Molecular Analysis of Short- versus Long-Term Survivors of High-Grade Serous Ovarian Carcinoma
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients
2.2. Cell Lines and Culture
2.3. Molecular Analyses
2.3.1. DNA Sequencing
2.3.2. RNA Sequencing
2.3.3. Estimates of Immune Contextures
2.3.4. Reverse-Phase Protein Array (RPPA)
2.4. Plasmid Constructs and Delivery
2.5. Transient Overexpression
2.6. cDNA Synthesis and Quantitative Real-Time Reverse-Transcriptase Polymerase Chain Reaction
2.7. Apoptosis, Proliferation, and Cell Cycle Assay
2.8. Immunohistochemistry Analysis
2.9. In Vivo Experiments
2.10. Statistical Analyses
3. Results
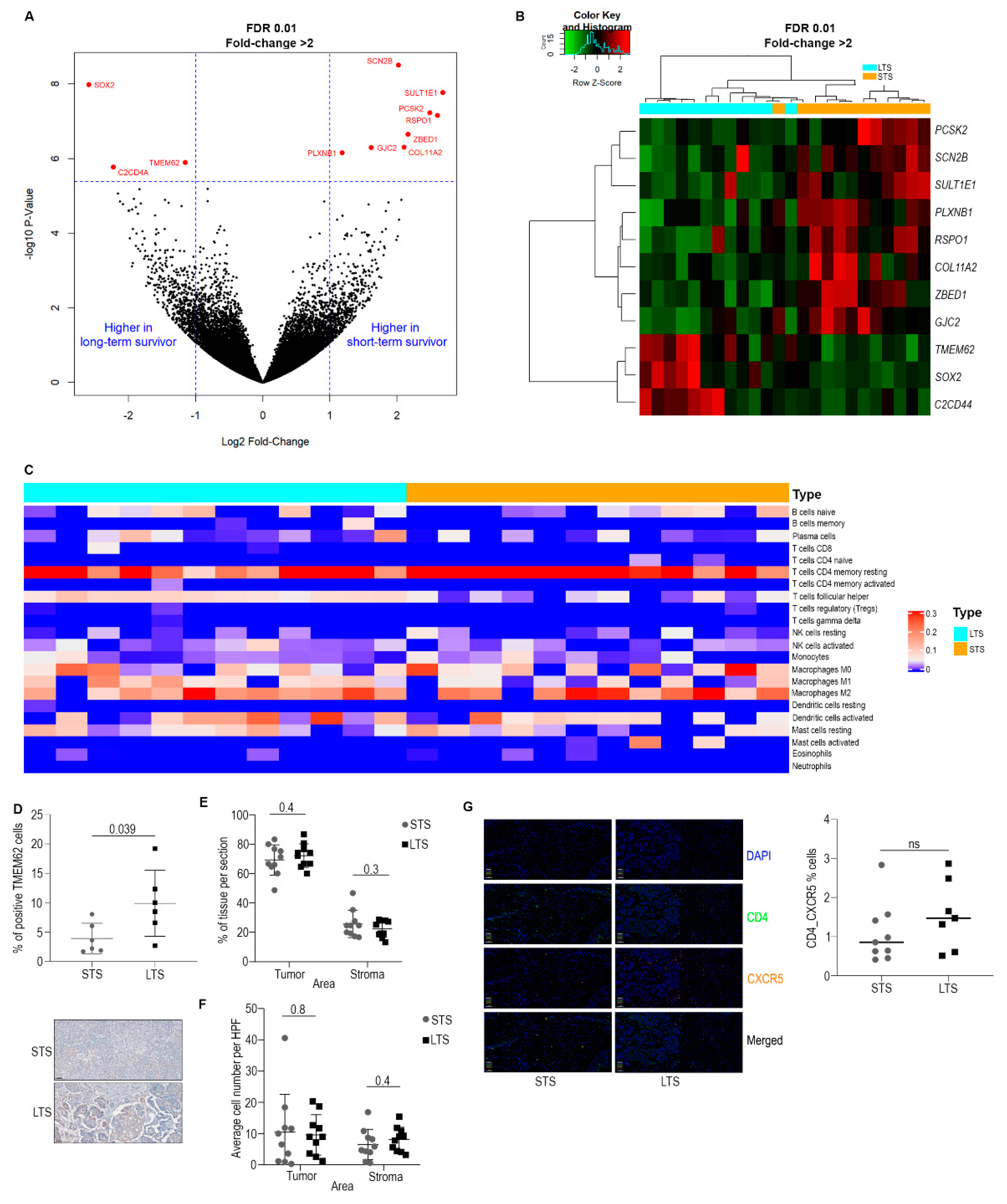
3.1. Omics Analysis
3.2. Characterization of Immune Cell Populations
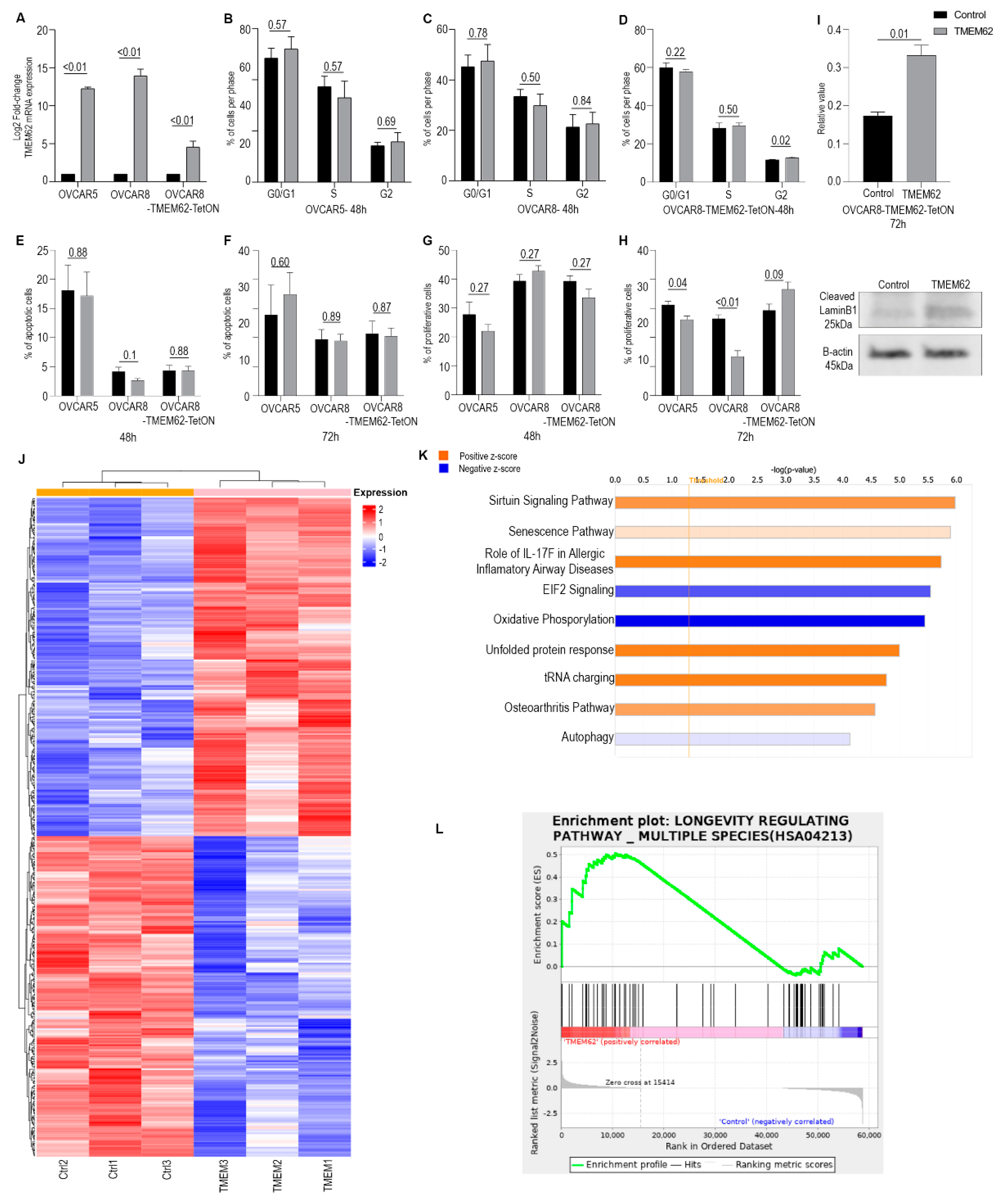
3.3. Effects of Changes in TMEM62 Expression
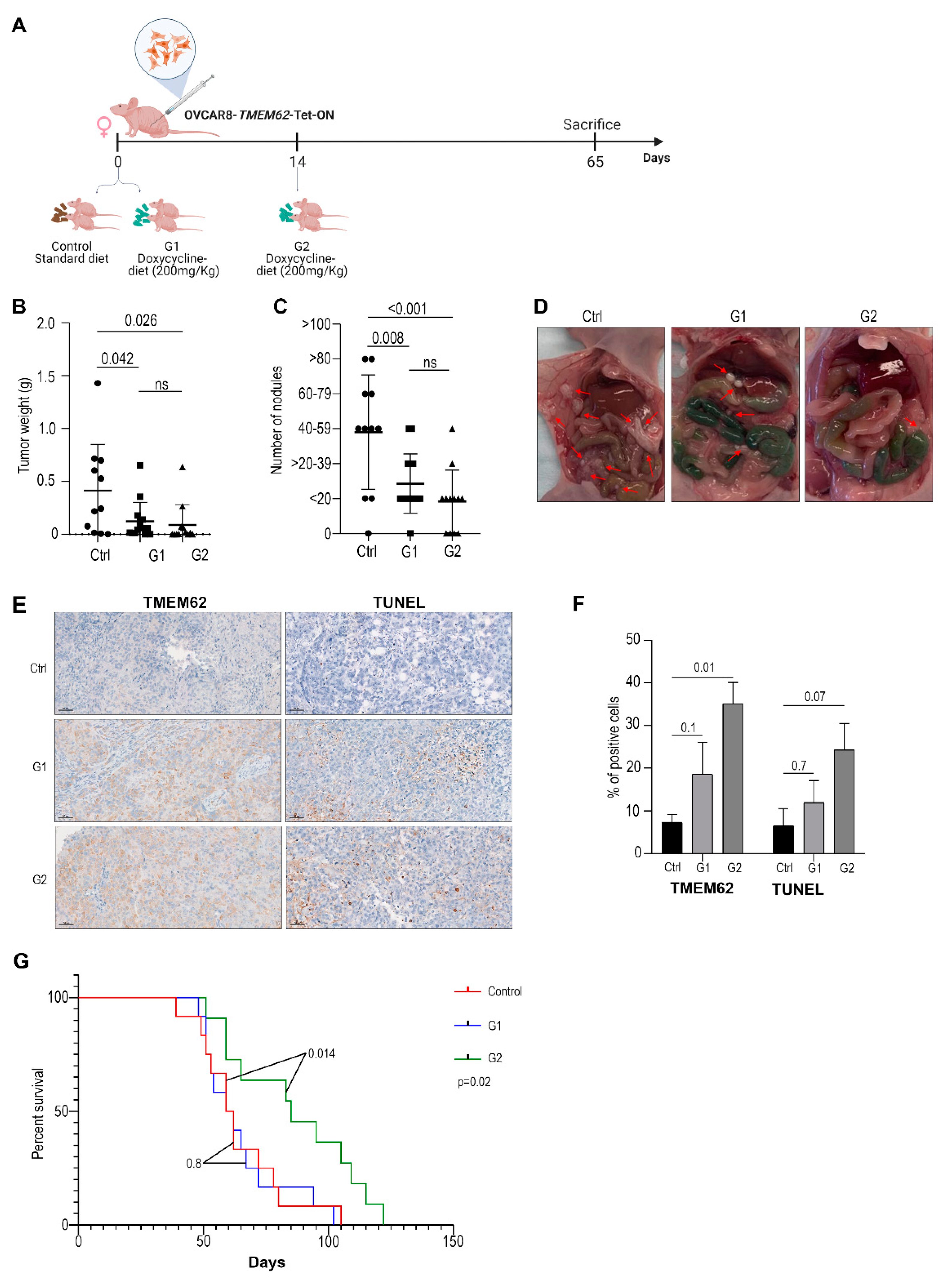
3.4. In Vivo Studies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Sauer, A.G. 2020 Cancer Facts & Figures 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [PubMed]
- Matulonis, U.A.; Sood, A.K.; Fallowfield, L.; Howitt, B.E.; Sehouli, J.; Karlan, B.Y. Ovarian cancer. Nat. Rev. Dis. Primers 2016, 2, 16061. [Google Scholar] [CrossRef]
- Hoppenot, C.; Eckert, M.A.; Tienda, S.M.; Lengyel, E. Who are the long-term survivors of high grade serous ovarian cancer? Gynecol. Oncol. 2018, 148, 204–212. [Google Scholar] [CrossRef] [PubMed]
- Berchuck, A.; Iversen, E.S.; Lancaster, J.M.; Pittman, J.; Luo, J.; Lee, P.; Murphy, S.K.; Dressman, H.K.; Febbo, P.G.; West, M.; et al. Patterns of gene expression that characterize long-term survival in advanced stage serous ovarian cancers. Clin. Cancer Res. 2005, 11, 3686–3696. [Google Scholar] [CrossRef] [PubMed]
- Bast, R.C., Jr.; Matulonis, U.A.; Sood, A.K.; Ahmed, A.A.; Amobi, A.E.; Balkwill, F.R.; Wielgos-Bonvallet, M.; Bowtell, D.D.L.; Brenton, J.D.; Brugge, J.S.; et al. Critical questions in ovarian cancer research and treatment: Report of an American Association for Cancer Research Special Conference. Cancer 2019, 125, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Landrum, L.M.; Java, J.; Mathews, C.A.; Lanneau, G.S., Jr.; Copeland, L.J.; Armstrong, D.K.; Walker, J.L. Prognostic factors for stage III epithelial ovarian cancer treated with intraperitoneal chemotherapy: A Gynecologic Oncology Group study. Gynecol. Oncol. 2013, 130, 12–18. [Google Scholar] [CrossRef]
- Hamilton, C.; Miller, A.; Casablanca, Y.; Horowitz, N.; Rungruang, B.; Krivak, T.; Richard, S.; Rodriguez, N.; Birrer, M.; Backes, F.; et al. Clinicopathologic characteristics associated with long-term survival in advanced epithelial ovarian cancer: An NRG Oncology/Gynecologic Oncology Group ancillary data study. Gynecol. Oncol. 2018, 148, 275–280. [Google Scholar] [CrossRef]
- Cress, R.D.; Chen, Y.S.; Morris, C.R.; Petersen, M.; Leiserowitz, G.S. Characteristics of Long-Term Survivors of Epithelial Ovarian Cancer. Obstet. Gynecol. 2015, 126, 491–497. [Google Scholar] [CrossRef]
- Dao, F.; Schlappe, B.A.; Tseng, J.; Lester, J.; Nick, A.M.; Lutgendorf, S.K.; McMeekin, S.; Coleman, R.L.; Moore, K.N.; Karlan, B.Y.; et al. Characteristics of 10-year survivors of high-grade serous ovarian carcinoma. Gynecol. Oncol. 2016, 141, 260–263. [Google Scholar] [CrossRef]
- Peres, L.C.; Sinha, S.; Townsend, M.K.; Fridley, B.L.; Karlan, B.Y.; Lutgendorf, S.K.; Shinn, E.; Sood, A.K.; Tworoger, S.S. Predictors of survival trajectories among women with epithelial ovarian cancer. Gynecol. Oncol. 2020, 156, 459–466. [Google Scholar] [CrossRef]
- Nikas, J.B.; Boylan, K.L.; Skubitz, A.P.; Low, W.C. Mathematical prognostic biomarker models for treatment response and survival in epithelial ovarian cancer. Cancer Inform. 2011, 10, 233–247. [Google Scholar] [CrossRef] [PubMed]
- Mlynska, A.; Vaišnorė, R.; Rafanavičius, V.; Jocys, S.; Janeiko, J.; Petrauskytė, M.; Bijeikis, S.; Cimmperman, P.; Intaitė, B.; Žilionytė, K.; et al. A gene signature for immune subtyping of desert, excluded, and inflamed ovarian tumors. Am. J. Reprod. Immunol. 2020, 84, e13244. [Google Scholar] [CrossRef]
- Dabi, Y.; Huchon, C.; Ouldamer, L.; Bendifallah, S.; Collinet, P.; Bricou, A.; Daraï, E.; Ballester, M.; Lavoue, V.; Haddad, B.; et al. Patients with stage IV epithelial ovarian cancer: Understanding the determinants of survival. J. Transl. Med. 2020, 18, 134. [Google Scholar] [CrossRef] [PubMed]
- Van Altena, A.M.; Karim-Kos, H.E.; de Vries, E.; Kruitwagen, R.F.; Massuger, L.F.; Kiemeney, L.A. Trends in therapy and survival of advanced stage epithelial ovarian cancer patients in the Netherlands. Gynecol. Oncol. 2012, 125, 649–654. [Google Scholar] [CrossRef] [PubMed]
- Timmermans, M.; Sonke, G.S.; Van de Vijver, K.K.; van der Aa, M.A.; Kruitwagen, R. No improvement in long-term survival for epithelial ovarian cancer patients: A population-based study between 1989 and 2014 in the Netherlands. Eur. J. Cancer 2018, 88, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Yarema, R.; Fetsych capital Te, C.; Volodko, N.; Capital, O.C.M.; Glehen, O. Complete cytoreduction for ovarian cancer: Is it enough for long-term survival? J. Surg. Oncol. 2018, 118, 593–594. [Google Scholar] [CrossRef]
- Ye, K.; Schulz, M.H.; Long, Q.; Apweiler, R.; Ning, Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics 2009, 25, 2865–2871. [Google Scholar] [CrossRef]
- Ha, G.; Roth, A.; Lai, D.; Bashashati, A.; Ding, J.; Goya, R.; Giuliany, R.; Rosner, J.; Oloumi, A.; Shumansky, K.; et al. Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res. 2012, 22, 1995–2007. [Google Scholar] [CrossRef]
- Olshen, A.B.; Venkatraman, E.S.; Lucito, R.; Wigler, M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics 2004, 5, 557–572. [Google Scholar] [CrossRef]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J.; et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, R80. [Google Scholar] [CrossRef] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Pounds, S.; Morris, S.W. Estimating the occurrence of false positives and false negatives in microarray studies by approximating and partitioning the empirical distribution of p-values. Bioinformatics 2003, 19, 1236–1242. [Google Scholar] [CrossRef]
- Macintyre, G.; Goranova, T.E.; De Silva, D.; Ennis, D.; Piskorz, A.M.; Eldridge, M.; Sie, D.; Lewsley, L.-A.; Hanif, A.; Wilson, C.; et al. Copy number signatures and mutational processes in ovarian carcinoma. Nat. Genet. 2018, 50, 1262–1270. [Google Scholar] [CrossRef]
- Park, S.S.; Choi, Y.W.; Kim, J.H.; Kim, H.S.; Park, T.J. Senescent tumor cells: An overlooked adversary in the battle against cancer. Exp. Mol. Med. 2021, 53, 1834–1841. [Google Scholar] [CrossRef]
- Ou, H.L.; Hoffmann, R.; Gonzalez-Lopez, C.; Doherty, G.J.; Korkola, J.E.; Munoz-Espin, D. Cellular senescence in cancer: From mechanisms to detection. Mol. Oncol. 2021, 15, 2634–2671. [Google Scholar] [CrossRef]
- Wyld, L.; Bellantuono, I.; Tchkonia, T.; Morgan, J.; Turner, O.; Foss, F.; George, J.; Danson, S.; Kirkland, J.L. Senescence and Cancer: A Review of Clinical Implications of Senescence and Senotherapies. Cancers 2020, 12, 2134. [Google Scholar] [CrossRef]
- Gire, V.; Dulic, V. Senescence from G2 arrest, revisited. Cell Cycle 2015, 14, 297–304. [Google Scholar] [CrossRef]
- Zhao, E.; Hou, J.; Ke, X.; Abbas, M.N.; Kausar, S.; Zhang, L.; Cui, H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers 2019, 11, 1949. [Google Scholar] [CrossRef]
- Bosch-Presegue, L.; Vaquero, A. The dual role of sirtuins in cancer. Genes Cancer 2011, 2, 648–662. [Google Scholar] [CrossRef]
- Chan, J.K.; Urban, R.; Cheung, M.K.; Osann, K.; Husain, A.; Teng, N.N.; Kapp, D.S.; Berek, J.S.; Leiserowitz, G.S. Ovarian cancer in younger vs older women: A population-based analysis. Br. J. Cancer 2006, 95, 1314–1320. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.Y.C.; Lheureux, S.; Karakasis, K.; Burnier, J.V.; Bruce, J.P.; Clouthier, D.L.; Danesh, A.; Quevedo, R.; Dowar, M.; Hanna, Y.; et al. Landscape of genomic alterations in high-grade serous ovarian cancer from exceptional long- and short-term survivors. Genome Med. 2018, 10, 81. [Google Scholar] [CrossRef] [PubMed]
- Aoudjit, F.; Vuori, K. Integrin signaling in cancer cell survival and chemoresistance. Chemother. Res. Pract. 2012, 2012, 283181. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Layseca, P.; Streuli, C.H. Signalling pathways linking integrins with cell cycle progression. Matrix Biol. 2014, 34, 144–153. [Google Scholar] [CrossRef] [PubMed]
- Mezu-Ndubuisi, O.J.; Maheshwari, A. The role of integrins in inflammation and angiogenesis. Pediatr. Res. 2021, 89, 1619–1626. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P. Integrin-Mediated Delivery of Drugs and Nucleic Acids for Anti-Angiogenic Cancer Therapy: Current Landscape and Remaining Challenges. Bioengineering 2018, 5, 76. [Google Scholar] [CrossRef]
- Li, N.; Zhan, X. Signaling pathway network alterations in human ovarian cancers identified with quantitative mitochondrial proteomics. EPMA J. 2019, 10, 153–172. [Google Scholar] [CrossRef]
- Mammadova-Bach, E.; Gil-Pulido, J.; Sarukhanyan, E.; Burkard, P.; Shityakov, S.; Schonhart, C.; Stegner, D.; Remer, K.; Nurden, P.; Nurden, A.T.; et al. Platelet glycoprotein VI promotes metastasis through interaction with cancer cell-derived galectin-3. Blood 2020, 135, 1146–1160. [Google Scholar] [CrossRef]
- Crotty, S. T follicular helper cell differentiation, function, and roles in disease. Immunity 2014, 41, 529–542. [Google Scholar] [CrossRef]
- Qin, L.; Waseem, T.; Sahoo, A.; Bieerkehazhi, S.; Zhou, H.; Galkina, E.V.; Nurieva, R. Insights Into the Molecular Mechanisms of T Follicular Helper-Mediated Immunity and Pathology. Front. Immunol. 2018, 9, 1884. [Google Scholar] [CrossRef]
- Gu-Trantien, C.; Loi, S.; Garaud, S.; Equeter, C.; Libin, M.; De Wind, A.; Ravoet, M.; Le Buanec, H.; Sibille, C.; Manfouo-Foutsop, G.; et al. CD4(+) follicular helper T cell infiltration predicts breast cancer survival. J. Clin. Investig. 2013, 123, 2873–2892. [Google Scholar] [CrossRef] [Green Version]
- Gu-Trantien, C.; Willard-Gallo, K. Tumor-infiltrating follicular helper T cells: The new kids on the block. Oncoimmunology 2013, 2, e26066. [Google Scholar] [CrossRef]
- Li, L.; Ma, Y.; Xu, Y. Follicular regulatory T cells infiltrated the ovarian carcinoma and resulted in CD8 T cell dysfunction dependent on IL-10 pathway. Int. Immunopharmacol. 2019, 68, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Ma, Y.; Xu, Y.; Maerkeya, K. TIM-3 expression identifies a distinctive PD-1(+) follicular helper T cell subset, with reduced interleukin 21 production and B cell help function in ovarian cancer patients. Int. Immunopharmacol. 2018, 57, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Conejo-Garcia, J.R.; Biswas, S.; Chaurio, R. Humoral immune responses: Unsung heroes of the war on cancer. Semin. Immunol. 2020, 49, 101419. [Google Scholar] [CrossRef] [PubMed]
- Truxova, I.; Kasikova, L.; Hensler, M.; Škapa, P.; Laco, J.; Pecen, L.; Belicova, L.; Praznovec, I.; Halaska, M.J.; Brtnicky, T.; et al. Mature dendritic cells correlate with favorable immune infiltrate and improved prognosis in ovarian carcinoma patients. J. Immunother. Cancer 2018, 6, 139. [Google Scholar] [CrossRef]
- Kroeger, D.R.; Milne, K.; Nelson, B.H. Tumor-Infiltrating Plasma Cells Are Associated with Tertiary Lymphoid Structures, Cytolytic T-Cell Responses, and Superior Prognosis in Ovarian Cancer. Clin. Cancer Res. 2016, 22, 3005–3015. [Google Scholar] [CrossRef]
- GeneCards. 2021 TMEM62 Gene. Available online: https://www.genecards.org/cgi-bin/carddisp.pl?gene=TMEM62 (accessed on 9 August 2022).
- Atlas, T.H.P. 2021 TMEM62. Available online: https://www.proteinatlas.org/ENSG00000137842-TMEM62/tissue (accessed on 9 August 2022).
- Schmit, K.; Michiels, C. TMEM Proteins in Cancer: A Review. Front. Pharmacol. 2018, 9, 1345. [Google Scholar] [CrossRef]
- UniProt. 2021 UniProtKB-Q0P6H9 (TMM62_HUMAN). Available online: https://www.uniprot.org/uniprot/Q0P6H9 (accessed on 9 August 2022).
- Jadoon, A.; Cunningham, P.; McDermott, L.C. Arachidonic acid metabolism in the human placenta: Identification of a putative lipoxygenase. Placenta 2014, 35, 422–424. [Google Scholar] [CrossRef]
- Crooks, S.W.; Stockley, R.A. Leukotriene B4. Int. J. Biochem. Cell Biol. 1998, 30, 173–178. [Google Scholar] [CrossRef]
- Seo, J.M.; Park, S.; Kim, J.H. Leukotriene B4 receptor-2 promotes invasiveness and metastasis of ovarian cancer cells through signal transducer and activator of transcription 3 (STAT3)-dependent up-regulation of matrix metalloproteinase 2. J. Biol. Chem. 2012, 287, 13840–13849. [Google Scholar] [CrossRef] [Green Version]
| Sample ID | Sample Type | Age | Site | Stage |
|---|---|---|---|---|
| 1 | LT | 63 | Ovary | IIIC |
| 2 * | LT | 63 | Ovary | IIIC |
| 3 | LT | 53 | Ovary | IIIC |
| 4 | LT | 71 | Ovary | IIIC |
| 5 | LT | 49 | Ovary | IIIC |
| 6 | LT | 54 | Ovary | IIIC |
| 7 * | LT | 67 | Ovary | IIIC |
| 8 * | LT | 66 | Peritoneum | IV |
| 9 * | LT | 56 | Ovary | IIIC |
| 10 * | LT | 58 | Ovary | IIIC |
| 11 | LT | 69 | Ovary | IIIC |
| 12 * | LT | 63 | Ovary | IIIC |
| 13 | ST | 62 | Tube | IIIC |
| 14 | ST | 50 | Ovary | IIIC |
| 15 | ST | 59 | Ovary | IIIC |
| 16 | ST | 57 | Tube | IV |
| 17 * | ST | 64 | Ovary | IIIC |
| 18 * | ST | 74 | Peritoneum | IIIC |
| 19 | ST | 54 | Tube | IV |
| 20 * | ST | 78 | Ovary | IV |
| 21 * | ST | 60 | Tube | IIIC |
| 22 | ST | 59 | Tube | IIIC |
| 23 | ST | 53 | Ovary | IIIC |
| 24 | ST | 66 | Ovary | IIIC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Stur, E.; Bayraktar, E.; Dal Molin, G.Z.; Wu, S.Y.; Mangala, L.S.; Yao, H.; Wang, Y.; Ram, P.T.; Corvigno, S.; Chen, H.; et al. Molecular Analysis of Short- versus Long-Term Survivors of High-Grade Serous Ovarian Carcinoma. Cancers 2022, 14, 4198. https://doi.org/10.3390/cancers14174198
Stur E, Bayraktar E, Dal Molin GZ, Wu SY, Mangala LS, Yao H, Wang Y, Ram PT, Corvigno S, Chen H, et al. Molecular Analysis of Short- versus Long-Term Survivors of High-Grade Serous Ovarian Carcinoma. Cancers. 2022; 14(17):4198. https://doi.org/10.3390/cancers14174198
Chicago/Turabian StyleStur, Elaine, Emine Bayraktar, Graziela Zibetti Dal Molin, Sherry Y. Wu, Lingegowda S. Mangala, Hui Yao, Ying Wang, Prahlad T. Ram, Sara Corvigno, Hu Chen, and et al. 2022. "Molecular Analysis of Short- versus Long-Term Survivors of High-Grade Serous Ovarian Carcinoma" Cancers 14, no. 17: 4198. https://doi.org/10.3390/cancers14174198
APA StyleStur, E., Bayraktar, E., Dal Molin, G. Z., Wu, S. Y., Mangala, L. S., Yao, H., Wang, Y., Ram, P. T., Corvigno, S., Chen, H., Liang, H., Tworoger, S. S., Levine, D. A., Lutgendorf, S. K., Liu, J., Moore, K. N., Baggerly, K. A., Karlan, B. Y., & Sood, A. K. (2022). Molecular Analysis of Short- versus Long-Term Survivors of High-Grade Serous Ovarian Carcinoma. Cancers, 14(17), 4198. https://doi.org/10.3390/cancers14174198






