Lipoprotein Deprivation Reveals a Cholesterol-Dependent Therapeutic Vulnerability in Diffuse Glioma Metabolism
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture and Cellular Viability Assessment
2.2. Flow Cytometric Cell Cycle Analysis
2.3. Transcriptomics
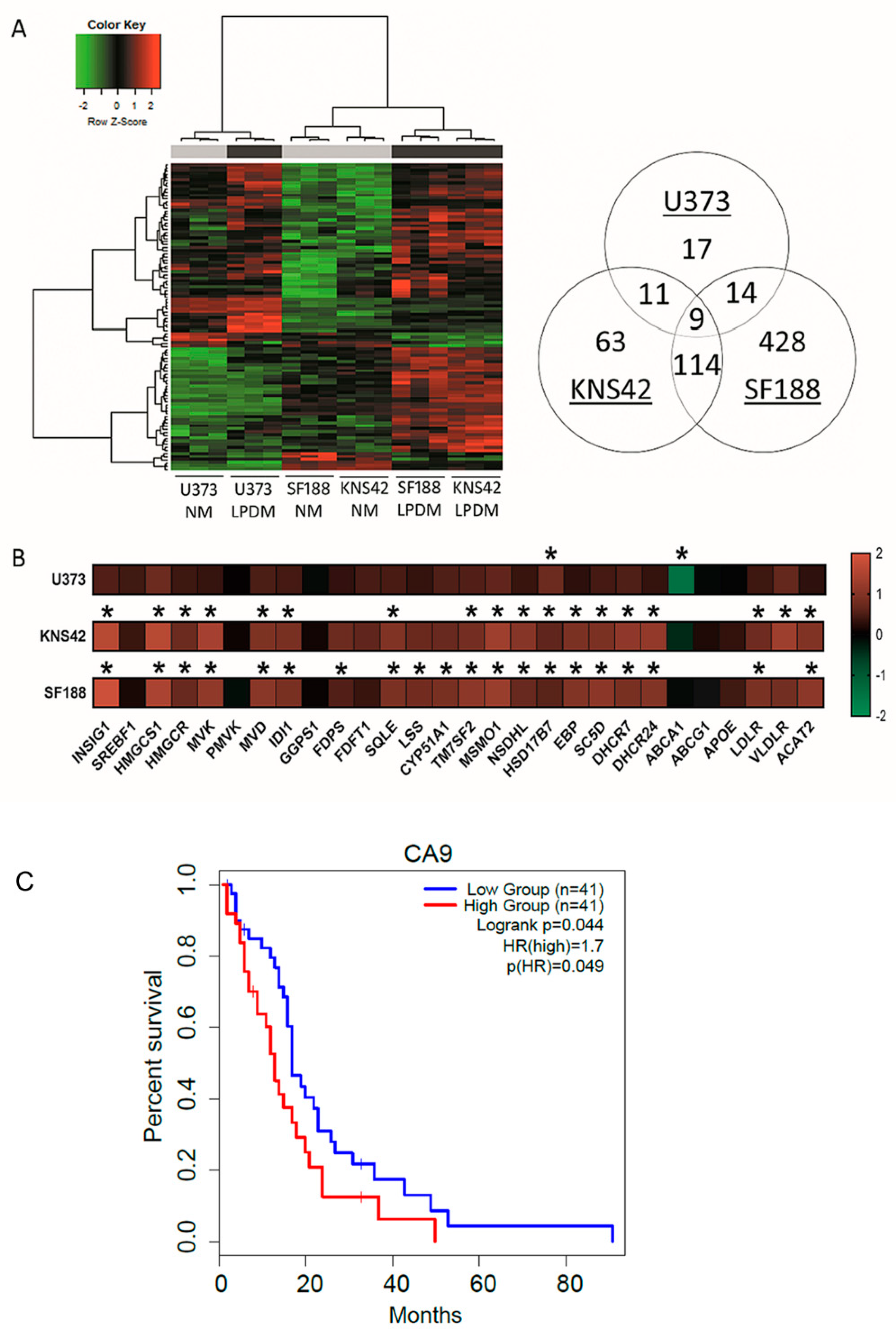
2.4. Survival Analyses
2.5. Metabolomics Data Acquisition and Analysis
2.6. Lipidomics Data Acquisition and Analysis
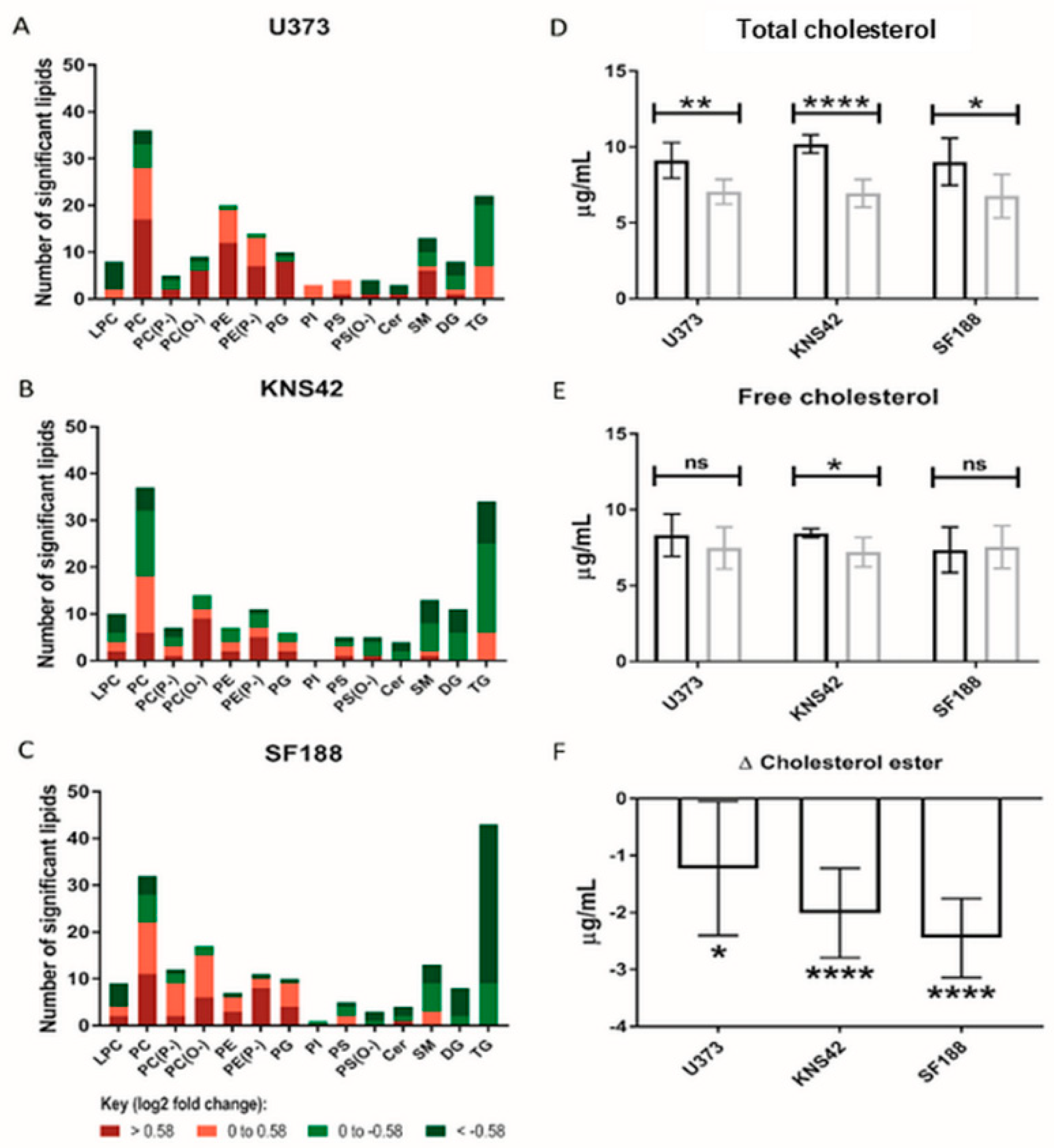
2.7. Cholesterol Assay
2.8. Statistical Analyses
3. Results
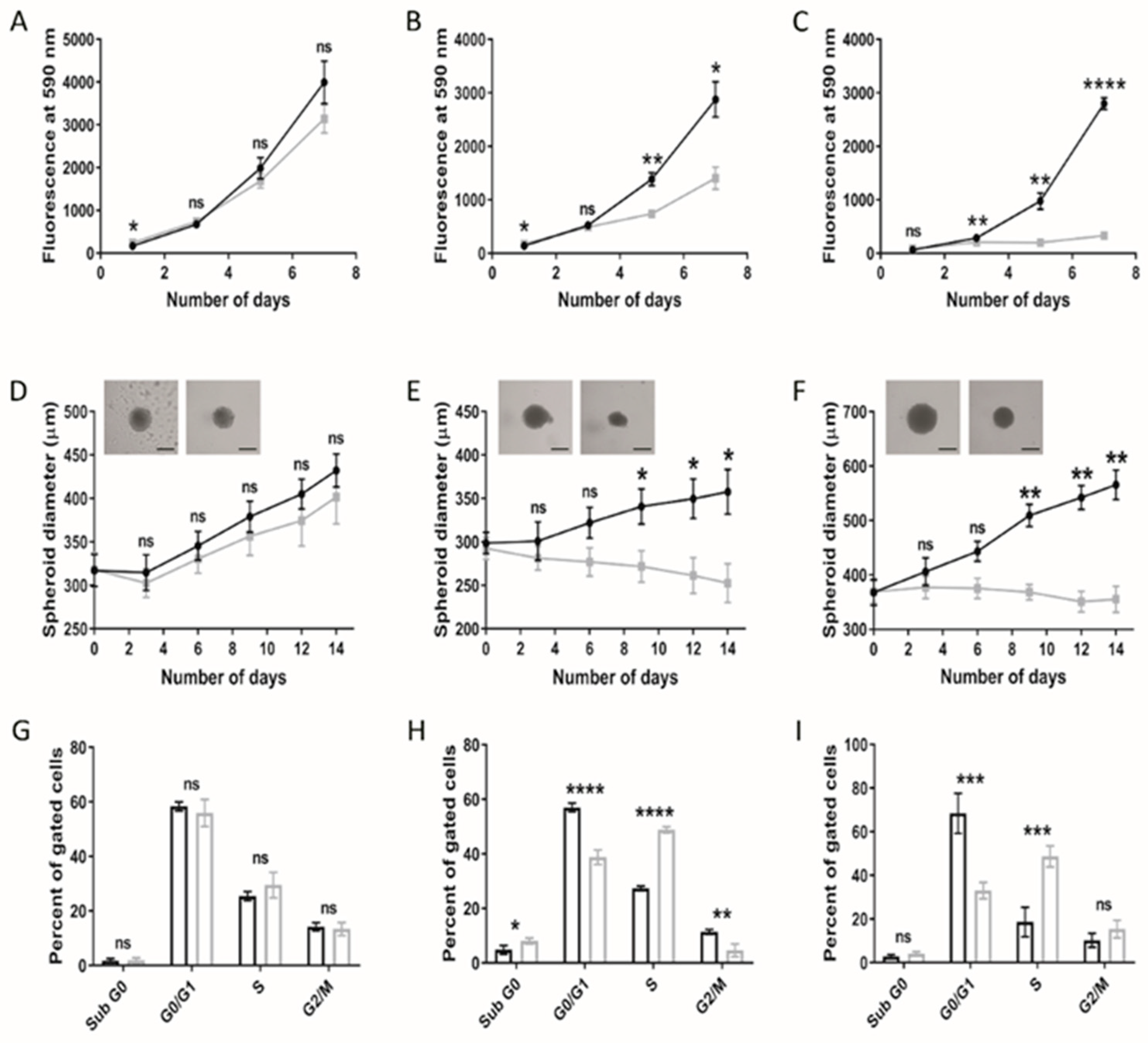
3.1. Paediatric Diffuse Glioma Cells Require Lipoproteins for Viability and Growth Maintenance
3.2. Cholesterol-Related Processes Define Diffuse Glioma Response to Lipoprotein Deprivation
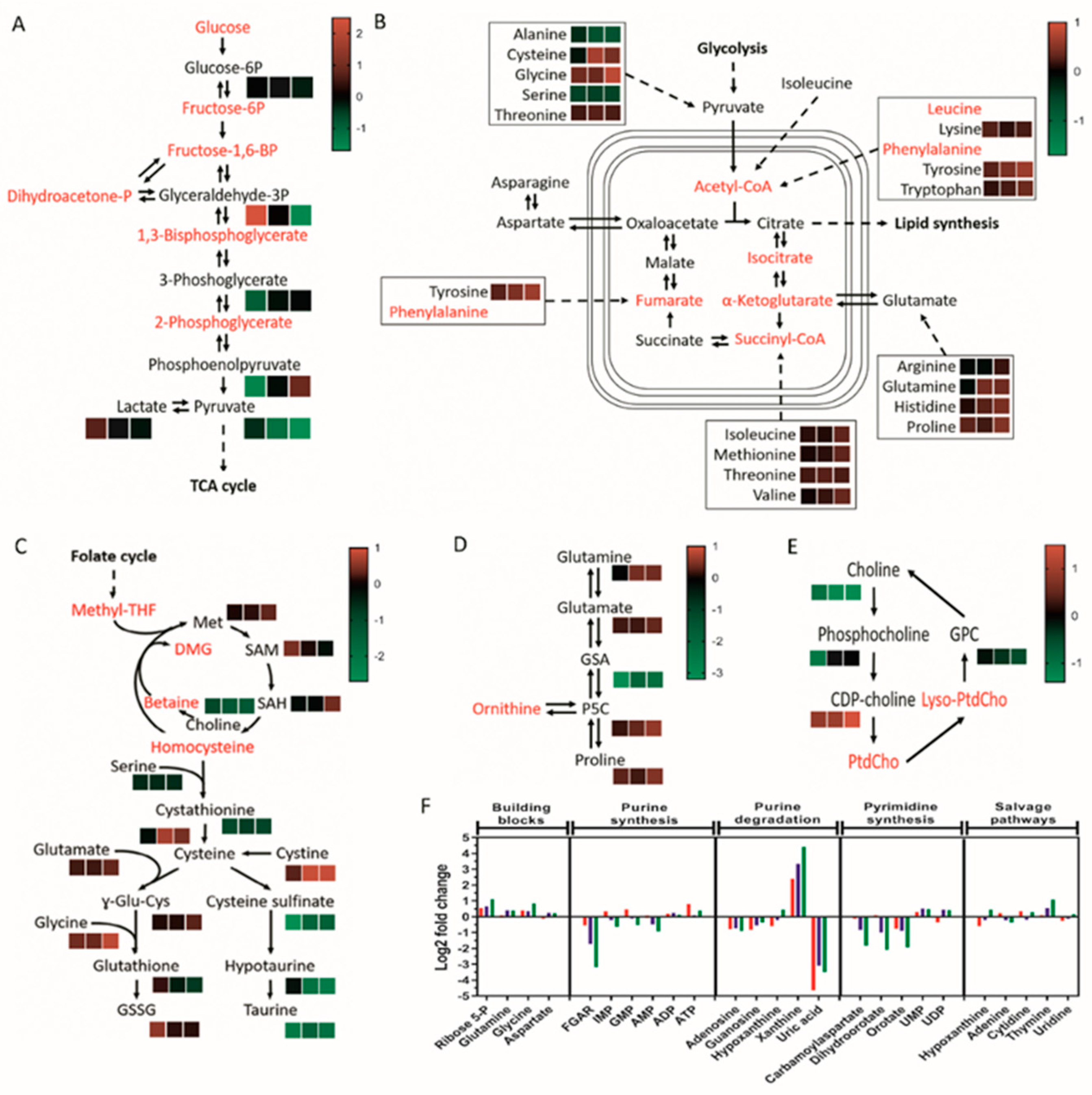
3.3. Taurine and Choline Metabolism Are Perturbed upon Removal of Exogenous Lipoproteins
3.4. Lipoprotein Starvation Induced Disturbances to Lipid and Cholesterol Species
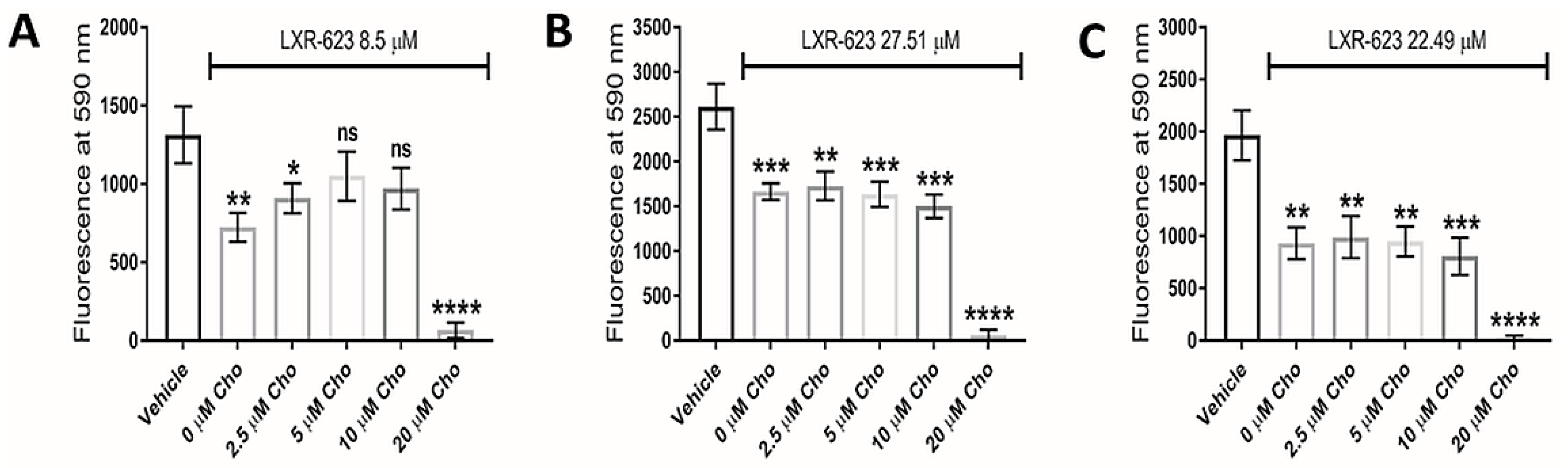
3.5. LXR Agonists
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ostrom, Q.T.; Gittleman, H.; Liao, P.; Vecchione-Koval, T.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary brain and other central nervous system tumors diagnosed in the United States in 2010–2014. Neuro. Oncol. 2017, 19, v1–v88. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sturm, D.; Witt, H.; Hovestadt, V.; Khuong-Quang, D.A.; Jones, D.T.W.; Konermann, C.; Pfaff, E.; Tönjes, M.; Sill, M.; Bender, S.; et al. Hotspot Mutations in H3F3A and IDH1 Define Distinct Epigenetic and Biological Subgroups of Glioblastoma. Cancer Cell 2012, 22, 425–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bastien, J.I.L.; McNeill, K.A.; Fine, H.A. Molecular characterizations of glioblastoma, targeted therapy, and clinical results to date. Cancer 2015, 121, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.B.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef]
- Furnari, F.B.; Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Heterogeneity of epidermal growth factor receptor signalling networks in glioblastoma. Nat. Rev. Cancer 2015, 15, 302–310. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Prins, R.M.; Dang, J.; Kuga, D.; Iwanami, A.; Soto, H.; Lin, K.Y.; Huang, T.T.; Akhavan, D.; Hock, M.B.; et al. EGFR Signaling Through an Akt-SREBP-1-Dependent, Rapamycin-Resistant Pathway Sensitizes Glioblastomas to Antilipogenic Therapy. Sci. Signal. 2009, 2, ra82. [Google Scholar] [CrossRef] [Green Version]
- Guo, D.; Reinitz, F.; Youssef, M.; Hong, C.; Nathanson, D.; Akhavan, D.; Kuga, D.; Amzajerdi, A.N.; Soto, H.; Zhu, S.; et al. An LXR agonist promotes glioblastoma cell death through inhibition of an EGFR/AKT/SREBP-1/LDLR-dependent pathway. Cancer Discov. 2011, 1, 442–456. [Google Scholar] [CrossRef] [Green Version]
- Villa, G.R.; Hulce, J.J.; Zanca, C.; Bi, J.; Ikegami, S.; Cahill, G.L.; Gu, Y.; Lum, K.M.; Masui, K.; Yang, H.; et al. An LXR-Cholesterol Axis Creates a Metabolic Co-Dependency for Brain Cancers. Cancer Cell 2016, 30, 683–693. [Google Scholar] [CrossRef] [Green Version]
- Simons, K.; Ikonen, E. How cells handle cholesterol. Science 2000, 290, 1721–1726. [Google Scholar] [CrossRef] [Green Version]
- Ho, Y.K.; Smith, R.G.; Brown, M.S.; Goldstein, J.L. Low-density lipoprotein (LDL) receptor activity in human acute myelogenous leukemia cells. Blood 1978, 52, 1099–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitols, S.; Gahrton, G.; Ost, A.; Peterson, C. Elevated low density lipoprotein receptor activity in leukemic cells with monocytic differentiation. Blood 1984, 63, 1186–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitols, S.; Angelin, B.; Ericsson, S.; Gahrton, G.; Juliusson, G.; Masquelier, M.; Paul, C.; Peterson, C.; Rudling, M.; Söderberg-Reid, K. Uptake of low density lipoproteins by human leukemic cells in vivo: Relation to plasma lipoprotein levels and possible relevance for selective chemotherapy. Proc. Natl. Acad. Sci. USA 1990, 87, 2598–2602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elegbede, J.A.; Elson, C.E.; Qureshi, A.; Dennis, W.H.; Yatvin, M.B. Increasing the thermosensitivity of a mammary tumor (CA755) through dietary modification. Eur. J. Cancer Clin. Oncol. 1986, 22, 607–615. [Google Scholar] [CrossRef]
- Brown, M.S.; Goldstein, J.L. A receptor-mediated pathway for cholesterol homeostasis. Science 1986, 232, 34–47. [Google Scholar] [CrossRef] [Green Version]
- Vance, J.E.; Hayashi, H. Formation and function of apolipoprotein E-containing lipoproteins in the nervous system. Biochim. Biophys. Acta 2010, 1801, 806–818. [Google Scholar] [CrossRef] [PubMed]
- Rudling, M.J.; Angelin, B.; Peterson, C.O.; Collins, V.P. Low density lipoprotein receptor activity in human intracranial tumors and its relation to the cholesterol requirement. Cancer Res. 1990, 50, 483–487. [Google Scholar]
- Ríos, M.; Foretz, M.; Viollet, B.; Prieto, A.; Fraga, M.; García-Caballero, T.; Costoya, J.A.; Señarís, R. Lipoprotein internalisation induced by oncogenic AMPK activation is essential to maintain glioblastoma cell growth. Eur. J. Cancer 2014, 50, 3187–3197. [Google Scholar] [CrossRef]
- Ivanov, D.P.; Parker, T.L.; Walker, D.A.; Alexander, C.; Ashford, M.B.; Gellert, P.R.; Garnett, M.C. Multiplexing spheroid volume, resazurin and acid phosphatase viability assays for high-throughput screening of tumour spheroids and stem cell neurospheres. PLoS ONE 2014, 9, e103817. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype-phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef]
- Xia, J.; Benner, M.J.; Hancock, R.E.W. NetworkAnalyst—Integrative approaches for protein-protein interaction network analysis and visual exploration. Nucleic Acids Res. 2014, 42, 167–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tautenhahn, R.; Böttcher, C.; Neumann, S. Highly sensitive feature detection for high resolution LC/MS. BMC Bioinform. 2008, 9, 504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheltema, R.A.; Jankevics, A.; Jansen, R.C.; Swertz, M.A.; Breitling, R. PeakML/mzMatch: A file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 2011, 83, 2786–2793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Creek, D.J.; Jankevics, A.; Breitling, R.; Watson, D.G.; Barrett, M.P.; Burgess, K.E.V. Toward global metabolomics analysis with hydrophilic interaction liquid chromatography-mass spectrometry: Improved metabolite identification by retention time prediction. Anal. Chem. 2011, 83, 8703–8710. [Google Scholar] [CrossRef] [Green Version]
- Creek, D.J.; Jankevics, A.; Burgess, K.E.V.; Breitling, R.; Barrett, M.P. IDEOM: An Excel interface for analysis of LC-MS-based metabolomics data. Bioinformatics 2012, 28, 1048–1049. [Google Scholar] [CrossRef] [PubMed]
- Sumner, L.W.; Amberg, A.; Barrett, D.; Beale, M.H.; Beger, R.; Daykin, C.A.; Fan, T.W.M.; Fiehn, O.; Goodacre, R.; Griffin, J.L.; et al. Proposed minimum reporting standards for chemical analysis: Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI). Metabolomics 2007, 3, 211–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumner, L.W.; Lei, Z.; Nikolau, B.J.; Saito, K.; Roessner, U.; Trengove, R. Proposed quantitative and alphanumeric metabolite identification metrics. Metabolomics 2014, 10, 1047–1049. [Google Scholar] [CrossRef]
- Robinet, P.; Wang, Z.; Hazen, S.L.; Smith, J.D. A simple and sensitive enzymatic method for cholesterol quantification in macrophages and foam cells. J. Lipid Res. 2010, 51, 3364–3369. [Google Scholar] [CrossRef] [Green Version]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Alexa, A.; Rahnenführer, J.; Lengauer, T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 2006, 22, 1600–1607. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Soufan, O.; Li, C.; Caraus, I.; Li, S.; Bourque, G.; Wishart, D.S.; Xia, J. MetaboAnalyst 4.0: Towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018, 46, W486–W494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppenol, W.H.; Bounds, P.L.; Dang, C.V. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat. Rev. Cancer 2011, 11, 325–337. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menendez, J.A.; Lupu, R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer 2007, 7, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Daniëls, V.W.; Smans, K.; Royaux, I.; Chypre, M.; Swinnen, J.V.; Zaidi, N. Cancer cells differentially activate and thrive on de novo lipid synthesis pathways in a low-lipid environment. PLoS ONE 2014, 9, 13–19. [Google Scholar] [CrossRef]
- Young, R.M.; Ackerman, D.; Quinn, Z.L.; Mancuso, A.; Gruber, M.; Liu, L.; Giannoukos, D.N.; Bobrovnikova-Marjon, E.; Diehl, J.A.; Keith, B.; et al. Dysregulated mTORC1 renders cells critically dependent on desaturated lipids for survival under tumor-like stress. Genes Dev. 2013, 27, 1115–1131. [Google Scholar] [CrossRef] [Green Version]
- Lewis, C.A.; Brault, C.; Peck, B.; Bensaad, K.; Griffiths, B.; Mitter, R.; Chakravarty, P.; East, P.; Dankworth, B.; Alibhai, D.; et al. SREBP maintains lipid biosynthesis and viability of cancer cells under lipid- and oxygen-deprived conditions and defines a gene signature associated with poor survival in glioblastoma multiforme. Oncogene 2015, 34, 5128–5140. [Google Scholar] [CrossRef]
- Angius, F.; Spolitu, S.; Uda, S.; Deligia, S.; Frau, A.; Banni, S.; Collu, M.; Accossu, S.; Madeddu, C.; Serpe, R.; et al. High-density lipoprotein contribute to G0-G1/S transition in Swiss NIH/3T3 fibroblasts. Sci. Rep. 2015, 5, 17812. [Google Scholar] [CrossRef] [Green Version]
- York, A.G.; Williams, K.J.; Argus, J.P.; Zhou, Q.D.; Brar, G.; Vergnes, L.; Gray, E.E.; Zhen, A.; Wu, N.C.; Yamada, D.H.; et al. Limiting Cholesterol Biosynthetic Flux Spontaneously Engages Type I IFN Signaling. Cell 2015, 163, 1716–1729. [Google Scholar] [CrossRef] [Green Version]
- Daniels, B.P.; Jujjavarapu, H.; Durrant, D.M.; Williams, J.L.; Green, R.R.; White, J.P.; Lazear, H.M.; Gale, M.; Diamond, M.S.; Klein, R.S. Regional astrocyte IFN signaling restricts pathogenesis during neurotropic viral infection. J. Clin. Investig. 2017, 127, 843–856. [Google Scholar] [CrossRef] [Green Version]
- Cheon, H.; Holvey-Bates, E.G.; Schoggins, J.W.; Forster, S.; Hertzog, P.; Imanaka, N.; Rice, C.M.; Jackson, M.W.; Junk, D.J.; Stark, G.R. IFNβ-dependent increases in STAT1, STAT2, and IRF9 mediate resistance to viruses and DNA damage. EMBO J. 2013, 32, 2751–2763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nan, J.; Wang, Y.; Yang, J.; Stark, G.R. IRF9 and unphosphorylated STAT2 cooperate with NF-κB to drive IL6 expression. Proc. Natl. Acad. Sci. USA 2018, 115, 3906–3911. [Google Scholar] [CrossRef] [Green Version]
- Inda, M.-M.; Bonavia, R.; Mukasa, A.; Narita, Y.; Sah, D.W.Y.; Vandenberg, S.; Brennan, C.; Johns, T.G.; Bachoo, R.; Hadwiger, P.; et al. Tumor heterogeneity is an active process maintained by a mutant EGFR-induced cytokine circuit in glioblastoma. Genes Dev. 2010, 24, 1731–1745. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boroughs, L.K.; DeBerardinis, R.J. Metabolic pathways promoting cancer cell survival and growth. Nat. Cell Biol. 2015, 17, 351–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villumsen, K.R.; Duelund, L.; Lambert, I.H. Acute cholesterol depletion leads to net loss of the organic osmolyte taurine in Ehrlich Lettré tumor cells. Amino Acids 2010, 39, 1521–1536. [Google Scholar] [CrossRef]
- Battelli, M.G.; Polito, L.; Bortolotti, M.; Bolognesi, A. Xanthine oxidoreductase in cancer: More than a differentiation marker. Cancer Med. 2016, 5, 546–557. [Google Scholar] [CrossRef]
- Sheikh, T.; Gupta, P.; Gowda, P.; Patrick, S.; Sen, E. Hexokinase 2 and nuclear factor erythroid 2-related factor 2 transcriptionally coactivate xanthine oxidoreductase expression in stressed glioma cells. J. Biol. Chem. 2018, 293, 4767–4777. [Google Scholar] [CrossRef] [Green Version]
- Bovenga, F.; Sabbà, C.; Moschetta, A. Uncoupling nuclear receptor LXR and cholesterol metabolism in cancer. Cell Metab. 2015, 21, 517–526. [Google Scholar] [CrossRef] [Green Version]
- Basseri, S.; Austin, R.C. Endoplasmic reticulum stress and lipid metabolism: Mechanisms and therapeutic potential. Biochem. Res. Int. 2012, 2012, 841362. [Google Scholar] [CrossRef]
- Röhrl, C.; Eigner, K.; Winter, K.; Korbelius, M.; Obrowsky, S.; Kratky, D.; Kovacs, W.J.; Stangl, H. Endoplasmic reticulum stress impairs cholesterol efflux and synthesis in hepatic cells. J. Lipid Res. 2014, 55, 94–103. [Google Scholar] [CrossRef] [Green Version]
- Clendening, J.W.; Penn, L.Z. Targeting tumor cell metabolism with statins. Oncogene 2012, 31, 4967–4978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaist, D.; Andersen, L.; Hallas, J.; Toft Sørensen, H.; Schrøder, H.D.; Friis, S. Use of statins and risk of glioma: A nationwide case–control study in Denmark. Br. J. Cancer 2013, 108, 715–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, T.T.T.; Ishida, C.T.; Shang, E.; Shu, C.; Torrini, C.; Zhang, Y.; Bianchetti, E.; Sanchez-Quintero, M.J.; Kleiner, G.; Quinzii, C.M.; et al. Activation of LXRβ inhibits tumor respiration and is synthetically lethal with Bcl-xL inhibition. EMBO Mol. Med. 2019, 11, e10769. [Google Scholar] [CrossRef] [PubMed]
- Candelaria, N.R.; Addanki, S.; Zheng, J.; Nguyen-Vu, T.; Karaboga, H.; Dey, P.; Gabbi, C.; Vedin, L.-L.; Liu, K.; Wu, W.; et al. Antiproliferative effects and mechanisms of liver X receptor ligands in pancreatic ductal adenocarcinoma cells. PLoS ONE 2014, 9, e106289. [Google Scholar] [CrossRef] [PubMed]
- Pencheva, N.; Buss, C.G.; Posada, J.; Merghoub, T.; Tavazoie, S.F. Broad-spectrum therapeutic suppression of metastatic melanoma through nuclear hormone receptor activation. Cell 2014, 156, 986–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]

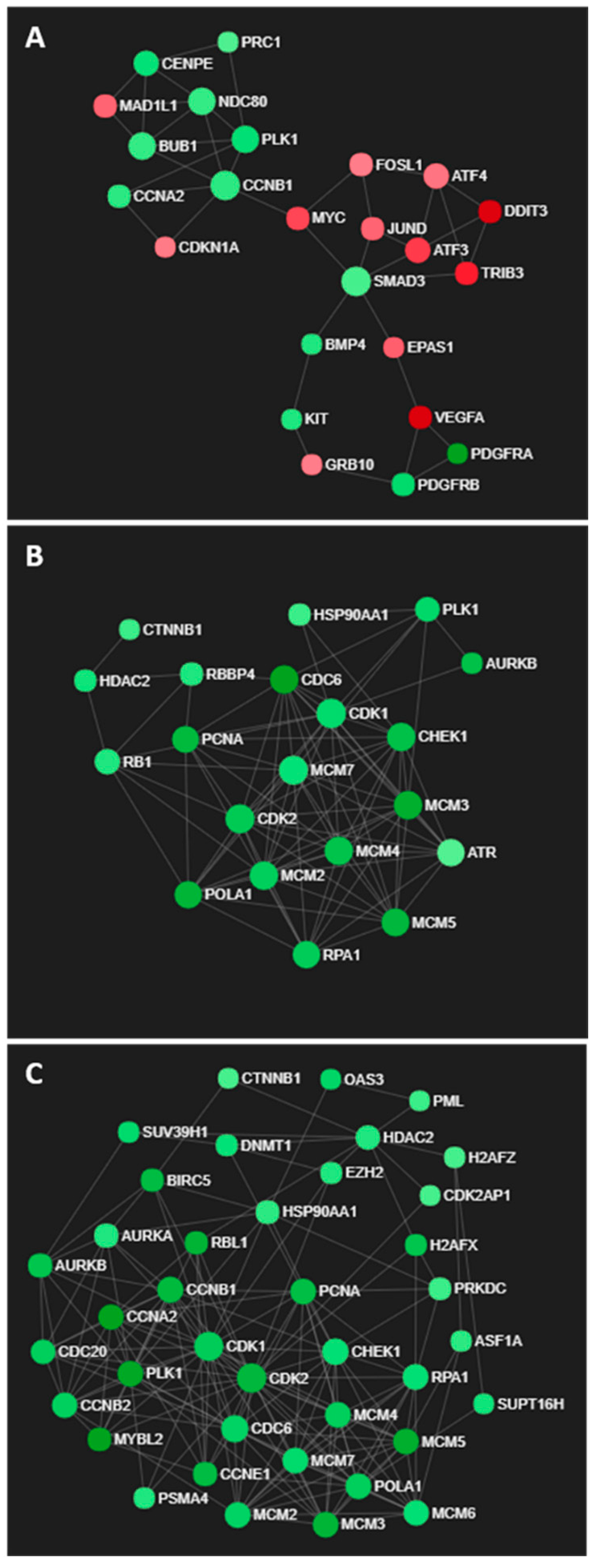
| GO.ID | Term | Annotated | Significant | Expected | classicFisher | Weight01KS | Genes |
|---|---|---|---|---|---|---|---|
| U373 | |||||||
| GO:0071456 | Cellular response to hypoxia | 98 | 5 | 0.53 | 1.70 × 104 | 8.80 × 104 | STC, CA9, BNIP3, VEGFA, NDNF |
| GO:0032376 | Positive regulation of cholesterol transport | 11 | 2 | 0.06 | 1.52 × 103 | 2.69 × 102 | ABCA1, LIPG |
| GO:0030823 | Regulation of cGMP metabolic process | 12 | 2 | 0.06 | 1.82 × 103 | 3.50 × 102 | VEGFA, PDE5A |
| GO:0030199 | Collagen fibril organisation | 15 | 3 | 0.08 | 6.40 × 105 | 4.81 × 102 | LOX, LUM, P4HA1 |
| KNS42 | |||||||
| GO:0060337 | Type 1 interferon signalling pathway | 52 | 16 | 0.99 | 8.00 × 10−16 | 2.10 × 109 | IFI6. OAS2, IRF7, OAS1, MX1, XAF1, IFI35, IFITM1, IRF9, IFI27, USP18, OAS3, ISG15, STAT2, IFIT3, IFIT2 |
| GO:0051607 | Defence response to virus | 107 | 17 | 2.05 | 1.30 × 10−11 | 2.00 × 105 | DDIT4, OAS2, IRF7, OAS1, IFI44L, MX1, IFITM1, IRF9, OAS3, ISG15, IFNE, CXCL10, STAT2, HTRA1, IFIT3, IFIT2, MICA |
| GO:0006695 | Cholesterol biosynthetic process | 42 | 15 | 0.8 | 5.10 × 10−16 | 8.00 × 103 | INSIG1, HMGCS1, MSM01, DHCR24, MVK, DHCR7, NSDHL, ACAT2, SQLE, MVD, SC5D, EBP, IDI1, TM7SF2, HMGCR |
| SF188 | |||||||
| GO:0060337 | Type 1 interferon signalling pathway | 55 | 20 | 3.08 | 4.90× 10−12 | 1.30 × 109 | IFI6, IFIT1, OAS2, XAF1, STAT2, MX1, IFIT2, IFIT3, IRF7, IRF9, MIR21, USP18, OAS3, ISG15, WNT5A, IFITM1, STAT1, SAMHD1, OASL, IFI35 |
| GO:0045540 | Regulation of cholesterol biosynthetic process | 27 | 14 | 1.51 | 2.50 × 10−11 | 1.10 × 107 | HMGCS1, MVK, SC5D, TM7SF2, DHCR7, MVD, SQLE, CYP51A1, IDI1, LSS, SCD, HMGCR, FDPS, FASN |
| GO:0051607 | Defence response to virus | 111 | 28 | 6.22 | 7.30 × 10−12 | 1.80 × 106 | IFIH1, IFIT1, OAS2, IL1B, STAT2, MX1, IFIT2, IFIT3, IL6, IRF7, IFI44L, PML, IRF9, HERC5, DDX58, OAS3, MICA, ISG15, GBP3, CXCL10, IFITM1, TNFAIP3, STAT1, HTRA1, SAMHD1, PARP9, OASL, EXOSC5 |
| GO:0006695 | Cholesterol biosynthetic process | 41 | 20 | 2.3 | 5.40× 10−15 | 2.80 × 103 | INSIG1, HMGCS1, MSMO1, MVK, ACAT2, SC5D, TM7SF2, EBP, DHCR7, MVD, SQLE, CYP51A1, DHCR24, IDI1, LSS, SCD, HMGCR, NSDHL, FDPS, FASN |
| GO:0016126 | Sterol biosynthetic process | 43 | 21 | 2.41 | 1.10 × 10−15 | 3.10 × 102 | INSIG1, HMGCS1, MSMO1, MVK, ACAT2, SC5D, TM7SF2, EBP, DHCR7, MVD, SQLE, CYP51A1, DHCR24, IDI1, LSS, CYB5R2, SCD, HMGCR, NSDHL, FDPS, FASN |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wood, J.; Abdelrazig, S.; Evseev, S.; Ortori, C.; Castellanos-Uribe, M.; May, S.T.; Barrett, D.A.; Diksin, M.; Chakraborty, S.; Kim, D.-H.; et al. Lipoprotein Deprivation Reveals a Cholesterol-Dependent Therapeutic Vulnerability in Diffuse Glioma Metabolism. Cancers 2022, 14, 3873. https://doi.org/10.3390/cancers14163873
Wood J, Abdelrazig S, Evseev S, Ortori C, Castellanos-Uribe M, May ST, Barrett DA, Diksin M, Chakraborty S, Kim D-H, et al. Lipoprotein Deprivation Reveals a Cholesterol-Dependent Therapeutic Vulnerability in Diffuse Glioma Metabolism. Cancers. 2022; 14(16):3873. https://doi.org/10.3390/cancers14163873
Chicago/Turabian StyleWood, James, Salah Abdelrazig, Sergey Evseev, Catherine Ortori, Marcos Castellanos-Uribe, Sean T. May, David A. Barrett, Mohammed Diksin, Sajib Chakraborty, Dong-Hyun Kim, and et al. 2022. "Lipoprotein Deprivation Reveals a Cholesterol-Dependent Therapeutic Vulnerability in Diffuse Glioma Metabolism" Cancers 14, no. 16: 3873. https://doi.org/10.3390/cancers14163873
APA StyleWood, J., Abdelrazig, S., Evseev, S., Ortori, C., Castellanos-Uribe, M., May, S. T., Barrett, D. A., Diksin, M., Chakraborty, S., Kim, D.-H., Grundy, R. G., & Rahman, R. (2022). Lipoprotein Deprivation Reveals a Cholesterol-Dependent Therapeutic Vulnerability in Diffuse Glioma Metabolism. Cancers, 14(16), 3873. https://doi.org/10.3390/cancers14163873








