinal fluid from patients with intracerebral haemorrhage | pathological condition of brain | Stylli, 2017 [30] | non-small cell lung cancer cells | overexpression is associated with better prognosis of NSCLC patients. | Liu S, 2020 [48] |
| hsa-miR-4539 | 0.066802 | Reported | atypical meningioma | downregulation is associated to radioresistance | Zhang, 2020 [32] | gastric cancer patients | not investigated | Zhang C, 2018 [49] |
| hsa-miR-4299 | 0.069597 | Reported | pediatric glioma stem cells exosomes | influence of tumor microenvironment/normal neural stem cells | Tuzesi, 2017 [26] | non-small cell lung cancer cells | overexpression inhibits the proliferation, migration and invasion in vitro | Yang, 2018 [50] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iannó, M.F.; Biassoni, V.; Schiavello, E.; Carenzo, A.; Boschetti, L.; Gandola, L.; Diletto, B.; Marchesi, E.; Vegetti, C.; Molla, A.; et al. A microRNA Prognostic Signature in Patients with Diffuse Intrinsic Pontine Gliomas through Non-Invasive Liquid Biopsy. Cancers 2022, 14, 4307. https://doi.org/10.3390/cancers14174307
Iannó MF, Biassoni V, Schiavello E, Carenzo A, Boschetti L, Gandola L, Diletto B, Marchesi E, Vegetti C, Molla A, et al. A microRNA Prognostic Signature in Patients with Diffuse Intrinsic Pontine Gliomas through Non-Invasive Liquid Biopsy. Cancers. 2022; 14(17):4307. https://doi.org/10.3390/cancers14174307
Chicago/Turabian StyleIannó, Maria F., Veronica Biassoni, Elisabetta Schiavello, Andrea Carenzo, Luna Boschetti, Lorenza Gandola, Barbara Diletto, Edoardo Marchesi, Claudia Vegetti, Alessandra Molla, and et al. 2022. "A microRNA Prognostic Signature in Patients with Diffuse Intrinsic Pontine Gliomas through Non-Invasive Liquid Biopsy" Cancers 14, no. 17: 4307. https://doi.org/10.3390/cancers14174307
APA StyleIannó, M. F., Biassoni, V., Schiavello, E., Carenzo, A., Boschetti, L., Gandola, L., Diletto, B., Marchesi, E., Vegetti, C., Molla, A., Kramm, C. M., van Vuurden, D. G., Gasparini, P., Gianno, F., Giangaspero, F., Modena, P., Bison, B., Anichini, A., Vennarini, S., ... De Cecco, L. (2022). A microRNA Prognostic Signature in Patients with Diffuse Intrinsic Pontine Gliomas through Non-Invasive Liquid Biopsy. Cancers, 14(17), 4307. https://doi.org/10.3390/cancers14174307







