Simple Summary
In this review, we present current knowledge of LncRNA NEAT1 in the development of malignant diseases. First, we illustrate the structure and functions of NEAT1 in cancer. Then we explore the interaction between NEAT1 and other molecules and analyze the impact of this interaction on cancer progression. Finally, we summarize the regulation of NEAT1 and the function of NEAT1-containing exosomes. Elucidating the molecular interaction of NEAT1 may shed light on future treatment of cancer.
Abstract
As one of the best-studied long noncoding RNAs, nuclear paraspeckle assembly transcript 1 (NEAT1) plays a pivotal role in the progression of cancers. NEAT1, especially its isoform NEAT1-1, facilitates the growth and metastasis of various cancers, excluding acute promyelocytic leukemia. NEAT1 can be elevated via transcriptional activation or stability alteration in cancers changing the aggressive phenotype of cancer cells. NEAT1 can also be secreted from other cells and be delivered to cancer cells through exosomes. Hence, elucidating the molecular interaction of NEAT1 may shed light on the future treatment of cancer. Herein, we review the molecular function of NEAT1 in cancer progression, and explain how NEAT1 interacts with RNAs, proteins, and DNA promoter regions to upregulate tumorigenic factors.
1. Introduction
Approximately 80% of the human genome is transcribed into RNA, but less than 2% can be translated into proteins [1]. Therefore, the majority of transcribed RNA belongs to noncoding RNAs (ncRNAs), of which those with lengths over 200 nucleotides are termed long noncoding RNAs (lncRNAs) [2].
According to various transcription patterns, lncRNAs can be categorized as enhancer lncRNAs, promoter upstream transcripts (PROMPTs), exon or intron sense-overlapping lncRNAs, long intergenic ncRNAs (lincRNAs), bidirectional lncRNAs, and natural antisense transcripts (NATs) (Figure 1A) [2,3]. Among the various types of lncRNAs, the best-studied are the lincRNAs transcribed from DNA sequences between two genes [3].
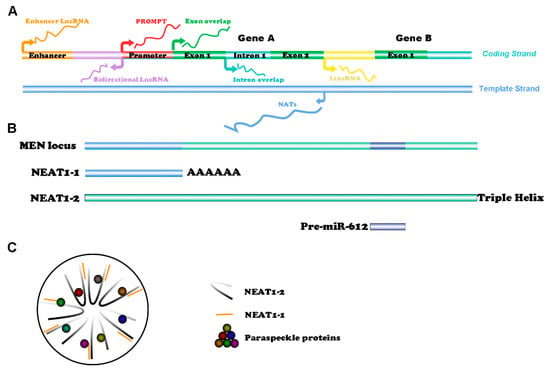
Figure 1.
Classification of lncRNAs and structure of NEAT1: (A) LncRNAs can be categorized as enhancer lncRNAs, promoter upstream transcripts (PROMPTs), exon or intron sense-overlapping lncRNAs, long intergenic ncRNAs (lincRNAs), bidirectional lncRNAs, and natural antisense transcripts (NATs); (B) Locations of NEAT1-1, NEAT1-2, and Pre-miR-612 at the MEN locus; (C) Cross-sectional structure of paraspeckles.
Nuclear paraspeckle assembly transcript 1 (NEAT1), which is transcribed from the multiple endocrine neoplasia (MEN) locus on chromosome 11q13.1, is a well-studied lincRNA in the field of cancer diseases [4,5]. Since NEAT1 belongs to the family of nucleotides, it has the ability to interact with other nucleotides, including mRNA, miRNA, or DNA, to change the expression of downstream proteins. In addition, the secondary structure of NEAT1 can capture some proteins and change their stability or molecular function [6].
Studies have shown that NEAT1 contributes to the progression of many cancers, including breast cancer, lung cancer, hepatocellular carcinoma, ovarian cancer, and prostate cancer [7,8,9,10]. A higher level of NEAT1 almost always indicates a poorer survival rate in cancer patients [11,12,13]. NEAT1 can also reduce cancers’ sensitivity to chemotherapy or radiotherapy [14,15]. However, NEAT1 overexpression seems to be a protective factor in acute myeloid leukemia (AML), and lower NEAT1 levels are responsible for differentiation disorders in AML [16,17].
Previous reviews of NEAT1 mainly depicted the interaction between NEAT1 and miRNAs and discussed other mechanisms in very short paragraphs. In our review, however, we describe the less-concerned DNA binding capacity of NEAT1 and place more emphasis on RNA-binding proteins. First, we illustrate the structure and functions of NEAT1 in cancer. Then, we explore the interaction between NEAT1 and other molecules and analyze the impact of this interaction on cancer progression. Finally, we summarize the regulation of NEAT1 and the function of NEAT1-containing exosomes. As an important diagnostic and therapeutic marker, further elucidation of the mechanisms of NEAT1 in cancer progression will undoubtedly benefit clinical work [18].
2. Structure of NEAT1 and Paraspeckles
NEAT1 has two transcription variants, namely, NEAT1-1 (3756 nt) and NEAT1-2 (22,743 nt) [19] (Figure 1B). Specifically, the shorter transcript NEAT1-1 (also known as MENepsilon) is stabilized by a canonical poly (A) tail at the 3′ end, while the longer transcript NEAT1-2 (also known as MENbeta) does not have a poly (A) tail and is stabilized by a triple helical structure at the 3′ end [20,21]. These two isoforms are produced by alternative 3′-end processing, and the sequence of NEAT1-1 can be regarded as part of NEAT1-2 [19]. Accordingly, siRNA, shRNA, and PCR primers designed to knockdown or detect NEAT1-1 actually target both of the isoforms of NEAT1, namely, NEAT1-1 and NEAT1-2. When the polyadenylation of NEAT1-1 is inhibited, the transcription process continues at the gene locus, forming the longer transcript NEAT1-2 [19,22]. Interestingly, the pre-miR-612 gene is also located at the MEN locus and shares an overlapping sequence with NEAT1-2. Mature miR-612 is a tumor-suppressive gene generated by alternative splicing, and the balance between the two NEAT1 transcripts and miR-612 can be regulated by polypyrimidine tract binding protein 3 (PTBP3) [23]. The two NEAT1 isoforms and certain RNA-binding proteins (RBPs) together form a pivotal nuclear structure named a paraspeckle (Figure 1C). NEAT1-2 plays a more crucial role in the formation of paraspeckles, while NEAT1-1 has been predicted to bind with the 3′ end of NEAT1-2 due to their complementary base pairs [24]. More than 50 RBPs, including paraspeckle component 1 (PSPC1), non-POU domain-containing octamer-binding protein (NONO/p54nrb), and splicing factor proline/glutamine-rich (SFPQ/PSF), can be members of paraspeckles [25,26].
Paraspeckles exert their functions mainly through the retention or sequestration of RNAs or proteins [27]. Paraspeckles can capture A-to-I–edited mRNAs, limiting their export from the nucleus [28]. Zhang et al. showed that multiple I residues in the edited RNA can form a complex with PSF, NONO, and matrin 3, resulting in the retention of an RNP complex in the nucleus [29]. Generally, dsRNA-dependent adenosine deaminases (ADARs) can convert adenosine (A) into inosine (I), which will be identified as guanosine (G) in the translation process, forming a different peptide if this is not a synonymous substitution [30]. Even though there is a synonymous substitution, which does not change the type of coding proteins, it can affect the degradation velocity of mRNAs, subsequently regulating the expression level of the corresponding proteins [31]. Hence, paraspeckles can prevent the translation of these A-to-I–edited mRNAs to a certain extent. In addition, the formation or unraveling of paraspeckles will change the amount of paraspeckle component proteins (PSPs), such as PSPC1, SFPQ, and NONO, sequestered in the paraspeckles [25]. For instance, a lower number of paraspeckles sequester less SFPQ; thus, the released SFPQ will relocate to the promoter region of the IL-8 gene locus, inhibiting the transcription of IL-8 mRNA in HeLa cells [32]. Paraspeckles can also promote the immune escape of hepatocellular carcinoma by sequestrating IFNGR-1 mRNA, inhibiting IFN-γ induced T-cell killing effects [33].
3. NEAT1 Sponges microRNAs and Stabilizes mRNAs
NEAT1 can bind to various microRNAs (miRNAs/miRs) and sponged miRNAs cannot combine with mRNA in the RNA-induced silencing complex (RISC) [34]. If the 3′ untranslated region (3′UTR) of mRNA and miRNA are perfectly complementary, the resulting heterodimers can be degraded in the RISC [12]. In contrast, the sponged dysfunctional miRNA prevents the degradation of mRNA, causing larger amounts of mRNA to be translated into functional proteins [5]. This is the most common mechanism identified by researchers who have attempted to elucidate the functions of NEAT1 in cancer diseases.
As an endogenous competing RNA (ceRNA), NEAT1 can facilitate the progression of many cancers (Table 1). For instance, NEAT1 can sponge miR-302a-3p, which is a tumor-suppressive molecule that suppresses the translation of RELA mRNA, thereby upregulating RELA and increasing the migration and proliferation capacity of pancreatic cancer cells [12]. Notably, NEAT1 has two transcripts, and some miRNAs can only interact with NEAT1-2, such as miR-491 and miR-106b-5p, which promote the metastasis and growth of thyroid cancer [35]. In this section, we mainly discuss the ceRNA network in various cancers.

Table 1.
NEAT1-miRNA-mRNA network involved in progression of cancer diseases.
Breast cancer is the leading cause of cancer death in the female population all over the world and its incidence rates are gradually increasing [36]. The expression of NEAT1 is elevated in breast cancer cell lines, including MCF-7, MDA-MB-453, MDA-MB-231, and SKBR3 cells, compared with normal mammary epithelial cells such as MCF-10A [37]. Moreover, the expression of NEAT1 is higher in cancer tissues compared with the normal adjacent tissues [38]. NEAT1 can upregulate many oncogenic factors, such as KLF12, Cyclin D1, ZEB1, and HMGA2, which subsequently promote the proliferation and metastasis of breast cancer [37,38,39,40,41,42].
Colorectal cancer is an important cause of global cancer-related death due to its late diagnosis and rapid metastasis [13,43]. One of the biggest obstacles to treating colorectal cancer is the lack of a molecular biomarker. Studies have shown that NEAT1 expression is upregulated in colorectal cancer tissues, and a higher level of NEAT1 contributes to the poor overall survival and disease-free survival [44,45]. Moreover, NEAT1 can also promote the proliferation and metastasis of colorectal cancer in vivo and in vitro [44,45]. NEAT1 can elevate the expression of some growth factors and transcription factors such as IGF2, GDNF, and BACH1, which facilitate the progression of colorectal cancer [44,46,47].
Gastric cancer is the third leading cause of cancer-related death worldwide [48]. Although early treatment of gastric cancer has achieved significant success, early diagnosis is still difficult; most patients present with an advanced stage at the time of diagnosis, causing a poor survival rate [49,50,51]. Hence, identifying novel biomarkers is important to improve the diagnosis and treatment of gastric cancer. NEAT1 is significantly upregulated in human gastric cancer cells, including BGC823, SGC-7901, AGS, MGC803, and MKN28 cells, compared with normal gastric epithelial cells such as GES-1 [52]. Studies have shown that NEAT1 can upregulate the expression of ABCC4, JAG1, SOX11, COX-2, etc. These factors will finally promote the proliferation, migration, and invasion of gastric cancer [49,53,54,55].
Glioma is the most prevalent primary central nervous system tumor, which has a high postoperative recurrence and mortality rate [56]. Although glioma can be treated by surgery, it is always hard to excise completely [57]. Studies have shown that NEAT1 is upregulated in glioma tissues compared with adjacent normal tissues [57,58]. NEAT1 can elevate the expression of many molecules, including DNMT1, CCT6A, CDK6, SOX2, and c-MET in glioma cells. Therefore, a higher level of NEAT1 can promote the proliferation, migration, and epithelial–mesenchymal transition (EMT) process in glioma cells [56,57,58,59,60].
Hepatocellular carcinoma causes approximately 600,000 deaths each year. It is the sixth most common cancer all over the world and the third leading cause of cancer-related death [61,62]. Chronic hepatitis B or C virus infections are the pivotal development factors of hepatocellular carcinoma [63]. Frequent intrahepatic and extrahepatic metastases contribute to the low resectability and high recurrence rate even in the early stage of this disease [64]. NEAT1 is upregulated in hepatocellular carcinoma tissues and cell lines compared with normal adjacent tissues and normal cell lines [8]. NEAT1 can elevate the expression of many factors, including SMO, LAGE3, AKT2, TGF-β1, and STAT3, which can increase the proliferation and metastasis both in vivo and in vitro, accelerating the progression of hepatocellular carcinoma [61,63,64,65,66]. NEAT1 can also sponge miR-124-3p, elevating the expression of ATGL1, which promotes the lipolysis and cancer progression in hepatocellular carcinoma [67].
Lung cancer is one of the most prevalent malignant tumors and the leading cause of cancer-related death worldwide [68]. About 80% of lung cancers are non-small cell lung cancers (NSCLC) [69]. Moreover, the 5-year survival rate of NSCLC patients still remains unsatisfactory. Studies have shown that the 5-year survival rate of late NSCLC patients is less than 15% [70]. The upregulation of NEAT1 in lung cancer tissues has been observed in several studies [68,69,71]. NEAT1 can promote the proliferation and metastasis of NSCLC in vivo and in vitro by upregulating the expression of many oncogenes, including NAUK1, SULF1, SOX9, and E2F3 [68,69,70,71]. Higher NEAT1 levels are also associated with more lymph node metastasis, higher TNM grades, and a poor overall survival in patients [70].
Ovarian cancer is a common malignant tumor of the female reproductive system and has a high mortality rate [10]. As the early symptoms are not obvious, the disease onset is hidden, and metastasis occurs earlier. About 47% of women diagnosed with ovarian cancer can survive 5 years after diagnosis [72]. Yin et al. [73] reported that NEAT1 is upregulated in ovarian tumors compared with the adjacent tissues. Moreover, the expression level of NEAT1 is higher in ovarian cancer cell lines including ES2, A2780, HO8910, and SKOV3 than the normal ovarian epithelial cell line IOSE80. Clinical data showed that highly-expressed NEAT1 is closely correlated with a shorter survival rate, a poor differentiation degree, larger tumors, an advanced FIGO stage, and significant peritoneal metastasis in patients with ovarian cancer [10]. NEAT1 can upregulate the expression of TJP3, MEST, and ROCK1 in ovarian cancer cell lines, subsequently promoting the progression of ovarian cancer [10,72,73]. NEAT1 can also regulate glucose metabolism to promote ovarian cancer’s progression. For instance, the knockdown of NEAT1 has been shown to upregulate miR-4500 and downregulate BZW1, subsequently inhibiting the glycolysis of ovarian cancer [74].
Prostate cancer is the second most common malignant tumor and the fifth leading cause of cancer-related death among the male population worldwide [75]. Guo et al. [75] found that an overexpression of NEAT1 is correlated with an advanced clinical stage of disease. After detecting the expression of NEAT1 in tissues and cell lines, they observed that NEAT1 was significantly upregulated in prostate cancer tissues and cell lines compared with normal tissues and the normal prostate epithelial cell line. A study showed that NEAT1 can promote the proliferation and invasion of ovarian cancer cells via the upregulation of HMGA2 [75]. Moreover, NEAT1 can elevate the expression ACSL4, enhancing docetaxel resistance in PCa cells in vitro and in vivo [76]. Hence, NEAT1 promotes the progression of prostate cancer and may be a therapeutic biomarker in clinical studies.
The contribution of NEAT1 to cancer progression has also been studied in other cancers, including cervical cancer [77], cholangiocarcinoma [78], endometrial cancer [79], esophageal carcinoma [80], gallbladder cancer [81], glioblastoma [82], Hodgkin’s lymphoma [83], laryngeal carcinoma [84], melanoma [85], multiple myeloma [11], nasopharyngeal carcinoma [86], osteosarcoma [87], pancreatic carcinoma [88], retinoblastoma [89], thyroid cancer [90], and tongue carcinoma [91]. In those cancers, NEAT1 plays a pro-proliferative role and promotes the progression of cancer by upregulating the expression of some oncogenes or downregulating the expression of some tumor suppressor genes. For example, a study showed that the knockdown of NEAT1 can sensitize 5-FU resistance in cervical cancer and inhibit its glycolysis via the miR-34a/LDHA axis [92].
Interestingly, NEAT1 exerts an opposite function in acute myeloid leukemia. As an important hematologic malignant disease, acute myeloid leukemia is marked by the abnormal abundance of clonal myeloid progenitor cells in the bone marrow and the inhibition of normal hematopoiesis [16]. NEAT1 upregulation suppresses cell growth, migration, and invasion but enhances the apoptosis of acute myeloid leukemia cells [17]. Furthermore, the expression of NEAT1 is downregulated in acute myeloid tissues compared with normal tissues [93]. In patients with acute myeloid leukemia, NEAT1 is downregulated and subsequently decreases the expression of CREBRF, promoting the progression of the disease [17].
Interestingly, NEAT1 can directly interact with mRNA to affect its stability without the participation of miRNA. NEAT1 can bind and stabilize TNFRSF1B mRNA, contributing to the NF-κB signaling pathway [127]. Feng et al. [128] suggested that NEAT1 can also increase the stability of ELF3 mRNA by increasing its N6-methyladenosine (m6A) modification, promoting the proliferation and metastasis of pancreatic cancer.
4. NEAT1 Recruits Transcription Factors, Enzymes and Other Proteins
Due to the relatively few existing studies of NEAT1-binding proteins involved in cancers, we include several RBPs involved in other diseases to fully illustrate this mechanism. When LncRNA NEAT1 and some specific proteins together form a complex, the functions of NEAT1 can be divided into three types: a guide, decoy, and scaffold [18]. In this review, NEAT1-binding proteins are categorized as enzymes, transcription factors, and receptors (Figure 2).
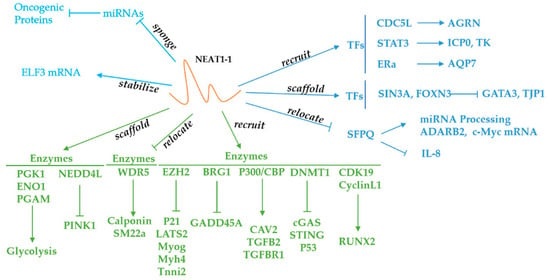
Figure 2.
Function and Signaling Pathways of NEAT1. NEAT1 can recruit enzymes and TFs to the genome locus. NEAT1 can also sponge miRs and relocate enzymes and SFPQ to suppress the function of these molecules. In addition, NEAT1 can form a scaffold bridge between different TFs, which allows the enzymes to bind more tightly to the substrates.
5. NEAT1-Binding Enzymes
Many RBPs can bind to NEAT1 to regulate gene expression, and they are categorized in Table 2. First, we will discuss the NEAT1-binding enzymes involved in cancer progression. NEAT1-binding enzymes mainly include DNA methyltransferase, histone methylase, and E3 ubiquitin protein ligase. Hence, epigenetic mechanisms play a pivotal role in the regulation of gene expression. For instance, NEAT1 can bind to DNA methyltransferase 1 (DNMT1) to facilitate the growth and metastasis of lung cancer by increasing DNA methylation in the promoter region of CGAS, STING, and P53 [129]. In addition, the knockdown of NEAT1 also promoted T cell infiltration in a xenograft tumor model [129]. EZH2, which is a subunit of the histone methylation transferase polycomb repressive complex 2 (PRC2), can also interact with NEAT1 [130]. EZH2 has the capacity to promote histone 3 lysine 27 trimethylation (H3K27me3), which is a transcriptional repressor of many tumor suppressor genes, such as GSK3B, ICAT, and Axin2 [131]. When NEAT1 recruits EZH2 to the promoter region of the genome locus, the repression of these genes contributes to the progression of cancers such as glioblastoma [131]. NEAT1 can also relocate EZH2 to the promoter regions of many other genes, such as P21, LATS2, MYOG, MYH4, and TNNI2 [130,132].

Table 2.
RNA-binding Proteins bound with NEAT1-regulating gene expression.
NEAT1 can also sequester WDR5, which is a component of the mixed-lineage leukemia (MLL) H3K4 methylase complex responsible for histone 3 lysine 4 trimethylation (H3K4me3) within the promoter regions of SM-specific genes [133]. Since H3K4me3 is an active histone modification, the upregulation of NEAT1 can inhibit the expression of SM-specific genes—such as SM22a and calponin—in smooth muscle cells via the sequestration of WDR5 [133]. Moreover, the authors found that a depletion of NEAT1 can increase histone 3 lysine 9 acetylation (H3K9ac) and decrease H3K27me3 modification within the SM22a and calponin promoter regions, promoting gene transcription. Another study showed that NEAT1 can interact with the P300/CBP complex to affect the histone modification of H3K27 [134]. The inhibition of NEAT1 decreased H3K27ac but increased histone 3 lysine 27 crotonylation (H3k27Cro), downregulating the expression of endocytosis-related genes and aggravating Alzheimer’s disease [134]. Similarly, BRG-1, which is a subunit of SWI/SNF, can interact with NEAT1; thus, BRG-1 can be recruited to the promoter region of GADD45A, increasing the level of H3K27me3 and decreasing the level of H3K4me3 [135]. As a result, the transcription of GADD45A is inactivated, and gastric cancer is aggravated [135].
TRAF6, which is an E3 ubiquitin protein ligase, can also be pulled down by NEAT1 [139]. The autoubiquitination of TRAF6 can be inhibited by NEAT1, and an elevated TRAF6 level alleviates acute-on-chronic liver failure [139]. Moreover, NEAT1 can interact with another E3 ubiquitin protein ligase, NEDD4L, promoting the ubiquitination-mediated degradation of PINK1, which plays a crucial role in the maintenance of mitochondrial integrity via mitophagy [138]. As a consequence, downregulated PINK1 impairs mitochondrial function and promotes Alzheimer’s disease [138]. NEAT1 can also stabilize STAT3 by inhibiting the ubiquitination-mediated degradation of STAT3 in CD4+ T cells [140].
Recently, NEAT1 has also been shown to interact with cyclin-dependent kinase and glucose metabolism-related enzymes. In prostate cancer, NEAT1 can bind with cyclinL1/CDK19 to promote cancer metastasis [137]. In breast cancer, NEAT1 can bind and form a scaffold bridge for the assembly of phosphoglycerate kinase (PGK), phosphoglycerate mutase (PGAM), and enolase (ENO), elevating glycolysis and cancer metastasis [136].
6. NEAT1-Binding Transcription Factors
NEAT1 is associated with many transcription factors and relocates them to or sequesters them from the promoter regions of specific genes, activating or inactivating gene transcription [137,142]. For example, NEAT1 can interact with CDC5L and recruit it to the promoter region of AGRN, facilitating prostate cancer’s progression [141]. Furthermore, NEAT1 can exert a bridge function between two proteins, and the junction between two proteins can be inhibited by RNase. For example, NEAT1 can form an indispensable bridge between SIN3A and FOXN3. FOXN3 only has a transcription-activating domain and cannot bind to the DNA promoter region, whereas SIN3A only has a DNA-binding domain [36]. Therefore, the combination of these two proteins is essential for normal transcription. The SIN3A–NEAT1–FOXN3 complex ultimately regulates the downstream transcription of GATA3 and TJP1, enhancing epithelial-to-mesenchymal transition (EMT) in breast cancer [36]. Nuclear receptors are considered a special type of transcription factor that translocate from the cellular membrane to the nucleus after pairing with ligands. NEAT1 can interact with the nuclear receptor ERa, and then the recruited ERa binds to the promoter region of AQP7, promoting steatosis in hepatic cancer cells [143].
Another important factor is SFPQ, which can bind to the promoter region of IL-8 and ADARB2 to downregulate the transcription of IL-8 or upregulate the transcription of ADARB2 [20,32]. SFPQ is a classical paraspeckle protein with two RNA recognition motifs (RRMs), and NEAT1 can sequester it from the promoter regions of genes, inhibiting its transcriptional function [32]. Lee et al. suggested that there is a putative DNA-binding domain in the structure of SFPQ, but the current studies are not sufficient to support this molecule as a transcription factor [149]. SFPQ can also promote miRNA processing by binding with Drosha/DGCR8 in the NEAT1 scaffold [144]. Strikingly, SFPQ can regulate mRNA translation. NEAT1 can sequester SFPQ and block its binding with the internal ribosome entry segment (IRES) of c-Myc mRNA, thus inhibiting mRNA translation [145]. Interestingly, the paraspeckle has more than 50 types of proteins, including SFPQ, NONO, and FUS/TLS, which means that these proteins can interact with NEAT1 in the nucleus. However, how this interaction contributes to cancer progression still needs further study [150]. Additionally, transcription coactivators can also interact with NEAT1. For instance, DDX5 can promote colorectal cancer metastasis by interacting with NEAT1 and β-catenin [13].
Finally, certain proteins related to the spliceosome, such as SRSF5 and U2AF65, can interact with NEAT1. After binding with NEAT1, SRSF5 can regulate PPAR-γ mRNA splicing, increasing the PPAR-γ2 isoform expression in adipocytes [147]. U2AF65-mediated hnRNPA2 elevation also depends on the existence of NEAT1, facilitating the progression of hepatocellular carcinoma [148].
7. NEAT1 Interacts with DNA Sequences to Regulate Transcription
When we indicated that NEAT1 can recruit proteins such as EZH2 to the promoter regions of specific genes, we assumed that NEAT1 had a guide function. However, how NEAT1 guides EZH2 to the promoter region of specific genes has not been discussed. It is difficult to determine whether NEAT1 exerts a guidance function or only provides a scaffold and translocates with EZH2.
With designed biotin-labeled primer sets for NEAT1, the NEAT1-binding DNA sequence can be pulled down by streptavidin magnetic beads using many methods, such as chromatin isolation by RNA purification (CHIRP), the capture hybridization analysis of RNA targets (CHART), and RNA affinity purification (RAP) [151].
Researchers have suggested many NEAT1-binding elements after the sequencing DNA segments precipitated in this way. CHART-seq suggested that NEAT1 can bind to the transcriptional start sites and transcriptional termination sites of its target genes [151]. Moreover, Wen et al. predicted a binding site between NEAT1 and the promoter region of RUNX2 through an algorithm analyzing the triplex binding capacity of NEAT1 to DNA duplexes. Then, the predicted binding site was verified by CHIRP-qPCR using the predicted primers. CDK19 and cyclinL1 are responsible for the phosphorylation of RNA pol II to promote the transcription of genes [137]. However, this study showed that the junction of the RUNX2 promoter region with CDK19 or Cyclin L1 requires the existence of NEAT1, supporting the notion of a direct binding between NEAT1 and DNA sequences [137].
Similar to NEAT1, lncRNA HOTAIR can also recruit EZH2 to the promoter regions of many genes to regulate gene transcription. Its recruitment capacity was further explored by CHIRP-seq, and the analysis of HOTAIR binding sequences revealed an enrichment of GA-rich motifs [152]. After the EZH2 depletion, however, the profile of HOTAIR occupancy on the genome locus was not altered [152]. Therefore, HOTAIR can bind to DNA sequences without EZH2 and can directly bind to GA-rich motifs in DNA segments.
RNA–DNA interactions are rarely studied in relation to NEAT1, and future studies are still needed to explore the corresponding detailed mechanism.
8. Factors That Regulate NEAT1 Expression
The expression of NEAT1 is mainly regulated in three ways: transcriptional regulation, alternative splicing, and stability alteration. Studies have shown that some transcription factors can directly bind to the promoter region of NEAT1 to regulate its expression. Specifically, NF-κB, OCT4, HIF-2a, and c-Myc can promote the transcription of NEAT1 in pancreatic cancer, lung cancer, breast cancer, and colorectal cancer, respectively [12,45,153,154]. Subsequently, the elevation of NEAT1 contributes to cancer progression (Figure 3A). Moreover, STAT3 can upregulate the expression of NEAT1 in HeLa cells to increase the viral replication of herpes simplex virus-1 (HSV-1) [142]. In contrast, some transcription factors can inhibit the expression of NEAT1 in cancer cells and immune cells. Lo et al. showed that BRCA1 can downregulate the expression of NEAT1 to suppress the progression of breast cancer [96]. NEAT1 can also be inhibited by E2F1, which decreases the H3K27ac level at the NEAT1 promoter, finally inducing a tolerogenic phenotype of dendritic cells [155].
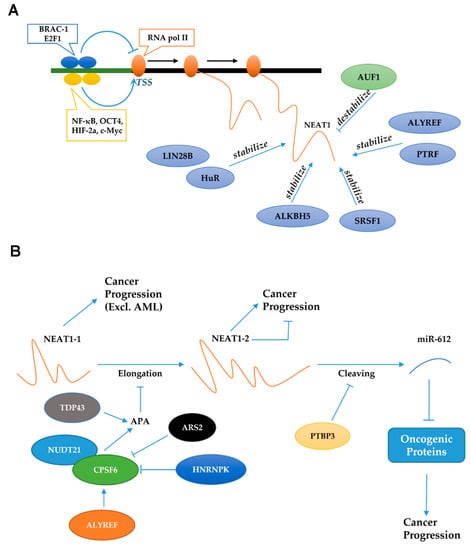
Figure 3.
Regulation of NEAT1 at transcriptional level (A) and posttranslational level (B). (A) After being regulated by transcriptional activator or repressor, NEAT1 can also be stabilized or destabilized by some proteins. (B) When the polyadenylation (APA) of NEAT1-1 is activated, the transcription site cannot cross the end of NEAT1-1. Consequently, the expression level of NEAT1-1 will be elevated, and the generation of NEAT1-2 will be inhibited. In addition, PYBP3 can inhibit the cleaving of NEAT1-2 to reduce the generation of miR-612, thereby promoting cancer progression.
In addition, some molecules can regulate the ratio of the two isoforms of NEAT1, namely, the long transcript NEAT1-2 and the short transcript NEAT1-1. When the polyadenylation of NEAT1-1 is activated, the transcription site cannot cross the end of NEAT1-1 (Figure 3B). Consequently, the expression level of NEAT1-1 will be elevated, and the generation of NEAT1-2 will be inhibited. Specifically, TDP-43 can increase polyadenylation levels, upregulating NEAT1-1, and downregulating NEAT1-2 in mouse embryonic cells. Finally, the formation of paraspeckles is repressed and pluripotent cells can maintain their pluripotency and embryonic patterning [156]. In mouse embryonic fibroblasts, CPSF and NUDT21 can form a heterodimer to promote alternative polyadenylation (APA); the formed poly-A tail can stabilize NEAT1-1 and inhibit the elongation of NEAT1-2 [150]. However, HNRNPK and ARS2 can inhibit this heterodimer and exert the opposite function [22,150]. After NEAT1-2 has been transcribed completely, RNase P can recognize the t-RNA-like structure and cleaves it to form the non-polyadenylated 3′-end of NEAT1-2 [157].
In breast cancer, ALYREF can stabilize CPSF to elevate the NEAT1-1 isoform, promoting carcinogenesis [158]. Furthermore, another important protein, PTBP3, can increase the expression of NEAT1-2 and total NEAT1 but decrease the induction of miR-612, which has an overlapping sequence with NEAT1-2, as mentioned above. Finally, PTBP3 promotes the growth and metastasis of hepatocellular carcinoma [23]. Obviously, NEAT1-1 almost always shows an oncogenic function in cancers excluding AML. However, the function of NEAT1-2 in cancer is controversial and there are only a few relevant studies. In colorectal cancer, a study showed that NEAT1-2 can inhibit caner proliferation [159]. In contrast, another study showed that a higher level of NEAT1-2 indicates a poor survival rate and can promote proliferation and glycolysis in hepatocellular carcinoma [160]. NEAT1-2 has also been shown to promote the metastasis of thyroid cancer [35].
Specific proteins are responsible for the stability of NEAT1, including HuR, LIN28B, ALKBH5, PTRF, ALYREF, and SRSF1 [158,161,162,163,164,165]. Specifically, HuR and LIN28B can promote ovarian cancer progression via stabilizing NEAT1 [161,164]. In glioblastoma, PTRF can increase the stability of NEAT1 to promote cancer metastasis [165]. In addition, ALKBH5-stabilized NEAT1 can significantly suppress hypoxia-induced tumor-associated macrophage (TAM) recruitment and immunosuppression in glioblastoma [163]. ALYREF and SRSF1 can also prolong the half-life of NEAT1, facilitating the progression of breast cancer and glioma, respectively [158,162]. In contrast, AUF1 can destabilize NEAT1 and accelerate its decay in HeLa cells [166]. Stability alterations can ultimately affect the expression level of NEAT1 transcripts, thereby regulating cancer progression.
9. NEAT1-Containing Exosomes in Cancer
Prokaryotic and eukaryotic cells can generate extracellular vesicles (EVs) and release them into the extracellular environment [167]. EVs can be divided into exosomes and ectosomes. Ectosomes are released via cellular membrane budding with a size range of 50 to 1000 nm, whereas exosomes are generated by endocytosis [168]. The sequential invagination of the cell membrane ultimately forms multivesicular bodies (MVBs), which can intersect with other intracellular vesicles and organelles. When MVBs fuse with the cell membrane, exosomes are released (size range 40 to 160 nm). Exosomes contain many components, including proteins, DNA, RNA, lipids, and metabolites [168].
Exosomal NEAT1 plays an important role in cancers and may serve as a diagnostic or treatment marker. In the exosomes extracted from patients with ovarian cancer, NEAT1 expression levels are upregulated and may contribute to cisplatin resistance [169]. Another study showed that NEAT1 expression is increased in exosomes derived from cancer-associated fibroblasts (CAFs) compared with exosomes derived from normal fibroblasts. Moreover, exosomal NEAT1 derived from cancer-associated fibroblasts (CAFs) promotes the progression of endometrial cancer [79]. Mo et al. found that prostate cancer cell-derived exosomes induce the osteogenesis of mesenchymal stem cells via their NEAT1 component [170]. Additionally, exosomes derived from mesenchymal stem cells pretreated with macrophage migration inhibitory factor (MIF), namely, exosomeMIF, can alleviate doxorubicin-induced cardiac senescence in chemotherapy [171]. Only a few studies have explored the functions of NEAT1 in exosomes; therefore, the detailed mechanisms of NEAT1 in cancers remain relatively ambiguous.
10. Conclusions and Perspectives
NEAT1 generally exerts oncogenic effects, promoting the progression of various cancers, excluding AML, in which NEAT1 facilitates the differentiation of leukocytes to mitigate the progression of the disease. NEAT1 can bind with miRNAs to upregulate the corresponding mRNAs in the cytosol via a ceRNA mechanism. Moreover, NEAT1 can interact with certain transcription factors, enzymes, or nuclear receptors to regulate transcription, histone modification, and protein stability. NEAT1 exerts its guide function by recruiting these proteins and exerts its decoy function by sequestering these factors. NEAT1 can also exert its scaffold function by providing a reactive platform for these molecules. Consequently, many tumorigenic factors are elevated, facilitating the progression of cancers.
There are many studies related to NEAT1, but how NEAT1 plays the role of ceRNA remains ambiguous. NEAT1 is transcribed in the nucleus and is involved in the assembly of paraspeckles. However, NEAT1 only exerts its sponge function after it is transported out of the nucleus. Further studies are needed to determine which proteins containing nuclear export sequences can interact with NEAT1. Consequently, NEAT1 is translocated through the nuclear pore complex under the influence of a specific exportin and enters the cytoplasm. In addition, there is a close relationship between paraspeckles and cell differentiation, but researchers scarcely focus on the formation of paraspeckles in cancers. Chen et al. reported that paraspeckles are absent in undifferentiated embryonic stem cells, but the number of paraspeckles gradually increases after the induction of differentiation [172]. The paraspeckles can retain many molecules related to cell stemness maintenance, thereby inhibiting cell stemness [156]. Therefore, not only the knockdown or knockout of NEAT1 but also the induction of paraspeckles may play an enlightening role in the future treatment of cancer.
Author Contributions
Conceptualization, J.G., B.Z. and R.A.; writing—original draft preparation, J.G., W.Q. and W.D.; writing—review and editing, L.H., Q.M. and Z.W. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by National Natural Science Foundation of China, grant number 82072699.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Kopp, F.; Mendell, J.T. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Yang, L.; Chen, L.L. The Diversity of Long Noncoding RNAs and Their Generation. Trends Genet. 2017, 33, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Mello, S.S.; Sinow, C.; Raj, N.; Mazur, P.K.; Bieging-Rolett, K.; Broz, D.K.; Imam, J.F.C.; Vogel, H.; Wood, L.D.; Sage, J.; et al. Neat1is a p53-inducible lincRNA essential for transformation suppression. Genes Dev. 2017, 31, 1095–1108. [Google Scholar] [CrossRef]
- Yu, X.; Li, Z.; Zheng, H.; Chan, M.T.; Wu, W.K. NEAT1: A novel cancer-related long non-coding RNA. Cell Prolif. 2017, 50, e12329. [Google Scholar] [CrossRef]
- Liang, J.; Liu, C.; Xu, D.; Xie, K.; Li, A. LncRNA NEAT1 facilitates glioma progression via stabilizing PGK1. J. Transl. Med. 2022, 20, 80. [Google Scholar] [CrossRef]
- Klec, C.; Prinz, F.; Pichler, M. Involvement of the long noncoding RNA NEAT1 in carcinogenesis. Mol. Oncol. 2019, 13, 46–60. [Google Scholar] [CrossRef]
- Fang, L.; Sun, J.; Pan, Z.; Song, Y.; Zhong, L.; Zhang, Y.; Liu, Y.; Zheng, X.; Huang, P. Long non-coding RNA NEAT1 promotes hepatocellular carcinoma cell proliferation through the regulation of miR-129-5p-VCP-IκB. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 313, G150–G156. [Google Scholar] [CrossRef]
- Xiong, W.; Huang, C.; Deng, H.; Jian, C.; Zen, C.; Ye, K.; Zhong, Z.; Zhao, X.; Zhu, L. Oncogenic non-coding RNA NEAT1 promotes the prostate cancer cell growth through the SRC3/IGF1R/AKT pathway. Int. J. Biochem. Cell Biol. 2018, 94, 125–132. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.; Fu, X.; Lu, Z. Long non-coding RNA NEAT1 promoted ovarian cancer cells’ metastasis through regulation of miR-382-3p/ROCK1 axial. Cancer Sci. 2018, 109, 2188–2198. [Google Scholar] [CrossRef]
- Yu, H.; Peng, S.; Chen, X.; Han, S.; Luo, J. Long non-coding RNA NEAT1 serves as a novel biomarker for treatment response and survival profiles via microRNA-125a in multiple myeloma. J. Clin. Lab. Anal. 2020, 34, e23399. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Yi, Z.J.; Ou, Z.L.; Han, T.; Wan, T.; Tang, Y.C.; Wang, Z.C.; Huang, F.Z. RELA/NEAT1/miR-302a-3p/RELA feedback loop modulates pancreatic ductal adenocarcinoma cell proliferation and migration. J. Cell Physiol. 2019, 234, 3583–3597. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Weng, W.; Zhang, Q.; Wu, Y.; Ni, S.; Tan, C.; Xu, M.; Sun, H.; Liu, C.; Wei, P.; et al. The lncRNA NEAT1 activates Wnt/beta-catenin signaling and promotes colorectal cancer progression via interacting with DDX5. J. Hematol. Oncol. 2018, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Wu, X.; Wang, X.; Huang, W.; Feng, Q. NEAT1 upregulates EGCG-induced CTR1 to enhance cisplatin sensitivity in lung cancer cells. Oncotarget 2016, 7, 43337–43351. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Jiang, G.; Ren, W.; Wang, B.; Yang, C.; Li, M. LncRNA NEAT1 regulates 5-fu sensitivity, apoptosis and invasion in colorectal cancer through the MiR-150-5p/CPSF4 axis. OncoTargets Ther. 2020, 13, 6373–6383. [Google Scholar] [CrossRef]
- Zeng, C.; Xu, Y.; Xu, L.; Yu, X.; Cheng, J.; Yang, L.; Chen, S.; Li, Y. Inhibition of long non-coding RNA NEAT1 impairs myeloid differentiation in acute promyelocytic leukemia cells. BMC Cancer 2014, 14, 693. [Google Scholar] [CrossRef]
- Feng, S.; Liu, N.; Chen, X.; Liu, Y.; An, J. Long non-coding RNA NEAT1/miR-338-3p axis impedes the progression of acute myeloid leukemia via regulating CREBRF. Cancer Cell Int. 2020, 20, 112. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Yue, J.; Hanley, S.J.B.; Kobayashi, N.; Todo, Y.; Watari, H. Long Non-coding RNA NEAT1: A Novel Target for Diagnosis and Therapy in Human Tumors. Front. Genet. 2018, 9, 471. [Google Scholar] [CrossRef]
- Naganuma, T.; Hirose, T. Paraspeckle formation during the biogenesis of long non-coding RNAs. RNA Biol. 2013, 10, 456–461. [Google Scholar] [CrossRef]
- Hirose, T.; Virnicchi, G.; Tanigawa, A.; Naganuma, T.; Li, R.; Kimura, H.; Yokoi, T.; Nakagawa, S.; Bénard, M.; Fox, A.H.; et al. NEAT1 long noncoding RNA regulates transcription via protein sequestration within subnuclear bodies. Mol. Biol. Cell 2014, 25, 169–183. [Google Scholar] [CrossRef]
- Wang, Z.; Li, K.; Huang, W. Long non-coding RNA NEAT1-centric gene regulation. Cell Mol. Life Sci. 2020, 77, 3769–3779. [Google Scholar] [CrossRef] [PubMed]
- Machitani, M.; Taniguchi, I.; Ohno, M. ARS2 Regulates Nuclear Paraspeckle Formation through 3′-End Processing and Stability of NEAT1 Long Noncoding RNA. Mol. Cell. Biol. 2020, 40, e00269–19. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Qu, S.; Wang, L.; Zhang, H.; Yang, Z.; Wang, J.; Dai, B.; Tao, K.; Shang, R.; Liu, Z.; et al. PTBP3 splicing factor promotes hepatocellular carcinoma by destroying the splicing balance of NEAT1 and pre-miR-612. Oncogene 2018, 37, 6399–6413. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Schmidt, B.F.; Bruchez, M.P.; McManus, C.J. Structural analyses of NEAT1 lncRNAs suggest long-range RNA interactions that may contribute to paraspeckle architecture. Nucleic Acids Res. 2018, 46, 3742–3752. [Google Scholar] [CrossRef] [PubMed]
- Prinz, F.; Kapeller, A.; Pichler, M.; Klec, C. The Implications of the Long Non-Coding RNA NEAT1 in Non-Cancerous Diseases. Int. J. Mol. Sci. 2019, 20, 627. [Google Scholar] [CrossRef] [PubMed]
- Nishimoto, Y.; Nakagawa, S.; Hirose, T.; Okano, H.J.; Takao, M.; Shibata, S.; Suyama, S.; Kuwako, K.; Imai, T.; Murayama, S.; et al. The long non-coding RNA nuclear-enriched abundant transcript 1_2 induces paraspeckle formation in the motor neuron during the early phase of amyotrophic lateral sclerosis. Mol. Brain 2013, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Taheri, M. Nuclear Enriched Abundant Transcript 1 (NEAT1): A long non-coding RNA with diverse functions in tumorigenesis. Biomed. Pharmacother. 2019, 111, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Prasanth, K.V.; Prasanth, S.G.; Xuan, Z.; Hearn, S.; Freier, S.M.; Bennett, C.F.; Zhang, M.Q.; Spector, D.L. Regulating gene expression through RNA nuclear retention. Cell 2005, 123, 249–263. [Google Scholar] [CrossRef]
- Zhang, Z.; Carmichael, G.G. The fate of dsRNA in the nucleus: A p54(nrb)-containing complex mediates the nuclear retention of promiscuously A-to-I edited RNAs. Cell 2001, 106, 465–475. [Google Scholar] [CrossRef]
- Xu, L.D.; Öhman, M. ADAR1 Editing and its Role in Cancer. Genes 2018, 10, 12. [Google Scholar] [CrossRef]
- Shen, X.; Song, S.; Li, C.; Zhang, J. Synonymous mutations in representative yeast genes are mostly strongly non-neutral. Nature 2022, 606, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Imamura, K.; Imamachi, N.; Akizuki, G.; Kumakura, M.; Kawaguchi, A.; Nagata, K.; Kato, A.; Kawaguchi, Y.; Sato, H.; Yoneda, M.; et al. Long noncoding RNA NEAT1-dependent SFPQ relocation from promoter region to paraspeckle mediates IL8 expression upon immune stimuli. Mol. Cell 2014, 53, 393–406. [Google Scholar] [CrossRef] [PubMed]
- Zan, J.; Zhao, X.; Deng, X.; Ding, H.; Wang, B.; Lu, M.; Wei, Z.; Huang, Z.; Wang, S. Paraspeckle Promotes Hepatocellular Carcinoma Immune Escape by Sequestering IFNGR1 mRNA. Cell. Mol. Gastroenterol. Hepatol. 2021, 12, 465–487. [Google Scholar] [CrossRef] [PubMed]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef]
- Sun, W.; Qin, Y.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Zhang, H. The NEAT1_2/miR-491 Axis Modulates Papillary Thyroid Cancer Invasion and Metastasis Through TGM2/NFκb/FN1 Signaling. Front. Oncol. 2021, 11, 610547. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Z.; Liu, X.; Cheng, X.; Zhang, Y.; Han, X.; Zhang, Y.; Liu, S.; Yang, J.; Xu, B.; et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Investig. 2017, 127, 3421–3440. [Google Scholar] [CrossRef]
- Jiang, X.; Zhou, Y.; Sun, A.-J.; Xue, J.-L. NEAT1 contributes to breast cancer progression through modulating miR-448 and ZEB1. J. Cell. Physiol. 2018, 233, 8558–8566. [Google Scholar] [CrossRef]
- Liu, X.; Yao, W.; Xiong, H.; Li, Q.; Li, Y. LncRNA NEAT1 accelerates breast cancer progression through regulating miR-410-3p/CCND1 axis. Cancer Biomark. 2020, 29, 277–290. [Google Scholar] [CrossRef]
- Li, X.; Wang, S.; Li, Z.; Long, X.; Guo, Z.; Zhang, G.; Zu, J.; Chen, Y.; Wen, L. The lncRNA NEAT1 facilitates cell growth and invasion via the miR-211/HMGA2 axis in breast cancer. Int. J. Biol. Macromol. 2017, 105, 346–353. [Google Scholar] [CrossRef]
- Yao, L.; Chen, L.; Zhou, H.; Duan, F.; Wang, L.; Zhang, Y. Long Noncoding RNA NEAT1 Promotes the Progression of Breast Cancer by Regulating miR-138-5p/ZFX Axis. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef]
- Zhou, D.P.; Gu, J.; Wang, Y.P.; Wu, H.G.; Cheng, W.; Wang, Q.P.; Zheng, G.P.; Wang, X.D. Long non-coding RNA NEAT1 transported by extracellular vesicles contributes to breast cancer development by sponging microRNA-141-3p and regulating KLF12. Cell Biosci. 2021, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Liu, Z.; Li, Z.; Wang, S.; Shen, N.; Xin, Y.; Huang, T. Long non-coding RNA nuclear paraspeckle assembly transcript 1 interacts with microRNA-107 to modulate breast cancer growth and metastasis by targeting carnitine palmitoyltransferase-1. Int. J. Oncol. 2019, 55, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Malhotra, A. Evaluation of Potential of Long Noncoding RNA NEAT1 in Colorectal Cancer. J. Environ. Pathol. Toxicol. Oncol. Off. Organ Int. Soc. Environ. Toxicol. Cancer 2020, 39, 101–111. [Google Scholar] [CrossRef] [PubMed]
- Zhong, F.; Zhang, W.; Cao, Y.; Wen, Q.; Cao, Y.; Lou, B.; Li, J.; Shi, W.; Liu, Y.; Luo, R.; et al. LncRNA NEAT1 promotes colorectal cancer cell proliferation and migration via regulating glial cell-derived neurotrophic factor by sponging miR-196a-5p. Acta Biochim. Biophys. Sin. 2018, 50, 1190–1199. [Google Scholar] [CrossRef]
- Liu, K.; Lei, S.; Kuang, Y.; Jin, Q.; Long, D.; Liu, C.; Jiang, Y.; Zhao, H.; Yao, H. A Novel Mechanism of the c-Myc/NEAT1 Axis Mediating Colorectal Cancer Cell Response to Photodynamic Therapy Treatment. Front. Oncol. 2021, 11, 652831. [Google Scholar] [CrossRef]
- Zhuang, S.T.; Cai, Y.J.; Liu, H.P.; Qin, Y.; Wen, J.F. LncRNA NEAT1/miR-185-5p/IGF2 axis regulates the invasion and migration of colon cancer. Mol. Genet. Genom. Med. 2020, 8, e1125. [Google Scholar] [CrossRef]
- Gao, Z.; Shi, Y.; Wang, J.; Li, W.; Bao, Y.; Wu, D.; Gu, Y. Long non-coding RNA NEAT1 absorbs let-7 g-5p to induce epithelial-mesenchymal transition of colon cancer cells through upregulating BACH1. Dig. Liver Dis. 2021. [Google Scholar] [CrossRef]
- Wu, D.; Li, H.; Wang, J.; Li, H.; Xiao, Q.; Zhao, X.; Huo, Z. LncRNA NEAT1 promotes gastric cancer progression via miR-1294/AKT1 axis. Open Med. 2020, 15, 1028–1038. [Google Scholar] [CrossRef]
- Gao, M.; Liu, L.; Zhang, D.; Yang, Y.; Chang, Z. Long non-coding rna neat1 serves as sponge for mir-365a-3p to promote gastric cancer progression via regulating abcc4. OncoTargets Ther. 2020, 13, 3977–3985. [Google Scholar] [CrossRef]
- Jiang, Y.; Jin, S.; Tan, S.; Xue, Y.; Cao, X. Long noncoding RNA NEAT1 regulates radio-sensitivity via microRNA-27b-3p in gastric cancer. Cancer Cell Int. 2020, 20, 581. [Google Scholar] [CrossRef]
- Zhou, Y.; Sha, Z.; Yang, Y.; Wu, S.; Chen, H. lncRNA NEAT1 regulates gastric carcinoma cell proliferation, invasion and apoptosis via the miR-500a-3p/XBP-1 axis. Mol. Med. Rep. 2021, 24, 503. [Google Scholar] [CrossRef]
- Tan, H.Y.; Wang, C.; Liu, G.; Zhou, X. Long noncoding RNA NEAT1-modulated miR-506 regulates gastric cancer development through targeting STAT3. J. Cell. Biochem. 2019, 120, 4827–4836. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chang, L.; Wu, Q.; Zuo, F. Long non-coding RNA NEAT1 increases the aggressiveness of gastric cancer by regulating the microRNA-142-5p/JAG1 axis. Exp. Ther. Med. 2021, 22, 862. [Google Scholar] [CrossRef] [PubMed]
- Zhai, S.; Gao, F.; Zhao, C.; Li, H.; Jiao, X. Lncrna neat1 enhances cell proliferation and invasion by regulating the MIR-221-5p/ SOX11 axis in gastric cancer. Basic Clin. Pharmacol. Toxicol. 2021, 128, 33–34. [Google Scholar]
- Rao, X.; Liu, X.; Liu, N.; Zhang, Y.; Zhang, Z.; Zhou, L.; Han, G.; Cen, R.; Shi, N.; Zhu, H.; et al. Long noncoding RNA NEAT1 promotes tumorigenesis in H. pylori gastric cancer by sponging miR-30a to regulate COX-2/BCL9 pathway. Helicobacter 2021, 26, e12847. [Google Scholar] [CrossRef]
- Li, B.; Lu, X.; Ma, C.; Sun, S.; Shu, X.; Wang, Z.; Sun, W. Long non-coding RNA NEAT1 promotes human glioma tumor progression via miR-152-3p/CCT6A pathway. Neurosci. Lett. 2020, 732, 135086. [Google Scholar] [CrossRef]
- Zhou, K.; Zhang, C.; Yao, H.; Zhang, X.; Zhou, Y.; Che, Y.; Huang, Y. Knockdown of long non-coding RNA NEAT1 inhibits glioma cell migration and invasion via modulation of SOX2 targeted by miR-132. Mol. Cancer 2018, 17, 105. [Google Scholar] [CrossRef]
- Yu, H.; Xu, A.; Wu, B.; Wang, M.; Chen, Z. Long noncoding RNA NEAT1 promotes progression of glioma as a ceRNA by sponging miR-185-5p to stimulate DNMT1/mTOR signaling. J. Cell. Physiol. 2021, 236, 121–130. [Google Scholar] [CrossRef]
- Li, Z.; Liu, Y.-H.; Diao, H.-Y.; Ma, J.; Yao, Y.-L. Long noncoding RNA NEAT1 promotes glioma pathogenesis by regulating miR-449b-5p/c-Met axis. Tumor Biol. 2016, 37, 673–683. [Google Scholar] [CrossRef]
- Wu, D.M.; Wang, S.; Wen, X.; Han, X.R.; Wang, Y.J.; Fan, S.H.; Zhang, Z.F.; Shan, Q.; Lu, J.; Zheng, Y.L. Long noncoding RNA nuclear enriched abundant transcript 1 impacts cell proliferation, invasion, and migration of glioma through regulating miR-139-5p/ CDK6. J. Cell. Physiol. 2019, 234, 5972–5987. [Google Scholar] [CrossRef]
- Zhang, X.N.; Zhou, J.; Lu, X.J. The long noncoding RNA NEAT1 contributes to hepatocellular carcinoma development by sponging miR-485 and enhancing the expression of the STAT3. J. Cell. Physiol. 2018, 233, 6733–6741. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hao, L.P.; Song, H.; Chu, X.Y.; Wang, R. The Potential Roles of Exosomal Non-Coding RNAs in Hepatocellular Carcinoma. Front. Oncol. 2022, 12, 790916. [Google Scholar] [CrossRef] [PubMed]
- Sun, C.; Xiao, T.; Xiao, Y.; Li, Y. Silencing of long non-coding RNA NEAT1 inhibits hepatocellular carcinoma progression by downregulating SMO by sponging microRNA-503. Mol. Med. Rep. 2020, 23, 168. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, X.; Zhou, Y.; Cheng, L.; Zhang, Y.; Zhang, Y. Long noncoding rna neat1 promotes cell proliferation and invasion and suppresses apoptosis in hepatocellular carcinoma by regulating mirna-22-3p/akt2 in vitro and in vivo. OncoTargets Ther. 2019, 12, 8991–9004. [Google Scholar] [CrossRef] [PubMed]
- Tu, J.; Zhao, Z.; Xu, M.; Lu, X.; Chang, L.; Ji, J. NEAT1 upregulates TGF-β1 to induce hepatocellular carcinoma progression by sponging hsa-mir-139-5p. J. Cell. Physiol. 2018, 233, 8578–8587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Huang, C.; Zhu, Z.; Hou, Y.; Huang, S.; Sun, C.; Tang, Y.; Zhang, Z.; Wang, L.; Chen, H.; et al. LncRNA NEAT1 regulates the proliferation and migration of hepatocellular carcinoma cells by acting as a miR-320a molecular sponge and targeting L antigen family member 3. Int. J. Oncol. 2020, 57, 1001–1012. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liang, Y.; Song, R.; Yang, G.; Han, J.; Lan, Y.; Pan, S.; Zhu, M.; Liu, Y.; Wang, Y.; et al. Long non-coding RNA NEAT1-modulated abnormal lipolysis via ATGL drives hepatocellular carcinoma proliferation. Mol. Cancer 2018, 17, 90. [Google Scholar] [CrossRef]
- Sun, C.; Li, S.; Zhang, F.; Xi, Y.; Wang, L.; Bi, Y.; Li, D. Long non-coding RNA NEAT1 promotes non-small cell lung cancer progression through regulation of miR-377-3p-E2F3 pathway. Oncotarget 2016, 7, 51784–51814. [Google Scholar] [CrossRef]
- Kong, X.; Zhao, Y.; Li, X.; Tao, Z.; Hou, M.; Ma, H. Overexpression of HIF-2α-dependent NEAT1 promotes the progression of non-small cell lung cancer through miR-101-3p/SOX9/Wnt/β-catenin signal pathway. Cell. Physiol. Biochem. 2019, 52, 368–381. [Google Scholar] [CrossRef]
- Zhao, M.M.; Ge, L.Y.; Yang, L.F.; Zheng, H.X.; Chen, G.; Wu, L.Z.; Shi, S.M.; Wang, N.; Hang, Y.P. Lncrna neat1/mir-204/nuak1 axis is a potential therapeutic target for non-small cell lung cancer. Cancer Manag. Res. 2020, 12, 13357–13368. [Google Scholar] [CrossRef]
- Chen, L.M.; Niu, Y.D.; Xiao, M.; Li, X.J.; Lin, H. LncRNA NEAT1 regulated cell proliferation, invasion, migration and apoptosis by targeting has-miR-376b-3p/SULF1 axis in non-small cell lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 4810–4821. [Google Scholar] [CrossRef]
- Luo, M.; Zhang, L.; Yang, H.; Luo, K.; Qing, C. Long non-coding RNA NEAT1 promotes ovarian cancer cell invasion and migration by interacting with miR-1321 and regulating tight junction protein 3 expression. Mol. Med. Rep. 2020, 22, 3429–3439. [Google Scholar] [CrossRef] [PubMed]
- Yin, L.; Wang, Y. Long non-coding RNA NEAT1 facilitates the growth, migration, and invasion of ovarian cancer cells via the let-7 g/MEST/ATGL axis. Cancer Cell Int. 2021, 21, 437. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Sun, X.; Huang, Y.; Si, Q.; Li, M. Long non-coding RNA NEAT1 modifies cell proliferation, colony formation, apoptosis, migration and invasion via the miR-4500/BZW1 axis in ovarian cancer. Mol. Med. Rep. 2020, 22, 3347–3357. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; He, C.; Yang, F.; Qin, L.; Lu, X.; Wu, J. Long non-coding RNA-NEAT1, a sponge for MIR-98-5p, promotes expression of oncogene HMGA2 in prostate cancer. Biosci. Rep. 2019, 39, BSR20190635. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Guo, S.; Zhang, Y.; Zhao, Y.; Li, X.; Jia, Y.; Xu, Y.; Ma, B. LncRNA NEAT1 promotes docetaxel resistance in prostate cancer by regulating ACSL4 via sponging miR-34a-5p and miR-204-5p. Cell. Signal. 2020, 65, 109422. [Google Scholar] [CrossRef] [PubMed]
- Dong, P.; Xu, D.; Konno, Y.; Ihira, K.; Watari, H. Lncrna neat1-mediated mir-361 downregulation contributes to EMT and sphere formation of cervical cancer cells via increasing hsp90 expression. Int. J. Gynecol. Cancer 2020, 30, A128–A129. [Google Scholar] [CrossRef]
- Li, O.; Jiang, B.; Yi, W.M.; Zhang, Y.; Yang, P.Z.; Guo, C.; Sun, Z.P.; Peng, C. LncRNA NEAT1 promotes cell proliferation, migration, and invasion via the miR-186-5p/PTP4A1 axis in cholangiocarcinoma. Kaohsiung J. Med. Sci. 2021, 37, 379–391. [Google Scholar] [CrossRef]
- Fan, J.T.; Zhou, Z.Y.; Luo, Y.L.; Luo, Q.; Chen, S.B.; Zhao, J.C.; Chen, Q.R. Exosomal lncRNA NEAT1 from cancer-associated fibroblasts facilitates endometrial cancer progression via miR-26a/b-5p-mediated STAT3/YKL-40 signaling pathway. Neoplasia 2021, 23, 692–703. [Google Scholar] [CrossRef]
- Li, Y.; Chen, D.; Gao, X.; Li, X.; Shi, G. LncRNA NEAT1 Regulates Cell Viability and Invasion in Esophageal Squamous Cell Carcinoma through the miR-129/CTBP2 Axis. Dis. Markers 2017, 2017, 5314649. [Google Scholar] [CrossRef]
- Yang, F.; Tang, Z.; Duan, A.; Yi, B.; Shen, N.; Bo, Z.; Yin, L.; Zhu, B.; Qiu, Y.; Li, J. Long noncoding RNA NEAT1 upregulates survivin and facilitates gallbladder cancer progression by sponging microRNA-335. OncoTargets Ther. 2020, 13, 2357–2367. [Google Scholar] [CrossRef] [PubMed]
- Lulli, V.; Buccarelli, M.; Ilari, R.; Castellani, G.; De Dominicis, C.; Di Giamberardino, A.; D′Alessandris, Q.G.; Giannetti, S.; Martini, M.; Stumpo, V.; et al. Mir-370-3p impairs glioblastoma stem-like cell malignancy regulating a complex interplay between HMGA2/HIF1A and the oncogenic long non-coding RNA (LncRNA) neat1. Int. J. Mol. Sci. 2020, 21, 3610. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.B.; Yan, X.H.; Tian, M.; Zhang, S.; Liu, J.L.; Sheng, Y.X.; Dong, L.; Zhang, W.L. Long non-coding RNA NEAT1 regulates Hodgkin’s lymphoma cell proliferation and invasion via miR-448 mediated regulation of DCLK1. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 6219–6227. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Zheng, C.; Wu, S. Long non-coding RNA NEAT1 promotes the malignancy of laryngeal squamous cell carcinoma by regulating the microRNA-204-5p/SEMA4B axis. Oncol. Lett. 2021, 22, 802. [Google Scholar] [CrossRef]
- Zhou, W.J.; Wang, H.Y.; Zhang, J.; Dai, H.Y.; Yao, Z.X.; Zheng, Z.; Meng-Yan, S.; Wu, K. NEAT1/miR-200b-3p/SMAD2 axis promotes progression of melanoma. Aging 2020, 12, 22759–22775. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, C.; Chen, C.; Wu, F.; Shen, P.; Zhang, P.; He, G.; Li, X. Long non-coding RNA NEAT1 regulates epithelial membrane protein 2 expression to repress nasopharyngeal carcinoma migration and irradiation-resistance through miR-101-3p as a competing endogenous RNA mechanism. Oncotarget 2017, 8, 70156–70171. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Li, J.; Xiao, J.K.; Xiao, L.; Xu, B.W.; Li, C. The lncRNA NEAT1 promotes the epithelial-mesenchymal transition and metastasis of osteosarcoma cells by sponging miR-483 to upregulate STAT3 expression. Cancer Cell Int. 2021, 21, 90. [Google Scholar] [CrossRef]
- Hu, H.; Chen, W.; Zhang, S.; Xue, Y.; He, Y.; Gu, Y. NEAT1/miR-101-dependent up-regulation of DNA-PKcs enhances malignant behaviors of pancreatic ductal adenocarcinoma cells. J. Cancer 2021, 12, 5622–5632. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, S.; Li, Q.; Xu, C.; Yu, Y.; Ge, H. LncRNA NEAT1 Knockdown Inhibits Retinoblastoma Progression by miR-3619-5p/LASP1 Axis. Front. Genet. 2020, 11, 574145. [Google Scholar] [CrossRef]
- Luo, Y.; Hao, T.; Zhang, J.; Zhang, M.; Sun, P.; Wu, L. MicroRNA-592 suppresses the malignant phenotypes of thyroid cancer by regulating lncRNA NEAT1 and downregulating NOVA1. Int. J. Mol. Med. 2019, 44, 1172–1182. [Google Scholar] [CrossRef]
- Li, Y.; Huang, W.Q.; Chen, L.L. LncRNA NEAT1 regulates proliferation, migration and invasion of tongue squamous cell carcinoma cells by regulating miR-339-5p/ITGA3 axis. Shanghai J. Stomatol. 2020, 29, 267–274. [Google Scholar]
- Shao, X.; Zheng, X.; Ma, D.; Liu, Y.; Liu, G. Inhibition of lncRNA-NEAT1 sensitizes 5-Fu resistant cervical cancer cells through de-repressing the microRNA-34a/LDHA axis. Biosci. Rep. 2021, 41, BSR20200533. [Google Scholar] [CrossRef] [PubMed]
- Rostami, M.; Kharajo, R.S.; Parsa-Kondelaji, M.; Ayatollahi, H.; Sheikhi, M.; Keramati, M.R. Altered expression of NEAT1 variants and P53, PTEN, and BCL-2 genes in patients with acute myeloid leukemia. Leuk. Res. 2022, 115, 106807. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Deng, S.; Pang, X.; Song, Y.; Luo, S.; Jin, L.; Pan, Y. LncRNA NEAT1 silenced miR-133b promotes migration and invasion of breast cancer cells. Int. J. Mol. Sci. 2019, 20, 3616. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Chen, Y.; Kong, W.; Li, Y.; Lu, F. Tumor protein D52 promotes breast cancer proliferation and migration via the long non-coding RNA NEAT1/microRNA-218-5p axis. Ann. Transl. Med. 2021, 9, 1008. [Google Scholar] [CrossRef]
- Lo, P.K.; Zhang, Y.; Wolfson, B.; Gernapudi, R.; Yao, Y.; Duru, N.; Zhou, Q. Dysregulation of the BRCA1/long non-coding RNA NEAT1 signaling axis contributes to breast tumorigenesis. Oncotarget 2016, 7, 65067–65089. [Google Scholar] [CrossRef]
- Shen, X.; Zhao, W.; Zhang, Y.; Liang, B. Long non-coding RNA-neat1 promotes cell migration and invasion via regulating MiR-124/NF-κB pathway in cervical cancer. OncoTargets Ther. 2020, 13, 3265–3276. [Google Scholar] [CrossRef]
- Yuan, L.Y.; Zhou, M.; Lv, H.; Qin, X.; Zhou, J.; Mao, X.; Li, X.; Xu, Y.; Liu, Y.; Xing, H. Involvement of NEAT1/miR-133a axis in promoting cervical cancer progression via targeting SOX4. J. Cell. Physiol. 2019, 234, 18985–18993. [Google Scholar] [CrossRef]
- Zhu, Z.; Du, S.; Yin, K.; Ai, S.; Yu, M.; Liu, Y.; Shen, Y.; Liu, M.; Jiao, R.; Chen, X.; et al. Knockdown long noncoding RNA nuclear paraspeckle assembly transcript 1 suppresses colorectal cancer through modulating miR-193a-3p/KRAS. Cancer Med. 2019, 8, 261–275. [Google Scholar] [CrossRef]
- Liu, Z.; Gu, Y.; Cheng, X.; Jiang, H.; Huang, Y.; Zhang, Y.; Yu, G.; Cheng, Y.; Zhou, L. Upregulation lnc-NEAT1 contributes to colorectal cancer progression through sponging miR-486-5p and activating NR4A1β-catenin pathway. Cancer Biomark. 2021, 30, 309–319. [Google Scholar] [CrossRef]
- Liu, H.; Li, A.; Sun, Z.; Zhang, J.; Xu, H. Long non-coding RNA NEAT1 promotes colorectal cancer progression by regulating miR-205-5p/VEGFA axis. Hum. Cell 2020, 33, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Chen, J.J.; Lv, Q.; Qin, J.; Huang, Y.Z.; Yu, M.H.; Zhong, M. Long non-coding RNA NEAT1 promotes colorectal cancer progression by competitively binding miR-34a with SIRT1 and enhancing the Wnt/β-catenin signaling pathway. Cancer Lett. 2019, 440–441, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Zhai, J.; Fu, Y. Overexpression of Nuclear Enriched Autosomal Transcript 1 Facilitates Cell Proliferation, Migration Invasion, and Suppresses Apoptosis in Endometrial Cancer by Targeting MicroRNA-202-3p/T Cell Immunoglobulin and Mucin Domain 4 Axis. Cancer Biother. Radiopharm. 2020. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Ge, L.; Xu, X.J.; Yang, T.; Yuan, Y.; Ma, X.L.; Zhang, X.H. LncRNA NEAT1 promotes endometrial cancer cell proliferation, migration and invasion by regulating the MIR-144-3p/EZH2 axis. Radiol. Oncol. 2019, 53, 434–442. [Google Scholar] [CrossRef]
- Dong, P.; Xiong, Y.; Yue, J.; Xu, D.; Ihira, K.; Konno, Y.; Kobayashi, N.; Todo, Y.; Watari, H. Long noncoding RNA NEAT1 drives aggressive endometrial cancer progression via miR-361-regulated networks involving STAT3 and tumor microenvironment-related genes. J. Exp. Clin. Cancer Res. 2019, 38, 295. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhao, X.; Guo, Z.; Ma, X.; Song, Y.; Guo, Y. Regulation of NEAT1/miR-214-3p on the growth, migration and invasion of endometrial carcinoma cells. Arch. Gynecol. Obstet. 2017, 295, 1469–1475. [Google Scholar] [CrossRef]
- Luo, J.; Xie, K.; Gao, X.; Yao, Y.; Wang, G.; Shao, C.; Li, X.; Xu, Y.; Ren, B.; Hu, L.; et al. Long Noncoding RNA Nuclear Paraspeckle Assembly Transcript 1 Promotes Progression and Angiogenesis of Esophageal Squamous Cell Carcinoma Through miR-590-3p/MDM2 Axis. Front. Oncol. 2020, 10, 618930. [Google Scholar] [CrossRef]
- Yang, L.; Wang, M.; He, P. Lncrna neat1 promotes the progression of gastric cancer through modifying the mir-1224-5p/rsf1 signaling axis. Cancer Manag. Res. 2020, 12, 11845–11855. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, M.; Sun, G. Long non-coding RNA NEAT1 regulates the proliferation, migration and invasion of gastric cancer cells via targeting miR-335-5p/ROCK1 axis. Pharmazie 2018, 73, 150–155. [Google Scholar] [CrossRef]
- Li, Y.; Ding, X.; Xiu, S.; Du, G.; Liu, Y. LncRNA neat1 promotes proliferation, migration and invasion via regulating miR-296-5p/CNN2 axis in hepatocellular carcinoma cells. OncoTargets Ther. 2019, 12, 9887–9897. [Google Scholar] [CrossRef]
- Xia, Y.; Zhou, Y.; Han, H.; Li, P.; Wei, W.; Lin, N. lncRNA NEAT1 facilitates melanoma cell proliferation, migration, and invasion via regulating miR-495-3p and E2F3. J. Cell. Physiol. 2019, 234, 19592–19601. [Google Scholar] [CrossRef] [PubMed]
- Ding, F.; Lai, J.; Gao, Y.; Wang, G.; Shang, J.; Zhang, D.; Zheng, S. NEAT1/miR-23a-3p/KLF3: A novel regulatory axis in melanoma cancer progression. Cancer Cell Int. 2019, 19, 217. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Fang, P.; Li, W.J.; Zhang, J.; Wang, G.P.; Jiang, D.F.; Chen, F.P. LncRNA NEAT1 sponges miR-214 to regulate M2 macrophage polarization by regulation of B7-H3 in multiple myeloma. Mol. Immunol. 2020, 117, 20–28. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, Y.; Fu, L.; Zhu, X. Mir-222 emerges as a cerna of lncrna neat1, and inhibits cell viability and migration of nasopharyngeal carcinoma cells via aldh1. Basic Clin. Pharmacol. Toxicol. 2021, 128, 94–95. [Google Scholar]
- Qi, L.; Liu, F.; Zhang, F.; Zhang, S.; Lv, L.; Bi, Y.; Yu, Y. lncRNA NEAT1 competes against let-7a to contribute to non-small cell lung cancer proliferation and metastasis. Biomed. Pharmacother. 2018, 103, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Mo, Q.; Wan, X.; Dan, J.; Hu, H. NEAT1/hsa-mir-98-5p/MAPK6 axis is involved in non–small-cell lung cancer development. J. Cell. Biochem. 2019, 120, 2836–2846. [Google Scholar] [CrossRef]
- Liu, J.; Li, K.; Wang, R.; Chen, S.; Wu, J.; Li, X.; Ning, Q.; Yang, G.; Pang, Y. The interplay between ATF2 and NEAT1 contributes to lung adenocarcinoma progression. Cancer Cell Int. 2020, 20, 594. [Google Scholar] [CrossRef]
- Yu, P.F.; Wang, Y.; Lv, W.; Kou, D.; Hu, H.L.; Guo, S.S.; Zhao, Y.J. LncRNA NEAT1/MIR-1224/KLF3 contributes to cell proliferation, apoptosis and invasion in lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8403–8410. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, X.Q.; Zhou, X.Q.; Liu, Q.B.; Chen, L.; Cai, F. NEAT1 induces osteosarcoma development by modulating the miR-339-5p/TGF-β1 pathway. J. Cell. Physiol. 2019, 234, 5097–5105. [Google Scholar] [CrossRef]
- Tan, H.; Zhao, L. lncRNA nuclear-enriched abundant transcript 1 promotes cell proliferation and invasion by targeting miR-186-5p/HIF-1α in osteosarcoma. J. Cell. Biochem. 2019, 120, 6502–6514. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Y.; Yang, J.; He, S.; Li, M.; Yan, S.; Chen, Y.; Qu, C.; Xu, L. NEAT1 regulates pancreatic cancer cell growth, invasion and migration though mircroRNA-335-5p/c-met axis. Am. J. Cancer Res. 2016, 6, 2361–2374. [Google Scholar] [PubMed]
- Luan, L.; Hu, Q.; Wang, Y.; Lu, L.; Ling, J. Knockdown of lncRNA NEAT1 expression inhibits cell migration, invasion and EMT by regulating the miR-24-3p/LRG1 axis in retinoblastoma cells. Exp. Ther. Med. 2021, 21, 367. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Zhang, Z.; Lu, Y.; Xiu, W.; Cui, J. Lncrna neat1 acts as an mir-148b-3p sponge to regulate rock1 inhibition of retinoblastoma growth. Cancer Manag. Res. 2021, 13, 5587–5597. [Google Scholar] [CrossRef] [PubMed]
- Zhong, W.; Yang, J.; Li, M.; Li, L.; Li, A. Long noncoding RNA NEAT1 promotes the growth of human retinoblastoma cells via regulation of miR-204/CXCR4 axis. J. Cell. Physiol. 2019, 234, 11567–11576. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cai, Y.; Zheng, L.; Zhang, Z.; Lin, X.; Jiang, N. Long noncoding RNA NEAT1 regulate papillary thyroid cancer progression by modulating miR-129-5p/KLK7 expression. J. Cell. Physiol. 2018, 233, 6638–6648. [Google Scholar] [CrossRef]
- Sun, W.; Lan, X.; Zhang, H.; Wang, Z.; Dong, W.; He, L.; Zhang, T.; Zhang, P.; Liu, J.; Qin, Y. NEAT1-2 functions as a competing endogenous RNA to regulate ATAD2 expression by sponging microRNA-106b-5p in papillary thyroid cancer. Cell Death Dis. 2018, 9, 380. [Google Scholar] [CrossRef]
- Pan, S.; Liu, R.; Wu, X.; Ma, K.; Luo, W.; Nie, K.; Zhang, C.; Meng, X.; Tong, T.; Chen, X.; et al. LncRNA NEAT1 mediates intestinal inflammation by regulating TNFRSF1B. Ann. Transl. Med. 2021, 9, 773. [Google Scholar] [CrossRef]
- Feng, Y.; Gao, L.; Cui, G.; Cao, Y. LncRNA NEAT1 facilitates pancreatic cancer growth and metastasis through stabilizing ELF3 mRNA. Am. J. Cancer Res. 2020, 10, 237–248. [Google Scholar]
- Ma, F.; Lei, Y.Y.; Ding, M.G.; Luo, L.H.; Xie, Y.C.; Liu, X.L. LncRNA NEAT1 Interacted With DNMT1 to Regulate Malignant Phenotype of Cancer Cell and Cytotoxic T Cell Infiltration via Epigenetic Inhibition of p53, cGAS, and STING in Lung Cancer. Front. Genet. 2020, 11, 250. [Google Scholar] [CrossRef]
- Wang, S.; Zuo, H.; Jin, J.; Lv, W.; Xu, Z.; Fan, Y.; Zhang, J.; Zuo, B. Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 2019, 10, 505. [Google Scholar] [CrossRef]
- Chen, Q.; Cai, J.; Wang, Q.; Wang, Y.; Liu, M.; Yang, J.; Zhou, J.; Kang, C.; Li, M.; Jiang, C. Long Noncoding RNA NEAT1, Regulated by the EGFR Pathway, Contributes to Glioblastoma Progression Through the WNT/beta-Catenin Pathway by Scaffolding EZH2. Clin. Cancer Res. 2018, 24, 684–695. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Liu, L.; Zhang, S.; Ming, Y.; Liu, S.; Cheng, K.; Zhao, Y. Long noncoding RNA NEAT1 suppresses hepatocyte proliferation in fulminant hepatic failure through increased recruitment of EZH2 to the LATS2 promoter region and promotion of H3K27me3 methylation. Exp. Mol. Med. 2020, 52, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, A.S.I.; Dong, K.; Liu, J.; Wen, T.; Yu, L.; Xu, F.; Kang, X.; Osman, I.; Hu, G.; Bunting, K.M.; et al. Long noncoding RNA NEAT1 (nuclear paraspeckle assembly transcript 1) is critical for phenotypic switching of vascular smooth muscle cells. Proc. Natl. Acad. Sci. USA 2018, 115, E8660–E8667. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, Y.; Xu, N.; Zhang, S.; Wang, S.; Mao, Y.; Zhu, Y.; Li, B.; Jiang, Y.; Tan, Y.; et al. NEAT1 regulates neuroglial cell mediating Abeta clearance via the epigenetic regulation of endocytosis-related genes expression. Cell Mol. Life Sci. 2019, 76, 3005–3018. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Pan, Y.; Yang, F.; Fang, Y.; Liu, W.; Zhao, C.; Yu, T.; Xie, M.; Jing, X.; Wu, X.; et al. KLF5-Modulated lncRNA NEAT1 Contributes to Tumorigenesis by Acting as a Scaffold for BRG1 to Silence GADD45A in Gastric Cancer. Mol. Nucleic Acids 2020, 22, 382–395. [Google Scholar] [CrossRef]
- Park, M.K.; Zhang, L.; Min, K.W.; Cho, J.H.; Yeh, C.C.; Moon, H.; Hormaechea-Agulla, D.; Mun, H.; Ko, S.; Lee, J.W.; et al. NEAT1 is essential for metabolic changes that promote breast cancer growth and metastasis. Cell Metab. 2021, 33, 2380–2397. [Google Scholar] [CrossRef]
- Wen, S.; Wei, Y.; Zen, C.; Xiong, W.; Niu, Y.; Zhao, Y. Long non-coding RNA NEAT1 promotes bone metastasis of prostate cancer through N6-methyladenosine. Mol. Cancer 2020, 19, 171. [Google Scholar] [CrossRef]
- Huang, Z.; Zhao, J.; Wang, W.; Zhou, J.; Zhang, J. Depletion of LncRNA NEAT1 Rescues Mitochondrial Dysfunction Through NEDD4L-Dependent PINK1 Degradation in Animal Models of Alzheimer’s Disease. Front. Cell. Neurosci. 2020, 14, 28. [Google Scholar] [CrossRef]
- Xu, Y.; Cao, Z.; Ding, Y.; Li, Z.; Xiang, X.; Lai, R.; Sheng, Z.; Liu, Y.; Cai, W.; Hu, R.; et al. Long Non-coding RNA NEAT1 Alleviates Acute-on-Chronic Liver Failure Through Blocking TRAF6 Mediated Inflammatory Response. Front. Physiol. 2019, 10, 1503. [Google Scholar] [CrossRef]
- Shui, X.; Chen, S.; Lin, J.; Kong, J.; Zhou, C.; Wu, J. Knockdown of lncRNA NEAT1 inhibits Th17/CD4(+) T cell differentiation through reducing the STAT3 protein level. J. Cell Physiol. 2019, 234, 22477–22484. [Google Scholar] [CrossRef]
- Li, X.; Wang, X.; Song, W.; Xu, H.; Huang, R.; Wang, Y.; Zhao, W.; Xiao, Z.; Yang, X. Oncogenic Properties of NEAT1 in Prostate Cancer Cells Depend on the CDC5L-AGRN Transcriptional Regulation Circuit. Cancer Res. 2018, 78, 4138–4149. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, P.; Zhao, Y.; Zhang, S.; Lu, J.; Xie, W.; Jiang, Y.; Lei, F.; Xu, N.; Zhang, Y. NEAT1 modulates herpes simplex virus-1 replication by regulating viral gene transcription. Cell Mol. Life Sci. 2017, 74, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Fu, X.; Zhu, J.; Zhang, L.; Shu, J. Long non-coding RNA NEAT1 promotes steatosis via enhancement of estrogen receptor alpha-mediated AQP7 expression in HepG2 cells. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1782–1787. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Shao, C.; Wu, Q.J.; Chen, G.; Zhou, J.; Yang, B.; Li, H.; Gou, L.T.; Zhang, Y.; Wang, Y.; et al. NEAT1 scaffolds RNA-binding proteins and the Microprocessor to globally enhance pri-miRNA processing. Nat. Struct. Mol. Biol. 2017, 24, 816–824. [Google Scholar] [CrossRef]
- Shen, W.; Liang, X.H.; Sun, H.; De Hoyos, C.L.; Crooke, S.T. Depletion of NEAT1 lncRNA attenuates nucleolar stress by releasing sequestered P54nrb and PSF to facilitate c-Myc translation. PLoS ONE 2017, 12, e0173494. [Google Scholar] [CrossRef]
- Jen, H.W.; Gu, D.L.; Lang, Y.D.; Jou, Y.S. PSPC1 Potentiates IGF1R Expression to Augment Cell Adhesion and Motility. Cells 2020, 9, 1490. [Google Scholar] [CrossRef]
- Cooper, D.R.; Carter, G.; Li, P.; Patel, R.; Watson, J.E.; Patel, N.A. Long Non-Coding RNA NEAT1 Associates with SRp40 to Temporally Regulate PPARγ2 Splicing during Adipogenesis in 3T3-L1 Cells. Genes 2014, 5, 1050–1063. [Google Scholar] [CrossRef]
- Mang, Y.; Li, L.; Ran, J.; Zhang, S.; Liu, J.; Li, L.; Chen, Y.; Liu, J.; Gao, Y.; Ren, G. Long noncoding RNA NEAT1 promotes cell proliferation and invasion by regulating hnRNP A2 expression in hepatocellular carcinoma cells. Oncol. Targets 2017, 10, 1003–1016. [Google Scholar] [CrossRef]
- Lee, M.; Sadowska, A.; Bekere, I.; Ho, D.; Gully, B.S.; Lu, Y.; Iyer, K.S.; Trewhella, J.; Fox, A.H.; Bond, C.S. The structure of human SFPQ reveals a coiled-coil mediated polymer essential for functional aggregation in gene regulation. Nucleic Acids Res. 2015, 43, 3826–3840. [Google Scholar] [CrossRef]
- Naganuma, T.; Nakagawa, S.; Tanigawa, A.; Sasaki, Y.F.; Goshima, N.; Hirose, T. Alternative 3’-end processing of long noncoding RNA initiates construction of nuclear paraspeckles. EMBO J. 2012, 31, 4020–4034. [Google Scholar] [CrossRef]
- West, J.A.; Davis, C.P.; Sunwoo, H.; Simon, M.D.; Sadreyev, R.I.; Wang, P.I.; Tolstorukov, M.Y.; Kingston, R.E. The long noncoding RNAs NEAT1 and MALAT1 bind active chromatin sites. Mol. Cell 2014, 55, 791–802. [Google Scholar] [CrossRef] [PubMed]
- Chu, C.; Qu, K.; Zhong, F.L.; Artandi, S.E.; Chang, H.Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA-chromatin interactions. Mol. Cell 2011, 44, 667–678. [Google Scholar] [CrossRef] [PubMed]
- Jen, J.; Tang, Y.A.; Lu, Y.H.; Lin, C.C.; Lai, W.W.; Wang, Y.C. Oct4 transcriptionally regulates the expression of long non-coding RNAs NEAT1 and MALAT1 to promote lung cancer progression. Mol. Cancer 2017, 16, 104. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, H.; Albukhari, A.; Morotti, M.; Haider, S.; Moralli, D.; Smythies, J.; Schödel, J.; Green, C.M.; Camps, C.; Buffa, F.; et al. Tumor hypoxia induces nuclear paraspeckle formation through HIF-2α dependent transcriptional activation of NEAT1 leading to cancer cell survival. Oncogene 2015, 34, 4482–4490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zheng, Y.; Sun, Y.; Li, S.; Chen, L.; Jin, X.; Hou, X.; Liu, X.; Chen, Q.; Li, J.; et al. Knockdown of NEAT1 induces tolerogenic phenotype in dendritic cells by inhibiting activation of NLRP3 inflammasome. Theranostics 2019, 9, 3425–3442. [Google Scholar] [CrossRef]
- Modic, M.; Grosch, M.; Rot, G.; Schirge, S.; Lepko, T.; Yamazaki, T.; Lee, F.C.Y.; Rusha, E.; Shaposhnikov, D.; Palo, M.; et al. Cross-Regulation between TDP-43 and Paraspeckles Promotes Pluripotency-Differentiation Transition. Mol. Cell 2019, 74, 951–965. [Google Scholar] [CrossRef]
- Sunwoo, H.; Dinger, M.E.; Wilusz, J.E.; Amaral, P.P.; Mattick, J.S.; Spector, D.L. MEN epsilon/beta nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009, 19, 347–359. [Google Scholar] [CrossRef]
- Klec, C.; Knutsen, E.; Schwarzenbacher, D.; Jonas, K.; Pasculli, B.; Heitzer, E.; Rinner, B.; Krajina, K.; Prinz, F.; Gottschalk, B.; et al. ALYREF, a novel factor involved in breast carcinogenesis, acts through transcriptional and post-transcriptional mechanisms selectively regulating the short NEAT1 isoform. Cell Mol. Life Sci. 2022, 79, 391. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, L.; Zhao, J.; Li, C.; Nie, J.; Liu, F.; Zhuo, C.; Zheng, Y.; Li, B.; Wang, Z.; et al. Nuclear-enriched abundant transcript 1 as a diagnostic and prognostic biomarker in colorectal cancer. Mol. Cancer 2015, 14, 191. [Google Scholar] [CrossRef]
- Zhang, H.; Su, X.; Burley, S.K.; Zheng, X.F.S. mTOR regulates aerobic glycolysis through NEAT1 and nuclear paraspeckle-mediated mechanism in hepatocellular carcinoma. Theranostics 2022, 12, 3518–3533. [Google Scholar] [CrossRef]
- Chai, Y.; Liu, J.; Zhang, Z.; Liu, L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016, 5, 1588–1598. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Li, X.; Yu, L.; Wang, R.; Hua, D.; Shi, C.; Sun, C.; Luo, W.; Rao, C.; Jiang, Z.; et al. The RNA-binding protein SRSF1 is a key cell cycle regulator via stabilizing NEAT1 in glioma. Int. J. Biochem. Cell Biol. 2019, 113, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Qin, X.; Wang, B.; Li, Q.; Hu, J.; Cheng, X.; Guo, D.; Cheng, F.; Fang, C.; Tan, Y.; et al. ALKBH5 Facilitates Hypoxia-Induced Paraspeckle Assembly and IL8 Secretion to Generate an Immunosuppressive Tumor Microenvironment. Cancer Res. 2021, 81, 5876–5888. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Deng, Y.; Zhu, J.; Duan, Y.; Weng, W.; Xu, M.; Ju, X.; Wu, X. Long noncoding RNA NEAT1, regulated by LIN28B, promotes cell proliferation and migration through sponging miR-506 in high-grade serous ovarian cancer. Cell Death Dis. 2018, 9, 861. [Google Scholar] [CrossRef]
- Yi, K.; Cui, X.; Liu, X.; Wang, Y.; Zhao, J.; Yang, S.; Xu, C.; Yang, E.; Xiao, M.; Hong, B.; et al. PTRF/Cavin-1 as a Novel RNA-Binding Protein Expedites the NF-κB/PD-L1 Axis by Stabilizing lncRNA NEAT1, Contributing to Tumorigenesis and Immune Evasion in Glioblastoma. Front. Immunol. 2021, 12, 802795. [Google Scholar] [CrossRef]
- Yoon, J.H.; De, S.; Srikantan, S.; Abdelmohsen, K.; Grammatikakis, I.; Kim, J.; Kim, K.M.; Noh, J.H.; White, E.J.; Martindale, J.L.; et al. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat. Commun. 2014, 5, 5248. [Google Scholar] [CrossRef]
- Rao, Y.; Fang, Y.; Tan, W.; Liu, D.; Pang, Y.; Wu, X.; Zhang, C.; Li, G. Delivery of Long Non-coding RNA NEAT1 by Peripheral Blood Monouclear Cells-Derived Exosomes Promotes the Occurrence of Rheumatoid Arthritis via the MicroRNA-23a/MDM2/SIRT6 Axis. Front. Cell Dev. Biol. 2020, 8, 551681. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Jia, X.; Wei, L.; Zhang, Z. NEAT1 Overexpression Indicates a Poor Prognosis and Induces Chemotherapy Resistance via the miR-491-5p/SOX3 Signaling Pathway in Ovarian Cancer. Front. Genet. 2021, 12, 616220. [Google Scholar] [CrossRef]
- Mo, C.; Huang, B.; Zhuang, J.; Jiang, S.; Guo, S.; Mao, X. LncRNA nuclear-enriched abundant transcript 1 shuttled by prostate cancer cells-secreted exosomes initiates osteoblastic phenotypes in the bone metastatic microenvironment via miR-205-5p/runt-related transcription factor 2/splicing factor proline- and glutamine-rich/polypyrimidine tract-binding protein 2 axis. Clin. Transl. Med. 2021, 11, e493. [Google Scholar] [CrossRef]
- Zhuang, L.; Xia, W.; Chen, D.; Ye, Y.; Hu, T.; Li, S.; Hou, M. Exosomal LncRNA-NEAT1 derived from MIF-treated mesenchymal stem cells protected against doxorubicin-induced cardiac senescence through sponging miR-221-3p. J. Nanobiotechnol. 2020, 18, 157. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L.; Carmichael, G.G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: Functional role of a nuclear noncoding RNA. Mol. Cell 2009, 35, 467–478. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).