Live Biotherapeutic Lactococcus lactis GEN3013 Enhances Antitumor Efficacy of Cancer Treatment via Modulation of Cancer Progression and Immune System
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Cell Culture
2.3. Bacteria
2.4. Bacterial Sampling
2.5. Metabolic Profiling
2.6. MTT Assay
2.7. Colony Forming Assay
2.8. Wound Healing Assay
2.9. T Cell Proliferation Assay
2.10. Immune Cell Profiling
2.11. Quantitative PCR (qPCR) Analysis
2.12. Statistical Analysis
3. Results
3.1. Lactococcus lactis GEN3013 Induces Cell Death in Various Cancer Cell Types
3.2. L. lactis GEN3013 Administration Inhibits Tumor Growth by Directly Affecting Cancer Cells
3.3. L. lactis GEN3013 Enhances the Chemotherapy Therapeutic Effects
3.4. Combination of L. lactis GEN3013 and Chemotherapy Reprograms the Host Immune System
3.5. L. lactis GEN3013 Enhances the Therapeutic Effects of Immunotherapy
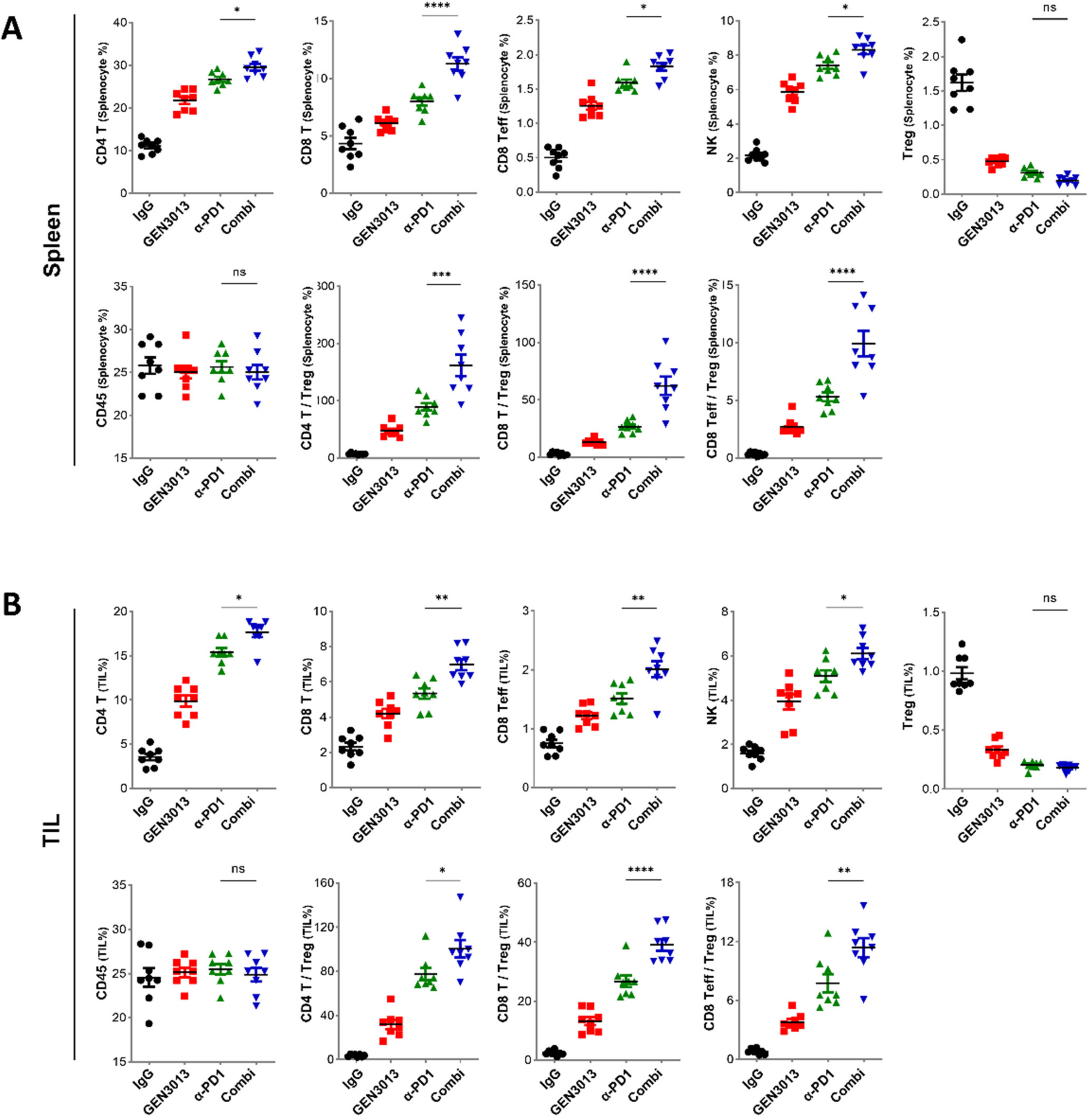
3.6. The Combination of L. lactis GEN3013 and Immunotherapy Reprograms the Host Immune System
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Yamashiro, Y. Gut microbiota in health and disease. Ann. Nutr. Metab. 2017, 71, 242–246. [Google Scholar] [CrossRef] [PubMed]
- Amsterdam, D.; Ostrov, B.E. The impact of the microbiome on immunosenescence. Immunol. Investig. 2018, 47, 801–811. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.; Zhang, J. Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 2017, 18, 2. [Google Scholar] [CrossRef]
- Lee, S.-H.; Cho, S.-Y.; Yoon, Y.; Park, C.; Sohn, J.; Jeong, J.-J.; Jeon, B.-N.; Jang, M.; An, C.; Lee, S. Bifidobacterium bifidum strains synergize with immune checkpoint inhibitors to reduce tumour burden in mice. Nat. Microbiol. 2021, 6, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Gill, H.S.; Rutherfurd, K.J.; Cross, M.L.; Gopal, P.K. Enhancement of immunity in the elderly by dietary supplementation with the probiotic Bifidobacterium lactis HN019. Am. J. Clin. Nutr. 2001, 74, 833–839. [Google Scholar] [CrossRef]
- Zitvogel, L.; Daillère, R.; Roberti, M.P.; Routy, B.; Kroemer, G. Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 2017, 15, 465–478. [Google Scholar] [CrossRef]
- Fulbright, L.E.; Ellermann, M.; Arthur, J.C. The microbiome and the hallmarks of cancer. PLoS Pathog. 2017, 13, e1006480. [Google Scholar] [CrossRef]
- Sethi, V.; Kurtom, S.; Tarique, M.; Lavania, S.; Malchiodi, Z.; Hellmund, L.; Zhang, L.; Sharma, U.; Giri, B.; Garg, B.; et al. Gut Microbiota Promotes Tumor Growth in Mice by Modulating Immune Response. Gastroenterology 2018, 155, 33–37.e6. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Helmink, B.A.; Spencer, C.N.; Reuben, A.; Wargo, J.A. The Influence of the Gut Microbiome on Cancer, Immunity, and Cancer Immunotherapy. Cancer Cell. 2018, 33, 570–580. [Google Scholar] [CrossRef]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377–388. [Google Scholar] [CrossRef]
- Peng, Z.; Cheng, S.; Kou, Y.; Wang, Z.; Jin, R.; Hu, H.; Zhang, X.; Gong, J.F.; Li, J.; Lu, M.; et al. The Gut Microbiome Is Associated with Clinical Response to Anti-PD-1/PD-L1 Immunotherapy in Gastrointestinal Cancer. Cancer Immunol. Res. 2020, 8, 1251–1261. [Google Scholar] [CrossRef] [PubMed]
- Rezasoltani, S.; Yadegar, A.; Aghdaei, H.A.; Zali, M.R. Modulatory effects of gut microbiome in cancer immunotherapy: A novel paradigm for blockade of immune checkpoint inhibitors. Cancer Med. 2021, 10, 1141–1154. [Google Scholar] [CrossRef] [PubMed]
- Gopalakrishnan, V.; Spencer, C.N.; Nezi, L.; Reuben, A.; Andrews, M.C.; Karpinets, T.V.; Prieto, P.A.; Vicente, D.; Hoffman, K.; Wei, S.C.; et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.M.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P.; et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Vivarelli, S.; Falzone, L.; Leonardi, G.C.; Salmeri, M.; Libra, M. Novel insights on gut microbiota manipulation and immune checkpoint inhibition in cancer. Int. J. Oncol. 2021, 59, 1–17. [Google Scholar] [CrossRef]
- Vivarelli, S.; Falzone, L.; Basile, M.S.; Nicolosi, D.; Genovese, C.; Libra, M.; Salmeri, M. Benefits of using probiotics as adjuvants in anticancer therapy. World Acad. Sci. J. 2019, 1, 125–135. [Google Scholar] [CrossRef]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Man Lei, Y.; Jabri, B.; Alegre, M.-L. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef]
- Azizpour, M.; Hosseini, S.D.; Jafari, P.; Akbary, N. Lactococcus lactis: A New Strategy for Vaccination. Avicenna J. Med. Biotechnol. 2017, 9, 163–168. [Google Scholar]
- Bohlul, E.; Hasanlou, F.; Taromchi, A.H.; Nadri, S. TRAIL-expressing recombinant Lactococcus lactis induces apoptosis in human colon adenocarcinoma SW480 and HCT116 cells. J. Appl. Microb. 2019, 126, 1558–1567. [Google Scholar] [CrossRef]
- Zhang, B.; Li, A.; Zuo, F.; Yu, R.; Zeng, Z.; Ma, H.; Chen, S. Recombinant Lactococcus lactis NZ9000 secretes a bioactive kisspeptin that inhibits proliferation and migration of human colon carcinoma HT-29 cells. Microb. Cell Fact. 2016, 15, 102. [Google Scholar] [CrossRef]
- Bermúdez-Humarán, L.G.; Cortes-Perez, N.G.; Ah-Leung, S.; Lefèvre, F.; Yang, G.; Pang, Q.; Wu, C.; Zeng, Y.; Adel-Patient, K.; Langella, P. Current prophylactic and therapeutic uses of a recombinant Lactococcus lactis strain secreting biologically active interleukin-12. J. Mol. Microb. Biotechnol. 2008, 14, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qi, L.; Lv, J.; Zhang, Z.; Zhou, P.; Ma, Z.; Wang, Y.; Zhang, Y.; Pan, L. The immune response to a recombinant Lactococcus lactis oral vaccine against foot-and-mouth disease virus in mice. Biotechnol. Lett. 2020, 42, 1907–1917. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, J.E.; Lee, K.W.; Lee, H.J. Lactococcus lactis ssp. lactis inhibits the proliferation of SNU-1 human stomach cancer cells through induction of G0/G1 cell cycle arrest and apoptosis via p53 and p21 expression. Ann. N. Y. Acad. Sci. 2009, 1171, 270–275. [Google Scholar] [CrossRef]
- Kim, J.Y.; Woo, H.J.; Kim, Y.S.; Kim, K.H.; Lee, H.J. Cell cycle dysregulation induced by cytoplasm of Lactococcus lactis ssp lactis in SNUC2A, a colon cancer cell line. Nutr. Cancer 2003, 46, 197–201. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Kim, S.Y.; Lee, K.W.; Lee, H.J. Arginine deiminase originating from Lactococcus lactis ssp. lactis American Type Culture Collection (ATCC) 7962 induces G1-phase cell-cycle arrest and apoptosis in SNU-1 stomach adenocarcinoma cells. Br. J. Nutr. 2009, 102, 1469–1476. [Google Scholar] [CrossRef] [PubMed]
- Han, K.J.; Lee, N.K.; Park, H.; Paik, H.D. Anticancer and Anti-Inflammatory Activity of Probiotic Lactococcus lactis NK34. J. Microb. Biotechnol. 2015, 25, 1697–1701. [Google Scholar] [CrossRef]
- Tay, R.E.; Richardson, E.K.; Toh, H.C. Revisiting the role of CD4+ T cells in cancer immunotherapy—New insights into old paradigms. Cancer Gene. Ther. 2021, 28, 5–17. [Google Scholar] [CrossRef] [PubMed]
- Raskov, H.; Orhan, A.; Christensen, J.P.; Gögenur, I. Cytotoxic CD8+ T cells in cancer and cancer immunotherapy. Br. J. Cancer 2021, 124, 359–367. [Google Scholar] [CrossRef]
- Jorgovanovic, D.; Song, M.; Wang, L.; Zhang, Y. Roles of IFN-γ in tumor progression and regression: A review. Biomark. Res. 2020, 8, 1–16. [Google Scholar] [CrossRef]
- Montfort, A.; Colacios, C.; Levade, T.; Andrieu-Abadie, N.; Meyer, N.; Ségui, B. The TNF paradox in cancer progression and immunotherapy. Front. Immunol. 2019, 10, 1818. [Google Scholar] [CrossRef]
- Jiang, T.; Zhou, C.; Ren, S. Role of IL-2 in cancer immunotherapy. Oncoimmunology 2016, 5, e1163462. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, S.E.; Keating, N.; Belz, G.T. Natural killer cells and anti-tumor immunity. Mol. Immunol. 2019, 110, 40–47. [Google Scholar] [CrossRef] [PubMed]
- Ben-Shmuel, A.; Biber, G.; Barda-Saad, M. Unleashing natural killer cells in the tumor microenvironment–the next generation of immunotherapy? Front. Immunol. 2020, 11, 275. [Google Scholar] [CrossRef] [PubMed]
- Jewett, A.; Kos, J.; Kaur, K.; Safaei, T.; Sutanto, C.; Chen, W.; Wong, P.; Namagerdi, A.K.; Fang, C.; Fong, Y. Natural killer cells: Diverse functions in tumor immunity and defects in pre-neoplastic and neoplastic stages of tumorigenesis. Mol. Ther. Oncolytics 2020, 16, 41–52. [Google Scholar] [CrossRef]
- Lim, J.-N.; Oh, J.-J.; Wang, T.; Lee, J.-S.; Kim, S.-H.; Kim, Y.-J.; Lee, H.-G. Trans-11 18: 1 vaccenic acid (TVA) has a direct anti-carcinogenic effect on MCF-7 human mammary adenocarcinoma cells. Nutrients 2014, 6, 627–636. [Google Scholar] [CrossRef]
- Mahmood, N.; Arakelian, A.; Muller, W.J.; Szyf, M.; Rabbani, S.A. An enhanced chemopreventive effect of methyl donor S-adenosylmethionine in combination with 25-hydroxyvitamin D in blocking mammary tumor growth and metastasis. Bone Res. 2020, 8, 1–12. [Google Scholar] [CrossRef]
- Jiang, L.; Wang, W.; He, Q.; Wu, Y.; Lu, Z.; Sun, J.; Liu, Z.; Shao, Y.; Wang, A. Oleic acid induces apoptosis and autophagy in the treatment of Tongue Squamous cell carcinomas. Sci. Rep. 2017, 7, 11277. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Y.; Wu, P. 5′-Methylthioadenosine and Cancer: Old molecules, new understanding. J. Cancer 2019, 10, 927. [Google Scholar] [CrossRef]
- Icard, P.; Coquerel, A.; Wu, Z.; Gligorov, J.; Fuks, D.; Fournel, L.; Lincet, H.; Simula, L. Understanding the central role of citrate in the metabolism of cancer cells and tumors: An update. Int. J. Mol. Sci. 2021, 22, 6587. [Google Scholar] [CrossRef]
- Kar, F.; Hacioglu, C.; Kacar, S.; Sahinturk, V.; Kanbak, G. Betaine suppresses cell proliferation by increasing oxidative stress–mediated apoptosis and inflammation in DU-145 human prostate cancer cell line. Cell Stress Chaperones 2019, 24, 871–881. [Google Scholar] [CrossRef]
- Li, Y.; Jia, Y.; Xu, Y.; Li, K. DMF Activates NRF2 to Inhibit the Pro-Invasion Ability of TAMs in Breast Cancer. Front. Oncol. 2021, 11, 706448. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Du, A.; Xiong, Y.; Jiang, J.; Zhang, Y.; Tian, Z.; Yan, H. γ-Aminobutyric acid inhibits the proliferation and increases oxaliplatin sensitivity in human colon cancer cells. Tumor. Biol. 2016, 37, 14885–14894. [Google Scholar] [CrossRef] [PubMed]
- Suarez-Arnedo, A.; Figueroa, F.T.; Clavijo, C.; Arbeláez, P.; Cruz, J.C.; Muñoz-Camargo, C. An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS ONE 2020, 15, e0232565. [Google Scholar]
- Tesniere, A.; Schlemmer, F.; Boige, V.; Kepp, O.; Martins, I.; Ghiringhelli, F.; Aymeric, L.; Michaud, M.; Apetoh, L.; Barault, L.; et al. Immunogenic death of colon cancer cells treated with oxaliplatin. Oncogene 2010, 29, 482–491. [Google Scholar] [CrossRef]
- Zhu, H.; Shan, Y.; Ge, K.; Lu, J.; Kong, W.; Jia, C. Oxaliplatin induces immunogenic cell death in hepatocellular carcinoma cells and synergizes with immune checkpoint blockade therapy. Cell Oncol. 2020, 43, 1203–1214. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, G.; Chen, Y.; Wang, H.; Hua, Y.; Cai, Z. Immunogenic cell death in cancer therapy: Present and emerging inducers. J. Cell Mol. Med. 2019, 23, 4854–4865. [Google Scholar] [CrossRef]
- Aaes, T.L.; Vandenabeele, P. The intrinsic immunogenic properties of cancer cell lines, immunogenic cell death, and how these influence host antitumor immune responses. Cell Death Differ. 2021, 28, 843–860. [Google Scholar] [CrossRef]
- Łuksza, M.; Riaz, N.; Makarov, V.; Balachandran, V.P.; Hellmann, M.D.; Solovyov, A.; Rizvi, N.A.; Merghoub, T.; Levine, A.J.; Chan, T.A. A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 2017, 551, 517–520. [Google Scholar] [CrossRef]
- Taube, J.M.; Klein, A.; Brahmer, J.R.; Xu, H.; Pan, X.; Kim, J.H.; Chen, L.; Pardoll, D.M.; Topalian, S.L.; Anders, R.A. Association of PD-1, PD-1 ligands, and other features of the tumor immune microenvironment with response to anti–PD-1 therapy. Clin. Cancer Res. 2014, 20, 5064–5074. [Google Scholar] [CrossRef]
- Hwang, S.; Kwon, A.-Y.; Jeong, J.-Y.; Kim, S.; Kang, H.; Park, J.; Kim, J.-H.; Han, O.J.; Lim, S.M.; An, H.J. Immune gene signatures for predicting durable clinical benefit of anti-PD-1 immunotherapy in patients with non-small cell lung cancer. Sci. Rep. 2020, 10, 643. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Oba, T.; Long, M.D.; Keler, T.; Marsh, H.C.; Minderman, H.; Abrams, S.I.; Liu, S.; Ito, F. Overcoming primary and acquired resistance to anti-PD-L1 therapy by induction and activation of tumor-residing cDC1s. Nat. Commun. 2020, 11, 5415. [Google Scholar] [CrossRef] [PubMed]
- Hurkmans, D.P.; Kuipers, M.E.; Smit, J.; van Marion, R.; Mathijssen, R.H.; Postmus, P.E.; Hiemstra, P.S.; Aerts, J.G.; von der Thüsen, J.H.; van der Burg, S.H. Tumor mutational load, CD8+ T cells, expression of PD-L1 and HLA class I to guide immunotherapy decisions in NSCLC patients. Cancer Immunol. Immunother. 2020, 69, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A.; Cruz, C.; Eder, J.P.; Braiteh, F.; Chung, C.; Tolaney, S.M.; Kuter, I.; Nanda, R.; Cassier, P.A.; Delord, J.-P. Long-term clinical outcomes and biomarker analyses of atezolizumab therapy for patients with metastatic triple-negative breast cancer: A phase 1 study. JAMA Oncol. 2019, 5, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Kagamu, H.; Kitano, S.; Yamaguchi, O.; Yoshimura, K.; Horimoto, K.; Kitazawa, M.; Fukui, K.; Shiono, A.; Mouri, A.; Nishihara, F. CD4+ T-cell immunity in the peripheral blood correlates with response to anti-PD-1 therapy. Cancer Immunol. Res. 2020, 8, 334–344. [Google Scholar] [CrossRef]
- Borst, J.; Ahrends, T.; Bąbała, N.; Melief, C.J.; Kastenmüller, W. CD4+ T cell help in cancer immunology and immunotherapy. Nat. Rev. Immunol. 2018, 18, 635–647. [Google Scholar] [CrossRef]
- WHO. Assessing National Capacity for the Prevention and Control of Noncommunicable Diseases: Report of the 2019 Global Survey; WHO: Geneva, Switzerland, 2020. [Google Scholar]
- Garrett, W.S. Cancer and the microbiota. Science 2015, 348, 80–86. [Google Scholar] [CrossRef]
- Tsilimigras, M.C.; Fodor, A.; Jobin, C. Carcinogenesis and therapeutics: The microbiota perspective. Nat. Microbiol. 2017, 2, 1–10. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillère, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef]
- Zheng, J.H.; Nguyen, V.H.; Jiang, S.-N.; Park, S.-H.; Tan, W.; Hong, S.H.; Shin, M.G.; Chung, I.-J.; Hong, Y.; Bom, H.-S. Two-step enhanced cancer immunotherapy with engineered Salmonella typhimurium secreting heterologous flagellin. Sci. Transl. Med. 2017, 9, eaak9537. [Google Scholar] [CrossRef]
- Kim, O.Y.; Park, H.T.; Dinh, N.T.H.; Choi, S.J.; Lee, J.; Kim, J.H.; Lee, S.-W.; Gho, Y.S. Bacterial outer membrane vesicles suppress tumor by interferon-γ-mediated antitumor response. Nat. Commun. 2017, 8, 626. [Google Scholar] [CrossRef] [PubMed]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Muller, P.Y.; Milton, M.N. The determination and interpretation of the therapeutic index in drug development. Nat. Rev. Drug Discov. 2012, 11, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Tamargo, J.; Le Heuzey, J.-Y.; Mabo, P. Narrow therapeutic index drugs: A clinical pharmacological consideration to flecainide. Eur. J. Clin. Pharmacol. 2015, 71, 549–567. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Chu, C.; Zhang, Y.-H.; Zheng, M.; Zhu, L.; Kong, X.; Huang, T. Identification of drug-drug interactions using chemical interactions. Curr. Bioinform. 2017, 12, 526–534. [Google Scholar] [CrossRef]
- Alcindor, T.; Beauger, N. Oxaliplatin: A review in the era of molecularly targeted therapy. Curr. Oncol. 2011, 18, 18–25. [Google Scholar] [CrossRef]
- Hanna, N.; Shepherd, F.A.; Fossella, F.V.; Pereira, J.R.; De Marinis, F.; Von Pawel, J.; Gatzemeier, U.; Tsao, T.C.Y.; Pless, M.; Muller, T. Randomized phase III trial of pemetrexed versus docetaxel in patients with non-small-cell lung cancer previously treated with chemotherapy. J. Clin. Oncol. 2004, 22, 1589–1597. [Google Scholar] [CrossRef]
- Pearce, A.; Haas, M.; Viney, R.; Pearson, S.-A.; Haywood, P.; Brown, C.; Ward, R. Incidence and severity of self-reported chemotherapy side effects in routine care: A prospective cohort study. PLoS ONE 2017, 12, e0184360. [Google Scholar]
- Montemurro, F.; Mittica, G.; Cagnazzo, C.; Longo, V.; Berchialla, P.; Solinas, G.; Culotta, P.; Martinello, R.; Foresto, M.; Gallizioli, S. Self-evaluation of adjuvant chemotherapy-related adverse effects by patients with breast cancer. JAMA Oncol. 2016, 2, 445–452. [Google Scholar] [CrossRef]
- Hou, X.; Zhang, P.; Du, H.; Chu, W.; Sun, R.; Qin, S.; Tian, Y.; Zhang, Z.; Xu, F. Akkermansia Muciniphila Potentiates the Antitumor Efficacy of FOLFOX in Colon Cancer. Front. Pharmacol. 2021, 12, 725583. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Paz-Ares, L.; Caro, R.B.; Zurawski, B.; Kim, S.-W.; Costa, E.C.; Park, K.; Alexandru, A.; Lupinacci, L.; de la Mora Jimenez, E. Nivolumab plus ipilimumab in advanced non–small-cell lung cancer. New. Engl. J. Med. 2019, 381, 2020–2031. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C. Nivolumab for recurrent squamous-cell carcinoma of the head and neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S. Pembrolizumab versus chemotherapy for PD-L1–positive non–small-cell lung cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef] [PubMed]
- Gandhi, L.; Garassino, M.C. Pembrolizumab plus chemotherapy in lung cancer. N. Engl. J. Med. 2018, 379, e18. [Google Scholar] [PubMed]
- Galluzzi, L.; Humeau, J.; Buqué, A.; Zitvogel, L.; Kroemer, G. Immunostimulation with chemotherapy in the era of immune checkpoint inhibitors. Nat. Rev. Clin. Oncol. 2020, 17, 725–741. [Google Scholar] [CrossRef] [PubMed]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef]
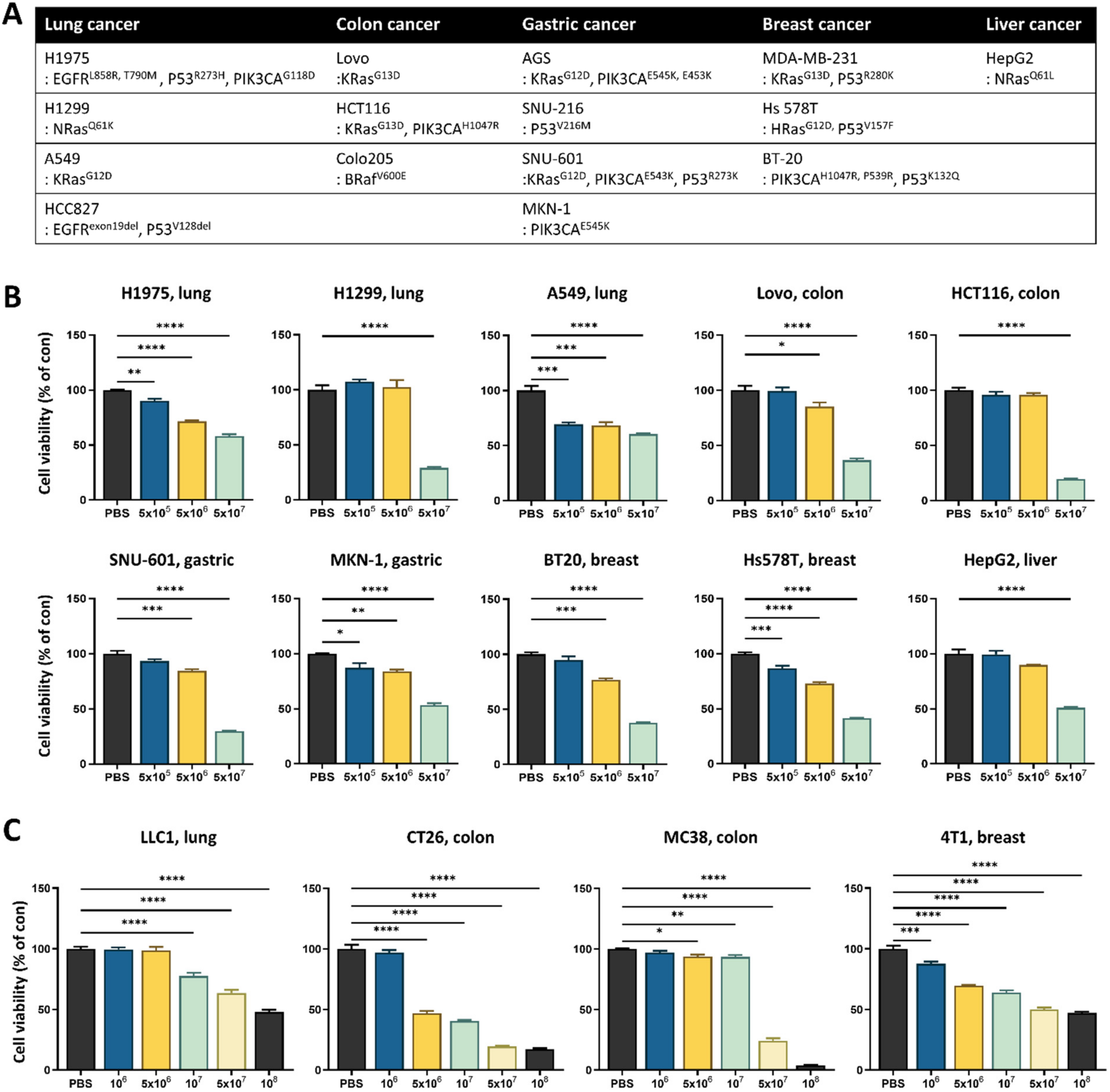
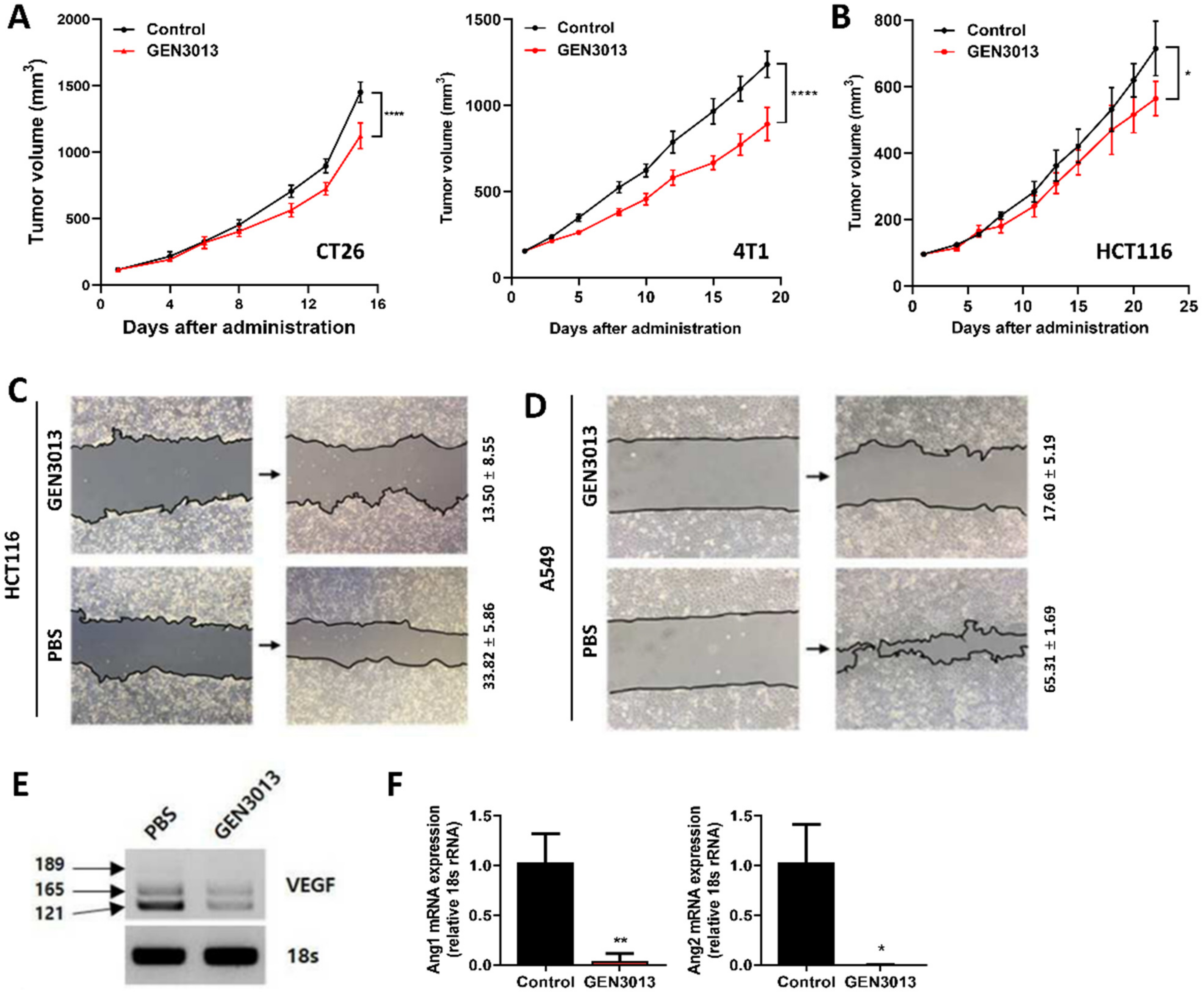
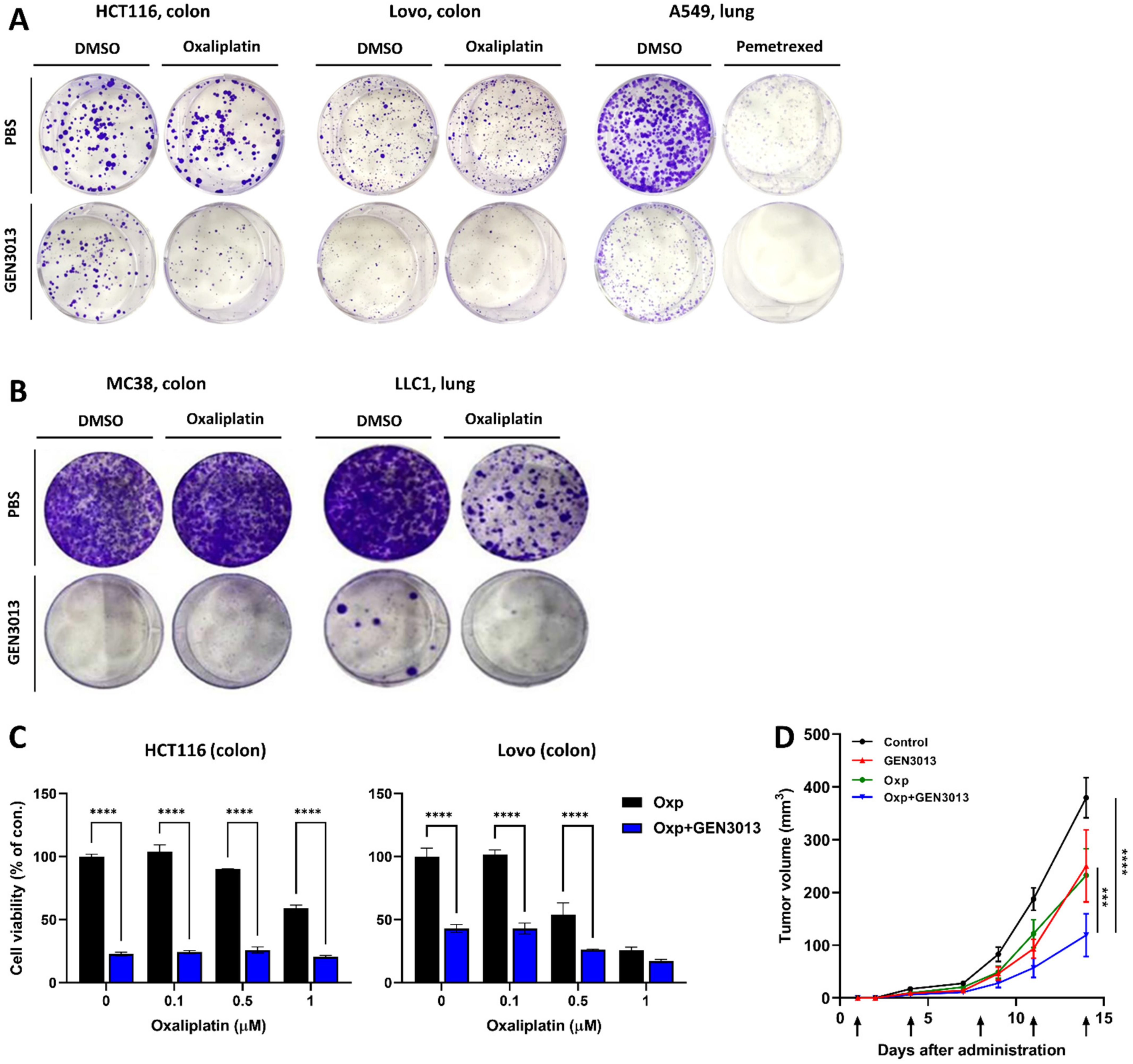

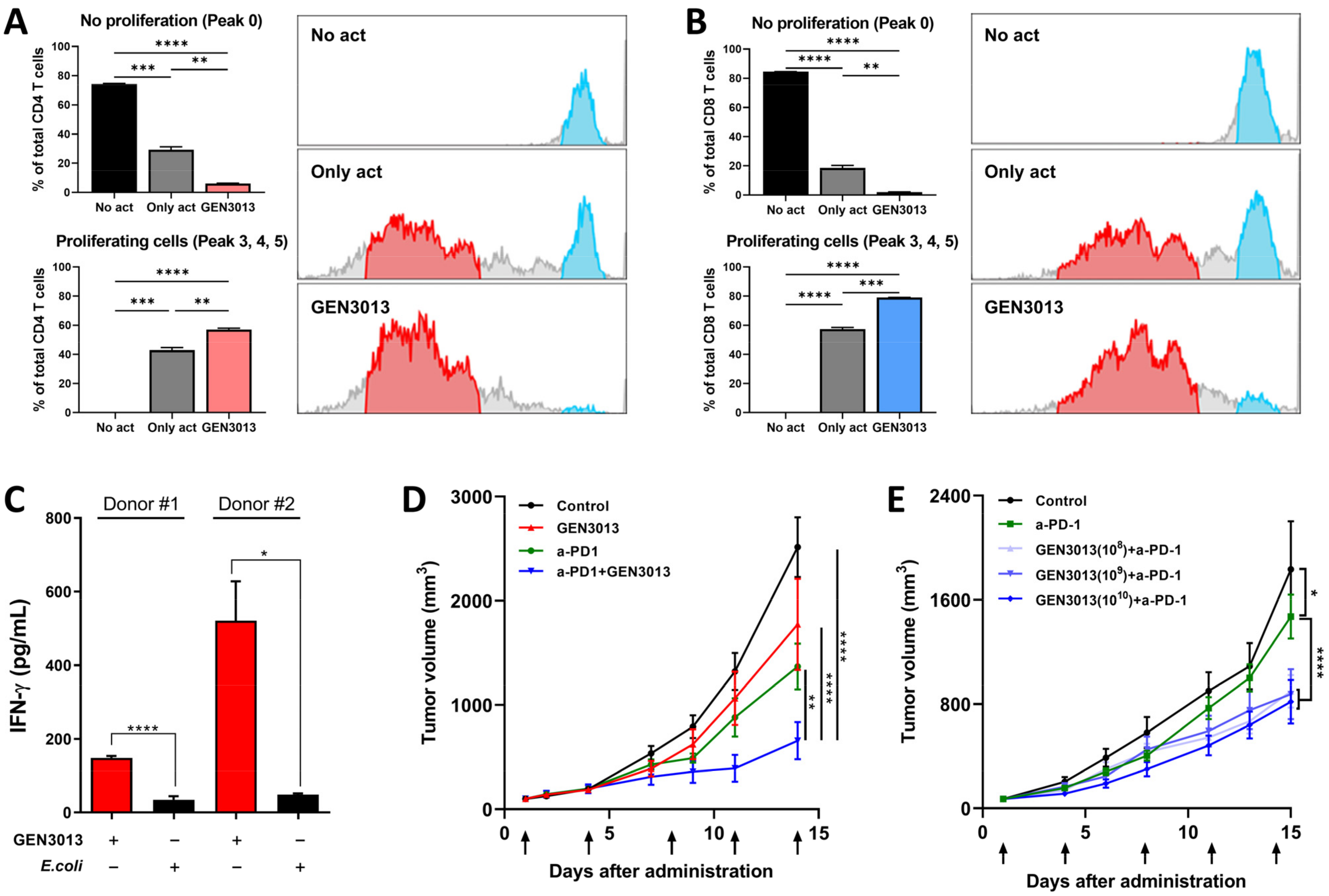
| Target | Conjugated Fluorescent | Company | Catalog Number | Usage Dilution |
|---|---|---|---|---|
| CD45 | APC/Cyanin7 | Biolegend | 103116 | 1/40 |
| CD3 | PerCP/Cyanine5.5 | Biolegend | 100218 | 1/20 |
| NK1.1 | PE | Biolegend | 108708 | 1/40 |
| CD49b | APC | Biolegend | 108910 | 1/40 |
| CD4 | PE/Cyanine7 | Biolegend | 100422 | 1/100 |
| CD25 | PE | Biolegend | 101904 | 1/100 |
| FOXP3 | APC | Invitrogen | 17-5773-82 | 1/40 |
| CD44 | PE | Biolegend | 103008 | 1/40 |
| CD62L | APC | Biolegend | 104412 | 1/40 |
| CD8a | FITC | Biolegend | 100706 | 1/40 |
| Target | Sequence | PCR Thermal Conditions | |||
|---|---|---|---|---|---|
| Ang1 | FW | 5′-CATTCTTCGCTGCCATTCTG-3′ | 95 °C | 1 min | Initial denaturation |
| Rev | 5′-GCACATTGCCCATGTTGAATC-3′ | 95 °C | 30 s | 35 cycles | |
| 60 °C | 30 s | ||||
| Ang2 | FW | 5′-TTAGCACAAAGGATTCGGACAAT-3′ | 72 °C | 1 min | |
| Rev | 5′-TTTTGTGGGTAGTACTGTCCATTCA-3′ | 72 °C | 1 min | Final extension | |
| Composition of MRS Media | Bacterial Lysate | p-Value | |
|---|---|---|---|
| Vaccenic acid | 0 ± 0 | 6.834 ± 0.023 | <0.001 |
| S-Adenosylmethionine | 0 ± 0 | 6.759 ± 0.050 | <0.001 |
| Oleic acid | 0 ± 0 | 5.014 ± 0.041 | <0.001 |
| N,N-Dimethylformamide | 0 ± 0 | 5.206 ± 0.057 | <0.001 |
| Methylthioadenosine | 0 ± 0 | 7.305 ± 0.027 | <0.001 |
| Indole lactic acid | 0 ± 0 | 7.442 ± 0.088 | <0.001 |
| GABA (gamma-aminobutyric acid) | 0 ± 0 | 5.425 ± 0.068 | <0.001 |
| Citrate | 0 ± 0 | 6.443 ± 0.085 | <0.001 |
| Betaine | 0 ± 0 | 5.802 ± 0.102 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.; Kim, Y.; Lee, S.; Kim, Y.; Jeon, B.; Kim, H.; Park, H. Live Biotherapeutic Lactococcus lactis GEN3013 Enhances Antitumor Efficacy of Cancer Treatment via Modulation of Cancer Progression and Immune System. Cancers 2022, 14, 4083. https://doi.org/10.3390/cancers14174083
Kim S, Kim Y, Lee S, Kim Y, Jeon B, Kim H, Park H. Live Biotherapeutic Lactococcus lactis GEN3013 Enhances Antitumor Efficacy of Cancer Treatment via Modulation of Cancer Progression and Immune System. Cancers. 2022; 14(17):4083. https://doi.org/10.3390/cancers14174083
Chicago/Turabian StyleKim, Sujeong, Yunjae Kim, Suro Lee, Yulha Kim, Byungkwan Jeon, Hyerim Kim, and Hansoo Park. 2022. "Live Biotherapeutic Lactococcus lactis GEN3013 Enhances Antitumor Efficacy of Cancer Treatment via Modulation of Cancer Progression and Immune System" Cancers 14, no. 17: 4083. https://doi.org/10.3390/cancers14174083
APA StyleKim, S., Kim, Y., Lee, S., Kim, Y., Jeon, B., Kim, H., & Park, H. (2022). Live Biotherapeutic Lactococcus lactis GEN3013 Enhances Antitumor Efficacy of Cancer Treatment via Modulation of Cancer Progression and Immune System. Cancers, 14(17), 4083. https://doi.org/10.3390/cancers14174083





