Possible Bias in Supervised Deep Learning Algorithms for CT Lung Nodule Detection and Classification
Abstract
:Simple Summary
Abstract
1. Introduction
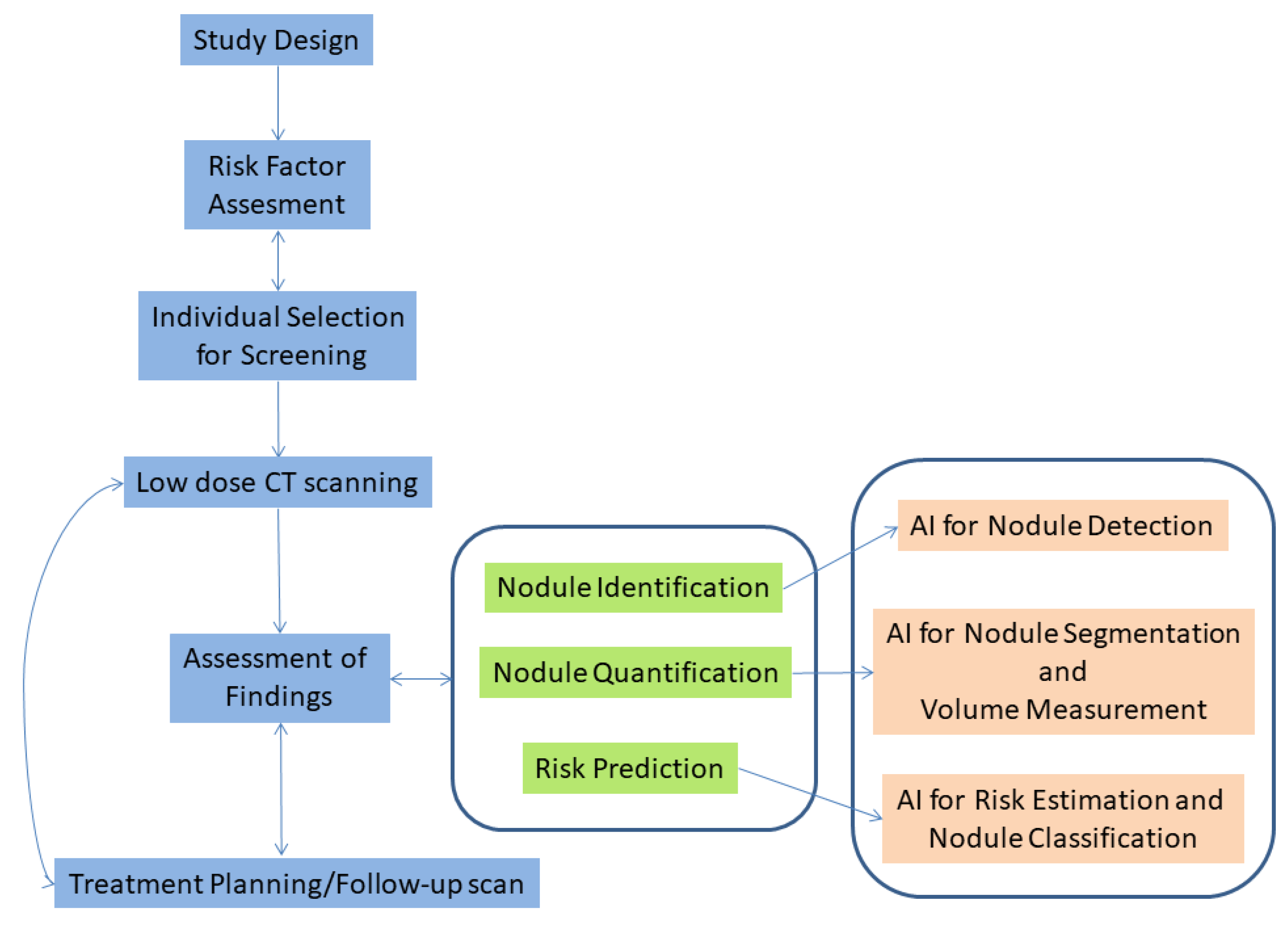
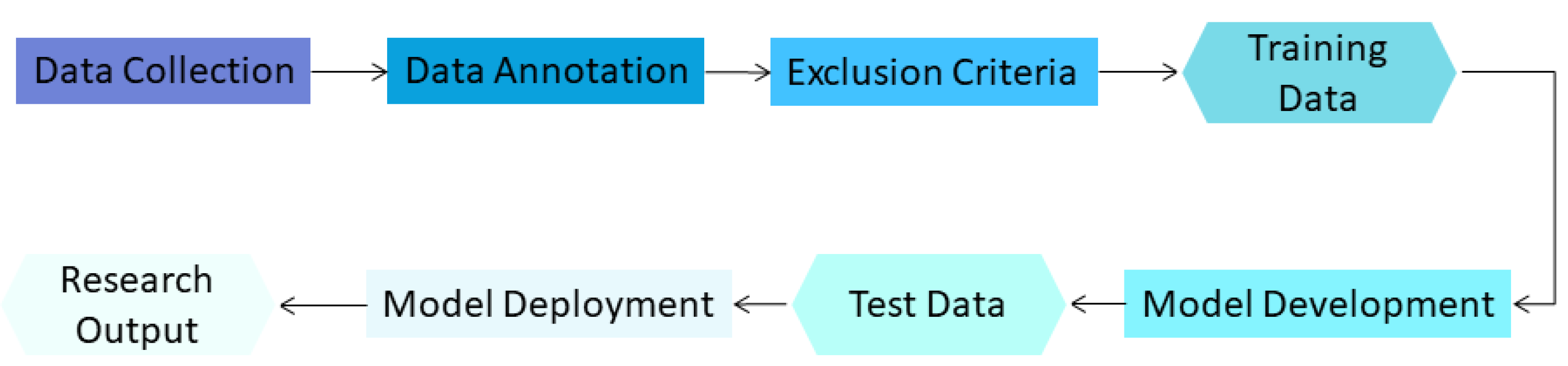
2. Materials and Methods
2.1. Overview
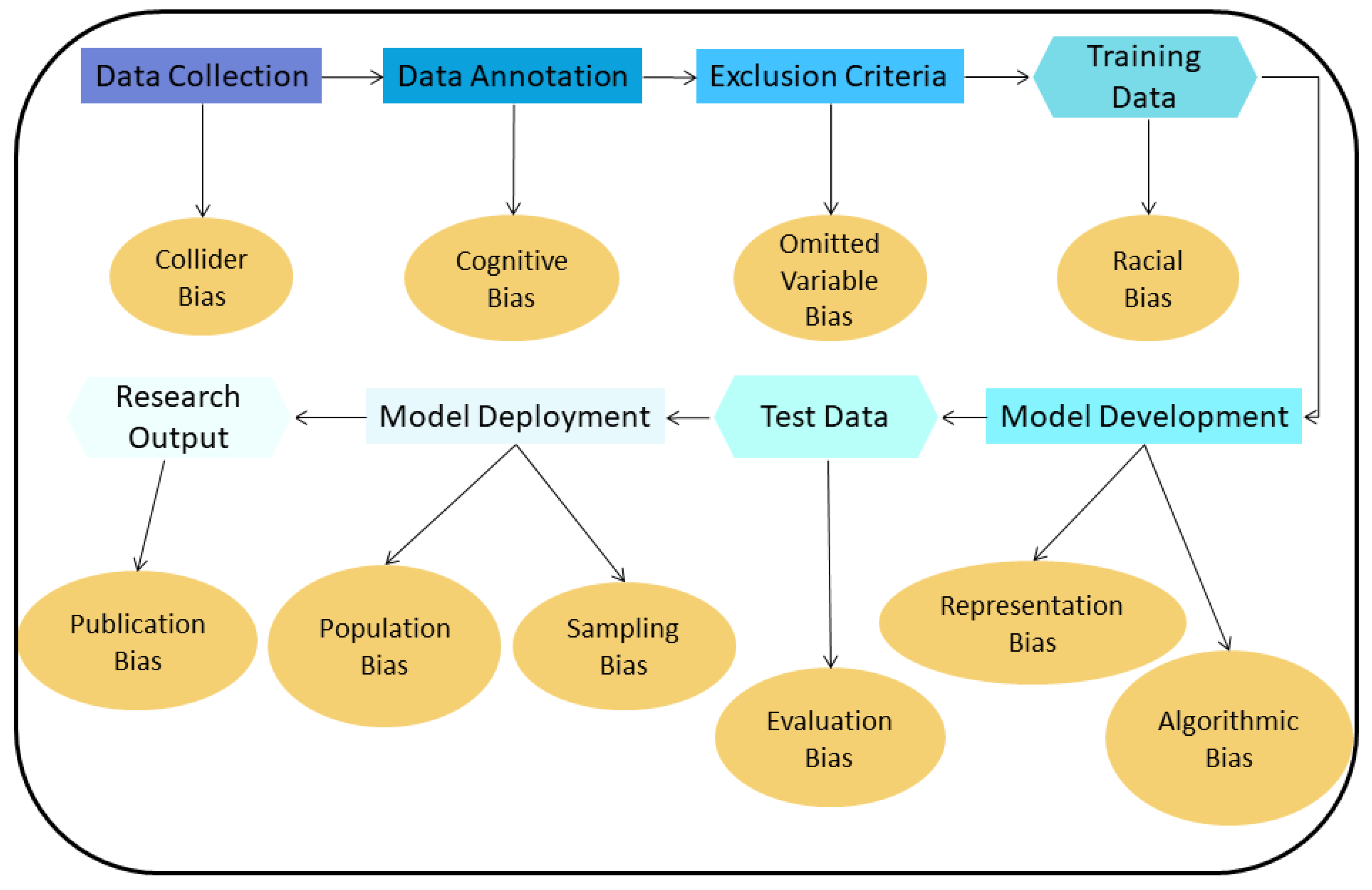
2.2. Types of Bias in Chest CT AI Algorithms
- 1
- Collider bias: A Collider is a variable that is the effect of two other variables. If it is assumed that these two variables influence how samples are selected, the association between these variables may become distorted, affecting the collider directly or indirectly. If then, the collider is used to draw conclusions about these associations, these conclusions will be invalid [19].
- 2
- Cognitive bias: In the training dataset used to develop an algorithm, radiologists are commonly responsible for finding and annotating nodules. If most radiologists missed a particular nodule and if a majority vote is used to decide if this finding is a nodule or not, then the resulting annotation will be incorrect for that finding (erroneously classified as non-nodule). The difference in interpretation and diagnosis by different radiologists results in cognitive bias [13,20].
- 3
- Omitted variable bias: When creating an AI algorithm, some important variables that can affect its performance may be deliberately left out, leading to this type of bias [21].
- 4
- Representation/Racial bias: Representation bias occurs when the dataset used to develop an AI algorithm is not diverse enough to represent many different population groups and/or characteristics. Sometimes, this type of bias is indistinguishable from racial bias; this could significant harm the underrepresented group [16].
- 5
- Algorithmic Bias: Most biases are assumed to be unintended, meaning they are unwanted and appear mostly during the data collection and data selection process. Algorithmic bias is an example of bias inserted on purpose and can be the result of the motives of the programmers/companies that develop the algorithm [22]. An AI algorithm can be deliberately designed to skew results f.e. with the goal to maximize profit.
- 6
- Evaluation bias: Such bias arises when an evaluation of the performance of an algorithm is conducted using an inappropriate dataset, like an internal one, which resembles the data used to train the algorithm, or even a contaminated evaluation dataset that contains samples from the training dataset [17]. This results in an overly optimistic estimate of the performance of the algorithm, compared to when this evaluation is performed using an external dataset collected, for example, from another hospital.
- 7
- Population bias: If an AI algorithm is developed using data acquired under specific conditions e.g., a specific population, and is applied to different clinical settings than those it was developed for, this type of bias emerges [17].
- 8
- Sampling bias: This bias can occur when an AI algorithm is developed using data sampled non-randomly, which may miss important cases and features of interest encountered during clinical practice [15].
- 9
- Publication bias: A common type of bias in many fields. This bias results from journals not willing to publish studies with negative or suboptimal findings, or studies that confirm or reject already published results (replication studies) [23].
3. Results
3.1. Bias in Chest CT Nodule Detection and Classification
- Collider Bias
- Cognitive Bias
- Omitted Variable Bias
- Representation/Racial Bias
- Algorithmic Bias
- Evaluation Bias
- Population Bias
- Sampling Bias
- Publication Bias
3.2. Ways to Mitigate Bias
- -
- How was the collection and annotation procedure of the dataset performed? Who contributed to that and on behalf of which entity?
- -
- Which characteristics/parameters were considered in the dataset creation process? How many examples of each of those characteristics/parameters were included in the dataset?
- -
- For which population groups and/or characteristics should the developed algorithm be used? Is the dataset representative of that population?
- -
- What are the characteristics of the dataset used for model validation?
- -
- Who funded the dataset creation process?
- -
- Was any preprocessing or cleaning of the dataset performed?
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- WorldHeathOrg. Cancer Fact Sheet No. 297. 2013. Available online: http://www.who.int/mediacentre/factsheets/fs297/en/ (accessed on 13 December 2021).
- International Early Lung Cancer Action Program Investigators. Survival of Patients with Stage I Lung Cancer Detected on CT Screening. N. Engl. J. Med. 2006, 355, 1763–1771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Birring, S.S.; Peake, M.D. Symptoms and the early diagnosis of lung cancer. Thorax 2005, 60, 268–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diederich, S.; Das, M. Solitary pulmonary nodule: Detection and management. Cancer Imaging Off. Publ. Int. Cancer Imaging Soc. 2006, 6, S42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Huang, S.; Zeng, Q.; Zhang, M.; Ni, Z.; Li, X.; Xu, X. A retrospective study analyzing missed diagnosis of lung metastases at their early stages on computed tomography. J. Thorac. Dis. 2019, 11, 3360. [Google Scholar] [CrossRef]
- Burzic, A.; O’Dowd, E.; Baldwin, D. The Future of Lung Cancer Screening: Current Challenges and Research Priorities. Cancer Manag. Res. 2022, 14, 637–645. [Google Scholar] [CrossRef]
- Obuchowski, N.; Bullen, J. Statistical Considerations for Testing an AI Algorithm Used for Prescreening Lung CT Images. Contemp. Clin. Trials Commun. 2019, 16, 100434. [Google Scholar] [CrossRef]
- Hwang, E.J.; Park, C.M. Clinical Implementation of Deep Learning in Thoracic Radiology: Potential Applications and Challenges. Korean J. Radiol. 2020, 21, 511. [Google Scholar] [CrossRef] [Green Version]
- Jiang, B.; Li, N.; Shi, X.; Zhang, S.; Li, J.; de Bock, G.H.; Vliegenthart, R.; Xie, X. Deep Learning Reconstruction Shows Better Lung Nodule Detection for Ultra–Low-Dose Chest CT. Radiology 2022, 303, 202–212. [Google Scholar] [CrossRef]
- Müller, F.C.; Raaschou, H.; Akhtar, N.; Brejnebøl, M.; Collatz, L.; Andersen, M.B. Impact of Concurrent Use of Artificial Intelligence Tools on Radiologists Reading Time: A Prospective Feasibility Study. Acad. Radiol. 2021, 29, 1085–1090. [Google Scholar] [CrossRef]
- Grand Challenge. AI Software for Radiology. 2022. Available online: https://grand-challenge.org/aiforradiology/ (accessed on 7 April 2022).
- Lachance, C.C.; Walter, M. Artificial Intelligence for Classification of Lung Nodules: A Review of Clinical Utility, Diagnostic Accuracy, Cost-Effectiveness, and Guidelines; Canadian Agency for Drugs and Technologies in Health: Ottawa, ON, Canada, 2020. [Google Scholar]
- Busby, L.P.; Courtier, J.L.; Glastonbury, C.M. Bias in Radiology: The How and Why of Misses and Misinterpretations. RadioGraphics 2018, 38, 236–247. [Google Scholar] [CrossRef] [Green Version]
- Arnold, M.; Bellamy, R.K.E.; Hind, M.; Houde, S.; Mehta, S.; Mojsilovic, A.; Nair, R.; Ramamurthy, K.N.; Reimer, D.; Olteanu, A.; et al. FactSheets: Increasing Trust in AI Services through Supplier’s Declarations of Conformity. IBM J. Res. Dev. 2018, 63. [Google Scholar] [CrossRef] [Green Version]
- Vokinger, K.; Feuerriegel, S.; Kesselheim, A. Mitigating bias in machine learning for medicine. Commun. Med. 2021, 1, 1–3. [Google Scholar] [CrossRef]
- Obermeyer, Z.; Powers, B.; Vogeli, C.; Mullainathan, S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019, 366, 447–453. [Google Scholar] [CrossRef] [Green Version]
- Mehrabi, N.; Morstatter, F.; Saxena, N.; Lerman, K.; Galstyan, A. A Survey on Bias and Fairness in Machine Learning. arXiv 2022, arXiv:1908.09635. [Google Scholar] [CrossRef]
- Suresh, H.; Guttag, J. A Framework for Understanding Sources of Harm throughout the Machine Learning Life Cycle. In Proceedings of the Equity and Access in Algorithms, Mechanisms, and Optimization, virtually, 5–9 October 2021; ACM: New York, NY, USA, 2021. [Google Scholar] [CrossRef]
- Griffith, G.; Morris, T.; Tudball, M.; Herbert, A.; Mancano, G.; Pike, L.; Sharp, G.; Sterne, J.; Palmer, T.; Davey Smith, G.; et al. Collider bias undermines our understanding of COVID-19 disease risk and severity. Nat. Commun. 2020, 11, 1–12. [Google Scholar] [CrossRef]
- Saposnik, G.; Redelmeier, D.; Ruff, C.; Tobler, P. Cognitive biases associated with medical decisions: A systematic review. BMC Med. Inform. Decis. Mak. 2016, 16, 138. [Google Scholar] [CrossRef] [Green Version]
- Wilms, R.; Mäthner, E.; Winnen, L.; Lanwehr, R. Omitted variable bias: A threat to estimating causal relationships. Methods Psychol. 2021, 5, 100075. [Google Scholar] [CrossRef]
- PatriciaHannon. Researchers Say Use of Artificial Intelligence in Medicine Raises Ethical Questions. 2018. Available online: https://med.stanford.edu/news/all-news/2018/03/researchers-say-use-of-ai-in-medicine-raises-ethical-questions.html (accessed on 7 April 2022).
- Montori, V.M.; Smieja, M.; Guyatt, G.H. Publication bias: A brief review for clinicians. Mayo Clin. Proc. Mayo Clin. 2001, 75, 1284–1288. [Google Scholar] [CrossRef] [Green Version]
- van Amsterdam, W.; Verhoeff, J.; Jong, P.; Leiner, T.; Eijkemans, M. Eliminating biasing signals in lung cancer images for prognosis predictions with deep learning. NPJ Digit. Med. 2019, 2, 122. [Google Scholar] [CrossRef] [Green Version]
- Cui, S.; Ming, S.; Lin, Y.; Chen, F.; Shen, Q.; Li, H.; Chen, G.; Gong, X.; Wang, H. Development and clinical application of deep learning model for lung nodules screening on CT images. Sci. Rep. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Nasrullah, N.; Sang, J.; Alam, M.S.; Mateen, M.; Cai, B.; Hu, H. Automated Lung Nodule Detection and Classification Using Deep Learning Combined with Multiple Strategies. Sensors 2019, 19, 3722. [Google Scholar] [CrossRef] [Green Version]
- Devaraj, A. Missed cancers in lung cancer screening–more than meets the eye. Eur. Radiol. 2015, 25, 89–91. [Google Scholar] [CrossRef] [Green Version]
- Veronesi, G.; Maisonneuve, P.; Spaggiari, L.; Rampinelli, C.; Pardolesi, A.; Bertolotti, R.; Filippi, N.; Bellomi, M. Diagnostic Performance of Low-Dose Computed Tomography Screening for Lung Cancer over Five Years. J. Thorac. Oncol. 2014, 9, 935–939. [Google Scholar] [CrossRef] [Green Version]
- Ciello, A.; Franchi, P.; Contegiacomo, A.; Cicchetti, G.; Bonomo, L.; Larici, A. Missed lung cancer: When, where, and why? Diagn. Interv. Radiol. 2017, 23, 118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Li, Q.; Ma, J.; Zhou, Z.; Sun, M.; Deng, Y.; Tu, W.; Wang, Y.; Fan, L.; Xia, C.; et al. Evaluating a Fully Automated Pulmonary Nodule Detection Approach and Its Impact on Radiologist Performance. Radiol. Artif. Intell. 2019, 1, e180084. [Google Scholar] [CrossRef]
- Maci, E.; Comito, F.; Frezza, A.M.; Tonini, G.; Pezzuto, A. Lung Nodule and Functional Changes in Smokers After Smoking Cessation Short-Term Treatment. Cancer Investig. 2014, 32, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Markaki, M.; Tsamardinos, I.; Langhammer, A.; Lagani, V.; Hveem, K.; Røe, O. A Validated Clinical Risk Prediction Model for Lung Cancer in Smokers of All Ages and Exposure Types: A HUNT Study. EBioMedicine 2018, 31, 36–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arslan, S.; Ünal, E. One of the many faces of COVID-19 infection: An irregularly shaped pulmonary nodule. Insights Into Imaging 2021, 12, 1–2. [Google Scholar] [CrossRef]
- Wachuła, E.; Szabłowska-Siwik, S.; Czyżewski, D.; Kozielski, J.; Adamek, M. Emphysema affects the number and appearance of solitary pulmonary nodules identified in chest low-dose computed tomography: A study on high risk lung cancer screenees recruited in Silesian District. Pol. Arch. Intern. Med. 2019, 130, 17–24. [Google Scholar] [CrossRef]
- Banerjee, I.; Bhimireddy, A.R.; Burns, J.L.; Celi, L.A.; Chen, L.C.; Correa, R.; Dullerud, N.; Ghassemi, M.; Huang, S.C.; Kuo, P.C.; et al. Reading Race: AI Recognises Patient’s Racial Identity In Medical Images. arXiv 2021, arXiv:2107.10356. [Google Scholar] [CrossRef]
- Ma, J.; Song, Y.; Tian, X.; Hua, Y.; Zhang, R.; Wu, J. Survey on deep learning for pulmonary medical imaging. Front. Med. 2020, 14, 450. [Google Scholar] [CrossRef] [Green Version]
- Hosny, A.; Parmar, C.; Coroller, T.; Grossmann, P.; Zeleznik, R.; Kumar, A.; Bussink, J.; Gillies, R.; Mak, R.; Aerts, H. Deep learning for lung cancer prognostication: A retrospective multi-cohort radiomics study. PLoS Med. 2018, 15, e1002711. [Google Scholar] [CrossRef] [Green Version]
- Gruetzemacher, R.; Gupta, A.; Paradice, D. 3D deep learning for detecting pulmonary nodules in CT scans. J. Am. Med. Inform. Assoc. 2018, 25, 1301–1310. [Google Scholar] [CrossRef]
- National Lung Screening Trial Research Team. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [Green Version]
- Yu, A.C.; Eng, J. One Algorithm May Not Fit All: How Selection Bias Affects Machine Learning Performance. RadioGraphics 2020, 40, 1932–1937. [Google Scholar] [CrossRef]
- Wang, J.; Chen, X.; Lu, H.; Zhang, L.; Pan, J.; Bao, Y.; Su, J.; Qian, D. Feature-shared adaptive-boost deep learning for invasiveness classification of pulmonary subsolid nodules in CT images. Med. Phys. 2020, 47, 1738–1749. [Google Scholar] [CrossRef]
- Larici, A.R.; Farchione, A.; Franchi, P.; Ciliberto, M.; Cicchetti, G.; Calandriello, L.; del Ciello, A.; Bonomo, L. Lung nodules: Size still matters. Eur. Respir. Rev. 2017, 26, 170025. [Google Scholar] [CrossRef]
- Fortunebusinessinsights. CT Scanners Market Size, Share and Covid-19 Impact Analysis, by Technology, by Application, by Modality, by End-Yse, Diagnostic and Regional Forecast 2020–2027; Fortune Business Insights: Pune, India, 2019. [Google Scholar]
- Willemink, M.J.; Koszek, W.A.; Hardell, C.; Wu, J.; Fleischmann, D.; Harvey, H.; Folio, L.R.; Summers, R.M.; Rubin, D.L.; Lungren, M.P. Preparing Medical Imaging Data for Machine Learning. Radiology 2020, 295, 4–15. [Google Scholar] [CrossRef]
- Li, D.; Mikela Vilmun, B.; Frederik Carlsen, J.; Albrecht-Beste, E.; Ammitzbøl Lauridsen, C.; Bachmann Nielsen, M.; Lindskov Hansen, K. The Performance of Deep Learning Algorithms on Automatic Pulmonary Nodule Detection and Classification Tested on Different Datasets That Are Not Derived from LIDC-IDRI: A Systematic Review. Diagnostics 2019, 9, 207. [Google Scholar] [CrossRef] [Green Version]
- Huang, G.; Wei, X.; Tang, H.; Bai, F.; Lin, X.; Xue, D. A systematic review and meta-analysis of diagnostic performance and physicians’ perceptions of artificial intelligence (AI)-assisted CT diagnostic technology for the classification of pulmonary nodules. J. Thorac. Dis. 2021, 13, 4797. [Google Scholar] [CrossRef]
- Lee, J.H.; Sun, H.Y.; Park, S.; Kim, H.; Hwang, E.J.; Goo, J.M.; Park, C.M. Performance of a Deep Learning Algorithm Compared with Radiologic Interpretation for Lung Cancer Detection on Chest Radiographs in a Health Screening Population. Radiology 2020, 297, 687–696. [Google Scholar] [CrossRef]
- Muşat, B.; Andonie, R. Semiotic Aggregation in Deep Learning. Entropy 2020, 22, 1365. [Google Scholar] [CrossRef]
- Nagaraj, Y.; Cornelissen, L.; Cai, J.; Rook, M.; Wisselink, H.; Veldhuis, R.; Oudkerk, M.; Vliegenthart, R.; Van Ooijen, P. An unsupervised anomaly detection model to identify emphysema in low-dose computed tomography. TechRxiv 2021. [Google Scholar] [CrossRef]
- Armato, S.G. Deep Learning Demonstrates Potential for Lung Cancer Detection in Chest Radiography. Radiology 2020, 297, 697–698. [Google Scholar] [CrossRef]
- Cobbe, J.; Lee, M.S.A.; Singh, J. Reviewable Automated Decision-Making: A Framework for Accountable Algorithmic Systems. In Proceedings of the 2021 ACM Conference on Fairness, Accountability, and Transparency, Virtual Event. Toronto, ON, Canada, 3–10 March 2021. [Google Scholar] [CrossRef]
- Gebru, T.; Morgenstern, J.; Vecchione, B.; Vaughan, J.W.; Wallach, H.; Daumé, H.; Crawford, K. Datasheets for Datasets. arXiv 2018, arXiv:1803.09010. [Google Scholar] [CrossRef]
- Kim, D.W.; Jang, H.; Kim, K.; Shin, Y.; Park, S. Design Characteristics of Studies Reporting the Performance of Artificial Intelligence Algorithms for Diagnostic Analysis of Medical Images: Results from Recently Published Papers. Korean J. Radiol. 2019, 20, 405. [Google Scholar] [CrossRef]
- Zhang, B.H.; Lemoine, B.; Mitchell, M. Mitigating Unwanted Biases with Adversarial Learning. arXiv 2018, arXiv:1801.07593. [Google Scholar] [CrossRef]
- Park, S.H.; Han, K. Methodologic Guide for Evaluating Clinical Performance and Effect of Artificial Intelligence Technology for Medical Diagnosis and Prediction. Radiology 2018, 286, 800–809. [Google Scholar] [CrossRef]
- Gu, D.; Liu, G.; Xue, Z. On the performance of lung nodule detection, segmentation and classification. Comput. Med. Imaging Graph. 2021, 89, 101886. [Google Scholar] [CrossRef]
- Helm, E.; Silva, C.T.; Roberts, H.C.; Manson, D.E.; Seed, M.; Amaral, J.G.; Babyn, P.S. Computer-aided detection for the identification of pulmonary nodules in pediatric oncology patients: Initial experience. Pediatr. Radiol. 2009, 39, 685–693. [Google Scholar] [CrossRef]
- Smuha, N. High-Level Expert Group on Artificial Intelligence. 2019. Available online: https://www.google.com.hk/url?sa=t&rct=j&q=&esrc=s&source=web&cd=&ved=2ahUKEwj93Mqhk7z5AhXYAKYKHRlOB3YQFnoECAoQAQ&url=https%3A%2F%2Fwww.aepd.es%2Fsites%2Fdefault%2Ffiles%2F2019-12%2Fai-definition.pdf&usg=AOvVaw3H9PZ6Ku1h_p2KFU2Zuqtb (accessed on 7 April 2022).
- Xia, C.; Rook, M.; Pelgrim, G.; Sidorenkov, G.; Wisselink, H.; van Bolhuis, J.; van Ooijen, P.; Guo, J.; Oudkerk, M.; Groen, H.; et al. Early imaging biomarkers of lung cancer, COPD and coronary artery disease in the general population: Rationale and design of the ImaLife (Imaging in Lifelines) Study. Eur. J. Epidemiol. 2020, 35, 75–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sourlos, N.; Wang, J.; Nagaraj, Y.; van Ooijen, P.; Vliegenthart, R. Possible Bias in Supervised Deep Learning Algorithms for CT Lung Nodule Detection and Classification. Cancers 2022, 14, 3867. https://doi.org/10.3390/cancers14163867
Sourlos N, Wang J, Nagaraj Y, van Ooijen P, Vliegenthart R. Possible Bias in Supervised Deep Learning Algorithms for CT Lung Nodule Detection and Classification. Cancers. 2022; 14(16):3867. https://doi.org/10.3390/cancers14163867
Chicago/Turabian StyleSourlos, Nikos, Jingxuan Wang, Yeshaswini Nagaraj, Peter van Ooijen, and Rozemarijn Vliegenthart. 2022. "Possible Bias in Supervised Deep Learning Algorithms for CT Lung Nodule Detection and Classification" Cancers 14, no. 16: 3867. https://doi.org/10.3390/cancers14163867
APA StyleSourlos, N., Wang, J., Nagaraj, Y., van Ooijen, P., & Vliegenthart, R. (2022). Possible Bias in Supervised Deep Learning Algorithms for CT Lung Nodule Detection and Classification. Cancers, 14(16), 3867. https://doi.org/10.3390/cancers14163867





