Advances in the Current Understanding of the Mechanisms Governing the Acquisition of Castration-Resistant Prostate Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
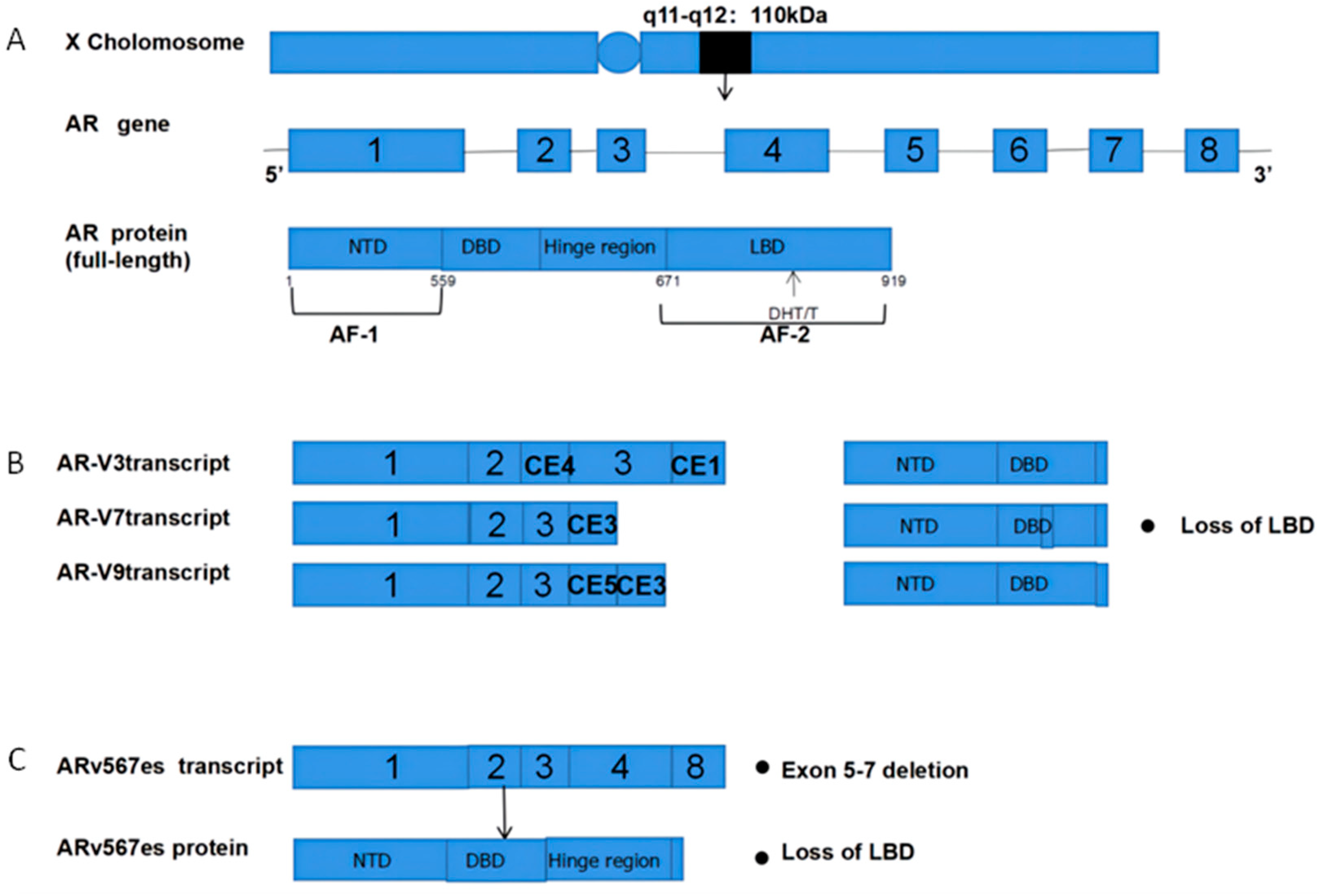
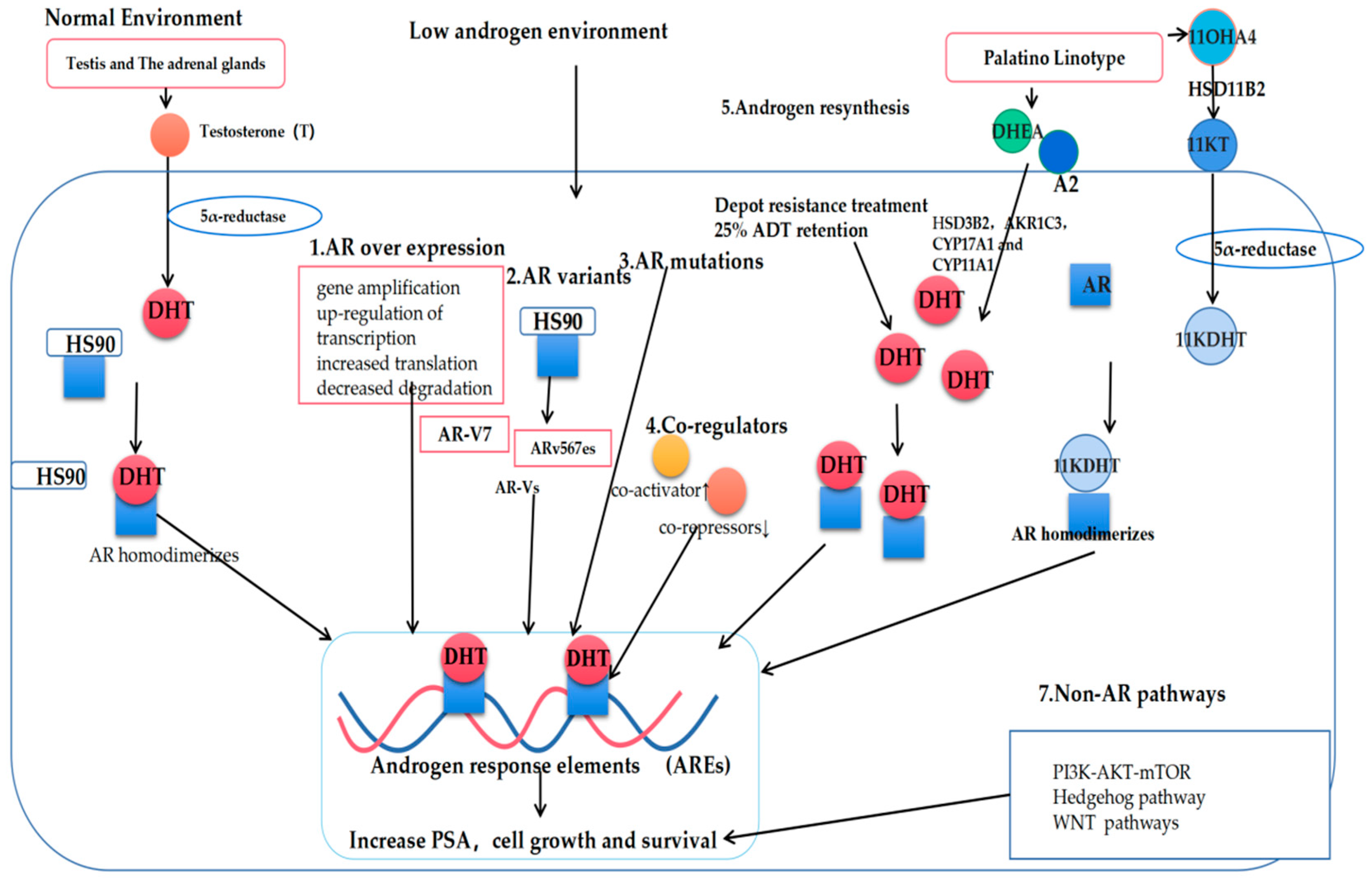
2. Androgen Receptor Pathways
3. Classic Mechanisms of AR Pathway Enhancement
3.1. AR Overexpression
3.2. De Novo Androgen Synthesis
3.3. AR Co-Regulatory Proteins
4. Abnormal AR Pathway Activation
4.1. Mutation of AR
4.2. Altered AR Splicing
5. AR Bypass Pathway: Glucocorticoid Receptor Induction
6. DNA Repair Pathway
7. Non-AR Pathways
7.1. Hedgehog Pathway Signaling
7.2. PI3K-AKT-mTOR Pathway Signaling
7.3. Wnt Pathway Activity
7.4. Non-Coding RNAs
7.4.1. miRNAs
7.4.2. LncRNAs
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Deng, T.; Cai, L.; Chen, Z.; Guo, J. Analysis of the burden of prostate cancer in China in 1990 and 2017. New Med. 2020, 30, 252–259. [Google Scholar]
- Fang, Y.; Zhou, X. Updates and interpretation of the 2020 guidelines on prostate cancer of European Association of Urology. Chin. J. Endourol. 2020, 14, 401–404. [Google Scholar]
- Cornford, P.; Bellmunt, J.; Bolla, M.; Briers, E.; de Santis, M.; Gross, T.; Henry, A.M.; Joniau, S.; Lam, T.B.; Mason, M.D.; et al. EAU-ESTRO-SIOG Guidelines on Prostate Cancer. Part II: Treatment of Relapsing, Metastatic, and Castration-Resistant Prostate Cancer. Eur. Urol. 2017, 71, 630–642. [Google Scholar] [CrossRef]
- Dehm, S.M.; Tindall, D.J. Androgen Receptor Structural and Functional Elements: Role and Regulation in Prostate Cancer. Mol. Endocrinol. 2007, 21, 2855–2863. [Google Scholar] [CrossRef]
- Agoulnik, I.U.; Weigel, N.L. Androgen receptor action in hormone-dependent and recurrent prostate cancer. J. Cell. Biochem. 2006, 99, 362–372. [Google Scholar] [CrossRef]
- Zarif, J.C.; Miranti, C.K. The importance of non-nuclear AR signaling in prostate cancer progression and therapeutic resistance. Cell Signal. 2016, 28, 348–356. [Google Scholar] [CrossRef] [Green Version]
- Kahn, B.; Collazo, J.; Kyprianou, N. Androgen receptor as a driver of therapeutic resistance in advanced prostate cancer. Int. J. Biol. Sci. 2014, 10, 588–595. [Google Scholar] [CrossRef] [Green Version]
- Auwerx, J.; Baulieu, E.; Beato, M.; Becker-Andre, M.; Burbach, P.H.; Camerino, G.; Chambon, P.; Cooney, A.; Dejean, A.; Dreyer, C.; et al. A Unified Nomenclature System for the Nuclear Receptor Superfamily. Cell 1999, 97, 161–163. [Google Scholar] [CrossRef] [Green Version]
- Claessens, F.; Denayer, S.; van Tilborgh, N.; Kerkhofs, S.; Helsen, C.; Haelens, A. Diverse roles of androgen receptor (AR) domains in AR-mediated signaling. Nucl. Recept. Signal. 2008, 6, e008. [Google Scholar] [CrossRef] [Green Version]
- Crowley, F.; Sterpi, M.; Buckley, C.; Margetich, L.; Handa, S.; Dovey, Z. A Review of the Pathophysiological Mechanisms Underlying Castration-resistant Prostate Cancer. Res. Rep. Urol. 2021, 13, 457–472. [Google Scholar] [CrossRef] [PubMed]
- Waltering, K.K.; Urbanucci, A.; Visakorpi, T. Androgen receptor (AR) aberrations in castration-resistant prostate cancer. Mol. Cell. Endocrinol. 2012, 360, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, J.C.; Tao, C.; Wu, S.; Liu, B.; Shu, Q.; Li, B.; Zhu, R. PAQR6 Upregulation Is Associated with AR Signaling and Unfavorite Prognosis in Prostate Cancers. Biomolecules 2021, 11, 1383. [Google Scholar] [CrossRef] [PubMed]
- Koivisto, P.A.; Helin, H.J. Androgen receptor gene amplification increases tissue PSA protein expression in hormone-refractory prostate carcinoma. J. Pathol. 1999, 189, 219–223. [Google Scholar] [CrossRef]
- Bubendorf, L.; Kononen, J.; Kallioniemi, O.-P.; Koivisto, P.; Gasser, T.C.; Schraml, P.; Willi, N.; Sauter, G. High-throughput survey of gene amplifications underlying prostate cancer progression using a novel tissue microarray (“tissue chip”) technology. J. Urol. 1999, 51, 72193671. [Google Scholar] [CrossRef]
- Linja, M.J.; Savinainen, K.J.; Saramäki, O.R.; Tammela, T.L.; Vessella, R.L.; Visakorpi, T. Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 2001, 61, 3550–3555. [Google Scholar]
- Cornford, P.A.; Dodson, A.R.; Parsons, K.F.; Desmond, A.D.; Woolfenden, A.; Fordham, M.; Neoptolemos, J.P.; Ke, Y.; Foster, C.S. Heat shock protein expression independently predicts clinical outcome in prostate cancer. Cancer Res. 2000, 60, 7099–7105. [Google Scholar]
- Ratajczak, W.; Lubkowski, M.; Lubkowska, A. Heat Shock Proteins in Benign Prostatic Hyperplasia and Prostate Cancer. Int. J. Mol. Sci. 2022, 23, 897. [Google Scholar] [CrossRef] [PubMed]
- Nishiyama, T.; Hashimoto, Y.; Takahashi, K. The Influence of Androgen Deprivation Therapy on Dihydrotestosterone Levels in the Prostatic Tissue of Patients with Prostate Cancer. Clin. Cancer Res. 2004, 10, 7121–7126. [Google Scholar] [CrossRef] [Green Version]
- Geller, J. Basis for hormonal management of advanced prostate cancer. Cancer 1993, 71, 1039–1045. [Google Scholar] [CrossRef]
- Geller, J.; Liu, J.; Albert, J.; Fay, W.; Berry, C.C.; Weis, P. Relationship between human prostatic epithelial cell protein synthesis and tissue dihydrotestosterone level. Clin. Endocrinol. 1987, 26, 155–161. [Google Scholar] [CrossRef] [PubMed]
- Petrylak, D.P. Current state of castration-resistant prostate cancer. Am. J. Manag. Care 2013, 19, S358–S365. [Google Scholar] [PubMed]
- Deslypere, J.P.; Young, M.; Wilson, J.D.; McPhaul, M.J. Testosterone and 5α-dihydrotestosterone interact differently with the androgen receptor to enhance transcription of the MMTV-CAT reporter gene. Mol. Cell. Endocrinol. 1992, 88, 15–22. [Google Scholar] [CrossRef]
- Sharifi, N.; Auchus, R.J. Steroid biosynthesis and prostate cancer. Steroids 2012, 77, 719–726. [Google Scholar] [CrossRef]
- Barnard, M.; Mostaghel, E.A.; Auchus, R.J.; Storbeck, K.-H. The role of adrenal derived androgens in castration resistant prostate cancer. J. Steroid Biochem. Mol. Biol. 2020, 197, 105506. [Google Scholar] [CrossRef]
- Nishiyama, T.; Ikarashi, T.; Hashimoto, Y.; Suzuki, K.; Takahashi, K. Association Between the Dihydrotestosterone Level in the Prostate and Prostate Cancer Aggressiveness Using the Gleason Score. J. Urol. 2006, 176, 1387–1391. [Google Scholar] [CrossRef]
- Nishiyama, T.; Ikarashi, T.; Hashimoto, Y.; Wako, K.; Takahashi, K. The Change in the Dihydrotestosterone Level in the Prostate Before and After Androgen Deprivation Therapy in Connection with Prostate Cancer Aggressiveness Using the Gleason Score. J. Urol. 2007, 178, 1282–1289. [Google Scholar] [CrossRef]
- Xiao, L.; Wang, Y.; Xu, K.; Hu, H.; Xu, Z.; Wu, D.; Wang, Z.; You, W.; Ng, C.-F.; Yu, S.; et al. Nuclear Receptor LRH-1 Functions to Promote Castration-Resistant Growth of Prostate Cancer via Its Promotion of Intratumoral Androgen Biosynthesis. Cancer Res. 2018, 78, 2205–2218. [Google Scholar] [CrossRef] [Green Version]
- Kaipainen, A.; Zhang, A.; Gil da Costa, R.M.; Lucas, J.; Marck, B.; Matsumoto, A.M.; Morrissey, C.; True, L.D.; Mostaghel, E.A.; Nelson, P.S. Testosterone accumulation in prostate cancer cells is enhanced by facilitated diffusion. Prostate 2019, 79, 1530–1542. [Google Scholar] [CrossRef]
- Hertzog, J.R.; Zhang, Z.; Bignan, G.; Connolly, P.J.; Heindl, J.E.; Janetopoulos, C.J.; Rupnow, B.A.; McDevitt, T.M. AKR1C3 mediates pan-AR antagonist resistance in castration-resistant prostate cancer. Prostate 2020, 80, 1223–1232. [Google Scholar] [CrossRef] [PubMed]
- Barnard, M.; Quanson, J.L.; Mostaghel, E.; Pretorius, E.; Snoep, J.L.; Storbeck, K.-H. 11-Oxygenated androgen precursors are the preferred substrates for aldo-keto reductase 1C3 (AKR1C3): Implications for castration resistant prostate cancer. J. Steroid Biochem. Mol. Biol. 2018, 183, 192–201. [Google Scholar] [CrossRef] [PubMed]
- Mostaghel, E.A.; Zhang, A.; Hernandez, S.; Marck, B.T.; Zhang, X.; Tamae, D.; Biehl, H.E.; Tretiakova, M.; Bartlett, J.; Burns, J.; et al. Contribution of Adrenal Glands to Intratumor Androgens and Growth of Castration-Resistant Prostate Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 426–439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Ma, T.; Zhou, J.; Gao, W.; Li, Y.; Yu, S.; Wang, Y.; Chan, F.L. Nuclear receptor ERRα contributes to castration-resistant growth of prostate cancer via its regulation of intratumoral androgen biosynthesis. Theranostics 2020, 10, 4201–4216. [Google Scholar] [CrossRef] [PubMed]
- Huhtaniemi, R.; Oksala, R.; Knuuttila, M.; Mehmood, A.; Aho, E.; Laajala, T.D.; Nicorici, D.; Aittokallio, T.; Laiho, A.; Elo, L.; et al. Adrenals Contribute to Growth of Castration-Resistant VCaP Prostate Cancer Xenografts. Am. J. Pathol. 2018, 188, 2890–2901. [Google Scholar] [CrossRef] [Green Version]
- Montgomery, R.B.; Mostaghel, E.A.; Vessella, R.; Hess, D.L.; Kalhorn, T.F.; Higano, C.S.; True, L.D.; Nelson, P.S. Maintenance of intratumoral androgens in metastatic prostate cancer: A mechanism for castration-resistant tumor growth. Cancer Res. 2008, 68, 4447–4454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maity, S.N.; Titus, M.A.; Gyftaki, R.; Wu, G.; Lu, J.-F.; Ramachandran, S.; Li-Ning-Tapia, E.M.; Logothetis, C.J.; Araujo, J.C.; Efstathiou, E. Targeting of CYP17A1 Lyase by VT-464 Inhibits Adrenal and Intratumoral Androgen Biosynthesis and Tumor Growth of Castration Resistant Prostate Cancer. Sci. Rep. 2016, 6, 35354. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swart, A.C.; Storbeck, K.-H. 11β-hydroxyandrostenedione: Downstream metabolism by 11βHSD, 17βHSD and SRD5A produces novel substrates in familiar pathways. Mol. Cell. Endocrinol. 2015, 408, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Turcu, A.F.; Nanba, A.T.; Auchus, R.J. The Rise, Fall, and Resurrection of 11-Oxygenated Androgens in Human Physiology and Disease. Horm. Res. Paediatr. 2018, 89, 284–291. [Google Scholar] [CrossRef]
- Rege, J.; Nakamura, Y.; Satoh, F.; Morimoto, R.; Kennedy, M.R.; Layman, L.C.; Honma, S.; Sasano, H.; Rainey, W.E. Liquid chromatography-tandem mass spectrometry analysis of human adrenal vein 19-carbon steroids before and after ACTH stimulation. J. Clin. Endocrinol. Metab. 2013, 98, 1182–1188. [Google Scholar] [CrossRef] [PubMed]
- Pretorius, E.; Africander, D.J.; Vlok, M.; Perkins, M.S.; Quanson, J.; Storbeck, K.-H. 11-Ketotestosterone and 11-Ketodihydrotestosterone in Castration Resistant Prostate Cancer: Potent Androgens Which Can No Longer Be Ignored. PLoS ONE 2016, 11, e0159867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swart, A.C.; Schloms, L.; Storbeck, K.-H.; Bloem, L.M.; du Toit, T.; Quanson, J.L.; Rainey, W.E.; Swart, P. 11β-Hydroxyandrostenedione, the product of androstenedione metabolism in the adrenal, is metabolized in LNCaP cells by 5α-reductase yielding 11β-hydroxy-5α-androstanedione. J. Steroid Biochem. Mol. Biol. 2013, 138, 132–142. [Google Scholar] [CrossRef]
- Barnard, L.; Schiffer, L.; Louw du-Toit, R.; Tamblyn, J.A.; Chen, S.; Africander, D.; Arlt, W.; Foster, P.A.; Storbeck, K.-H. 11-Oxygenated Estrogens are a Novel Class of Human Estrogens but Do not Contribute to the Circulating Estrogen Pool. Endocrinology 2021, 162, bqaa231. [Google Scholar] [CrossRef] [PubMed]
- Snaterse, G.; Mies, R.; van Weerden, W.M.; French, P.J.; Jonker, J.W.; Houtsmuller, A.B.; van Royen, M.E.; Visser, J.A.; Hofland, J. Androgen receptor mutations modulate activation by 11-oxygenated androgens and glucocorticoids. Prostate Cancer Prostatic Dis. 2022. [Google Scholar] [CrossRef] [PubMed]
- Van de Wijngaart, D.J.; Dubbink, H.J.; van Royen, M.E.; Trapman, J.; Jenster, G. Androgen receptor coregulators: Recruitment via the coactivator binding groove. Mol. Cell. Endocrinol. 2012, 352, 57–69. [Google Scholar] [CrossRef]
- DePriest, A.D.; Fiandalo, M.V.; Schlanger, S.; Heemers, F.; Mohler, J.L.; Liu, S.; Heemers, H.V. Regulators of Androgen Action Resource: A one-stop shop for the comprehensive study of androgen receptor action. Database 2016, 2016, bav125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heemers, H.V.; Tindall, D.J. Androgen Receptor (AR) Coregulators: A Diversity of Functions Converging on and Regulating the AR Transcriptional Complex. Endocr. Rev. 2007, 28, 778–808. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, C.; Heemers, H.; Sharifi, N. Androgen Signaling in Prostate Cancer. Cold Spring Harb. Perspect. Med. 2017, 7, a030452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, T.; Li, J. New Insights into the Binding Mechanism of Co-regulator BUD31 to AR AF2 Site: Structural Determination and Analysis of the Mutation Effect. Curr. Comput. Aided Drug Des. 2020, 16, 45–53. [Google Scholar] [CrossRef]
- Askew, E.B.; Bai, S.; Parris, A.B.; Minges, J.T.; Wilson, E.M. Androgen receptor regulation by histone methyltransferase Suppressor of variegation 3–9 homolog 2 and Melanoma antigen-A11. Mol. Cell. Endocrinol. 2017, 443, 42–51. [Google Scholar] [CrossRef] [Green Version]
- Chmelar, R.; Buchanan, G.; Need, E.F.; Tilley, W.; Greenberg, N.M. Androgen receptor coregulators and their involvement in the development and progression of prostate cancer. Int. J. Cancer 2006, 120, 719–733. [Google Scholar] [CrossRef]
- Gelman, I.H. Androgen receptor activation in castration-recurrent prostate cancer: The role of Src-family and Ack1 tyrosine kinases. Int. J. Biol. Sci. 2014, 10, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Karaca, M.; Zhang, Z.; Gioeli, D.; Earp, H.S.; Whang, Y.E. Dasatinib inhibits site-specific tyrosine phosphorylation of androgen receptor by Ack1 and Src kinases. Oncogene 2010, 29, 3208–3216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Silva, D.; Zhang, Z.; Liu, Y.; Parker, J.S.; Xu, C.; Cai, L.; Wang, G.G.; Earp, H.S.; Whang, Y.E. Interaction between androgen receptor and coregulator SLIRP is regulated by Ack1 tyrosine kinase and androgen. Sci. Rep. 2019, 9, 18637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, J.; Zhu, J.; Goodman, O.B.; Pestell, R.G.; Schlegel, P.N.; Nanus, D.M.; Shen, R. Activation of p300 histone acetyltransferase activity and acetylation of the androgen receptor by bombesin in prostate cancer cells. Oncogene 2006, 25, 2011–2021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Purayil, H.T.; Zhang, Y.; Black, J.B.; Gharaibeh, R.; Daaka, Y. Nuclear βArrestin1 regulates androgen receptor function in castration resistant prostate cancer. Oncogene 2021, 40, 2610–2620. [Google Scholar] [CrossRef]
- Chen, C.D.; Welsbie, D.S.; Tran, C.; Baek, S.H.; Chen, R.; Vessella, R.; Rosenfeld, M.G.; Sawyers, C.L. Molecular determinants of resistance to antiandrogen therapy. Nat. Med. 2003, 10, 33–39. [Google Scholar] [CrossRef]
- Senapati, D.; Kumari, S.; Heemers, H.V. Androgen receptor co-regulation in prostate cancer. Asian J. Urol. 2020, 7, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.-A.; Bai, S.; Grossman, G.; Titus, M.A.; Ford, O.H.; Pop, E.A.; Smith, G.J.; Mohler, J.L.; Wilson, E.M.; French, F.S. Mechanism of androgen receptor corepression by CKβBP2/CRIF1, a multifunctional transcription factor coregulator expressed in prostate cancer. Mol. Cell. Endocrinol. 2014, 382, 302–313. [Google Scholar] [CrossRef] [Green Version]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Gottlieb, B.; Beitel, L.K.; Nadarajah, A.; Paliouras, M.; Trifiro, M. The androgen receptor gene mutations database: 2012 update. Hum. Mutat. 2012, 33, 887–894. [Google Scholar] [CrossRef]
- Fujita, K.; Nonomura, N. Role of Androgen Receptor in Prostate Cancer: A Review. World J. Men’s Health 2019, 37, 288–295. [Google Scholar] [CrossRef] [PubMed]
- Prekovic, S.; van Royen, M.E.; Voet, A.R.D.; Geverts, B.; Houtman, R.; Melchers, D.; Zhang, K.Y.J.; van den Broeck, T.; Smeets, E.; Spans, L.; et al. The Effect of F877L and T878A Mutations on Androgen Receptor Response to Enzalutamide. Mol. Cancer Ther. 2016, 15, 1702–1712. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sumiyoshi, T.; Mizuno, K.; Yamasaki, T.; Miyazaki, Y.; Makino, Y.; Okasho, K.; Li, X.; Utsunomiya, N.; Goto, T.; Kobayashi, T.; et al. Clinical utility of androgen receptor gene aberrations in circulating cell-free DNA as a biomarker for treatment of castration-resistant prostate cancer. Sci. Rep. 2019, 9, 4030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Wang, L.; Tian, J.; Li, J.; Liu, H. Molecular Dynamics Studies on the Enzalutamide Resistance Mechanisms Induced by Androgen Receptor Mutations. J. Cell. Biochem. 2017, 118, 2792–2801. [Google Scholar] [CrossRef]
- Coleman, D.J.; van Hook, K.; King, C.J.; Schwartzman, J.; Lisac, R.; Urrutia, J.; Sehrawat, A.; Woodward, J.; Wang, N.J.; Gulati, R.; et al. Cellular androgen content influences enzalutamide agonism of F877L mutant androgen receptor. Oncotarget 2016, 7, 40690–40703. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Connolly, P.J.; Lim, H.K.; Pande, V.; Meerpoel, L.; Teleha, C.; Branch, J.R.; Ondrus, J.; Hickson, I.; Bush, T.; et al. Discovery of JNJ-63576253: A Clinical Stage Androgen Receptor Antagonist for F877L Mutant and Wild-Type Castration-Resistant Prostate Cancer (mCRPC). J. Med. Chem. 2021, 64, 909–924. [Google Scholar] [CrossRef]
- O’Neill, D.; Jones, D.; Wade, M.; Grey, J.; Nakjang, S.; Guo, W.; Cork, D.; Davies, B.R.; Wedge, S.R.; Robson, C.N.; et al. Development and exploitation of a novel mutant androgen receptor modelling strategy to identify new targets for advanced prostate cancer therapy. Oncotarget 2015, 6, 26029–26040. [Google Scholar] [CrossRef] [Green Version]
- Sridhar, S.S.; Freedland, S.J.; Gleave, M.E.; Higano, C.; Mulders, P.; Parker, C.; Sartor, O.; Saad, F. Castration-Resistant Prostate Cancer: From New Pathophysiology to New Treatment. Eur. Urol. 2014, 65, 289–299. [Google Scholar] [CrossRef]
- Bogner, J.; Zolghadr, K.; Hickson, I.; Romer, T.; Yurlova, L. The fluorescent two-hybrid assay for live-cell profiling of androgen receptor modulators. J. Steroid Biochem. Mol. Biol. 2017, 166, 45–53. [Google Scholar] [CrossRef]
- Steinkamp, M.P.; O’Mahony, O.A.; Brogley, M.; Rehman, H.; Lapensee, E.W.; Dhanasekaran, S.; Hofer, M.D.; Kuefer, R.; Chinnaiyan, A.; Rubin, M.A.; et al. Treatment-dependent androgen receptor mutations in prostate cancer exploit multiple mechanisms to evade therapy. Cancer Res. 2009, 69, 4434–4442. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Makino, T.; Izumi, K.; Mizokami, A. Undesirable Status of Prostate Cancer Cells after Intensive Inhibition of AR Signaling: Post-AR Era of CRPC Treatment. Biomedicines 2021, 9, 414. [Google Scholar] [CrossRef] [PubMed]
- Obinata, D.; Lawrence, M.G.; Takayama, K.; Choo, N.; Risbridger, G.P.; Takahashi, S.; Inoue, S. Recent Discoveries in the Androgen Receptor Pathway in Castration-Resistant Prostate Cancer. Front. Oncol. 2020, 10, 581515. [Google Scholar] [CrossRef] [PubMed]
- Park, H.K.; Lim, S.D.; Kwon, G.Y. mRNA expressions of androgen receptor and its variants in matched hormone-sensitive and castration-resistant prostate cancer. Scand. J. Urol. 2019, 53, 365–371. [Google Scholar] [CrossRef]
- Tsaur, I.; Becker, C.; Thelen, P.; Roos, F.C. Spleißvariante AR-V7. Der Urol. 2017, 56, 1164–1167. [Google Scholar] [CrossRef]
- Wach, S.; Taubert, H.; Cronauer, M. Role of androgen receptor splice variants, their clinical relevance and treatment options. World J. Urol. 2019, 38, 647–656. [Google Scholar] [CrossRef] [PubMed]
- Kallio, H.M.L.; Hieta, R.; Latonen, L.; Brofeldt, A.; Annala, M.; Kivinummi, K.; Tammela, T.L.; Nykter, M.; Isaacs, W.B.; Lilja, H.G.; et al. Constitutively active androgen receptor splice variants AR-V3, AR-V7 and AR-V9 are co-expressed in castration-resistant prostate cancer metastases. Br. J. Cancer 2018, 119, 347–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Messner, E.A.; Steele, T.M.; Tsamouri, M.M.; Hejazi, N.; Gao, A.C.; Mudryj, M.; Ghosh, P.M. The Androgen Receptor in Prostate Cancer: Effect of Structure, Ligands and Spliced Variants on Therapy. Biomedicines 2020, 8, 422. [Google Scholar] [CrossRef]
- Hu, R.; Dunn, T.A.; Wei, S.; Isharwal, S.; Veltri, R.W.; Humphreys, E.; Han, M.; Partin, A.W.; Vessella, R.L.; Isaacs, W.B.; et al. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009, 69, 16–22. [Google Scholar] [CrossRef] [Green Version]
- Lu, C.; Brown, L.C.; Antonarakis, E.S.; Armstrong, A.J.; Luo, J. Androgen receptor variant-driven prostate cancer II: Advances in laboratory investigations. Prostate Cancer Prostatic Dis. 2020, 23, 381–397. [Google Scholar] [CrossRef]
- Hu, R.; Lu, C.; Mostaghel, E.A.; Yegnasubramanian, S.; Gurel, M.; Tannahill, C.; Edwards, J.; Isaacs, W.B.; Nelson, P.S.; Bluemn, E.; et al. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012, 72, 3457–3462. [Google Scholar] [CrossRef] [Green Version]
- Kanayama, M.; Lu, C.; Luo, J.; Antonarakis, E.S. AR Splicing Variants and Resistance to AR Targeting Agents. Cancers 2021, 13, 2563. [Google Scholar] [CrossRef]
- Van der Steen, T.; Tindall, D.J.; Huang, H. Posttranslational modification of the androgen receptor in prostate cancer. Int. J. Mol. Sci. 2013, 14, 14833–14859. [Google Scholar] [CrossRef] [Green Version]
- Hillebrand, A.C.; Pizzolato, L.S.; Neto, B.S.; Branchini, G.; Brum, I.S. Androgen receptor isoforms expression in benign prostatic hyperplasia and primary prostate cancer. PLoS ONE 2018, 13, e0200613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efstathiou, E.; Titus, M.; Wen, S.; Hoang, A.; Karlou, M.; Ashe, R.; Tu, S.M.; Aparicio, A.; Troncoso, P.; Mohler, J.; et al. Molecular characterization of enzalutamide-treated bone metastatic castration-resistant prostate cancer. Eur. Urol. 2015, 67, 53–60. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Yang, R.; Henzler, C.M.; Ho, Y.; Passow, C.; Auch, B.; Carreira, S.; Nava Rodrigues, D.; Bertan, C.; Hwang, T.H.; et al. Diverse AR Gene Rearrangements Mediate Resistance to Androgen Receptor Inhibitors in Metastatic Prostate Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 1965–1976. [Google Scholar] [CrossRef]
- Nagandla, H.; Robertson, M.J.; Putluri, V.; Putluri, N.; Coarfa, C.; Weigel, N.L. Isoform-specific Activities of Androgen Receptor and its Splice Variants in Prostate Cancer Cells. Endocrinology 2021, 162, bqaa227. [Google Scholar] [CrossRef]
- Kiliccioglu, I.; Bilen, C.Y.; Sozen, S.; Konac, E. Upregulation of potential regulatory signaling molecules correlate with androgen receptor splice variants AR-V7 and AR-V567es in prostate cancer metastasis. Gene 2021, 772, 145377. [Google Scholar] [CrossRef] [PubMed]
- Sharp, A.; Coleman, I.; Yuan, W.; Sprenger, C.; Dolling, D.; Rodrigues, D.N.; Russo, J.W.; Figueiredo, I.; Bertan, C.; Seed, G.; et al. Androgen receptor splice variant-7 expression emerges with castration resistance in prostate cancer. J. Clin. Investig. 2019, 129, 192–208. [Google Scholar] [CrossRef] [Green Version]
- Tietz, K.T.; Dehm, S.M. Androgen receptor variants: RNA-based mechanisms and therapeutic targets. Hum. Mol. Genet. 2020, 29, R19–R26. [Google Scholar] [CrossRef] [PubMed]
- Kohli, M.; Ho, Y.; Hillman, D.W.; van Etten, J.L.; Henzler, C.; Yang, R.; Sperger, J.M.; Li, Y.; Tseng, E.; Hon, T.; et al. Androgen Receptor Variant AR-V9 Is Coexpressed with AR-V7 in Prostate Cancer Metastases and Predicts Abiraterone Resistance. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 4704–4715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simon, I.; Perales, S.; Casado-Medina, L.; Rodríguez-Martínez, A.; Garrido-Navas, M.D.C.; Puche-Sanz, I.; Diaz-Mochon, J.J.; Alaminos, C.; Lupiañez, P.; Lorente, J.A.; et al. Cross-Resistance to Abiraterone and Enzalutamide in Castration Resistance Prostate Cancer Cellular Models is Mediated by AR Transcriptional Reactivation. Cancers 2021, 13, 1483. [Google Scholar] [CrossRef] [PubMed]
- Fettke, H.; Kwan, E.M.; Docanto, M.M.; Bukczynska, P.; Ng, N.; Graham, L.-J.K.; Mahon, K.; Hauser, C.; Tan, W.; Wang, X.H.; et al. Combined Cell-free DNA and RNA Profiling of the Androgen Receptor: Clinical Utility of a Novel Multianalyte Liquid Biopsy Assay for Metastatic Prostate Cancer. Eur. Urol. 2020, 78, 173–180. [Google Scholar] [CrossRef]
- Sieuwerts, A.M.; Onstenk, W.; Kraan, J.; Beaufort, C.M.; Van, M.; de Laere, B.; Dirix, L.Y.; Hamberg, P.; Beeker, A.; Meulenbeld, H.J.; et al. AR splice variants in circulating tumor cells of patients with castration-resistant prostate cancer: Relation with outcome to cabazitaxel. Mol. Oncol. 2019, 13, 1795–1807. [Google Scholar] [CrossRef] [Green Version]
- Sun, S.; Sprenger, C.C.T.; Vessella, R.L.; Haugk, K.; Soriano, K.; Mostaghel, E.A.; Page, S.T.; Coleman, I.M.; Nguyen, H.M.; Sun, H.; et al. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J. Clin. Investig. 2010, 120, 2715–2730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontana, F.; Limonta, P. Dissecting the Hormonal Signaling Landscape in Castration-Resistant Prostate Cancer. Cells 2021, 10, 1133. [Google Scholar] [CrossRef] [PubMed]
- Thadani-Mulero, M.; Portella, L.; Sun, S.; Sung, M.; Matov, A.; Vessella, R.L.; Corey, E.; Nanus, D.M.; Plymate, S.R.; Giannakakou, P. Androgen receptor splice variants determine taxane sensitivity in prostate cancer. Cancer Res. 2014, 74, 2270–2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tagawa, S.T.; Antonarakis, E.S.; Gjyrezi, A.; Galletti, G.; Giannakakou, P. Clinical Significance of AR-V567es in Prostate Cancer—Response. Clin. Cancer Res. 2019, 25, 6010–6011. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Ledet, E.; Li, D.; Dotiwala, A.; Steinberger, A.; Feibus, A.; Li, J.; Qi, Y.; Silberstein, J.; Lee, B.; et al. A Whole Blood Assay for AR-V7 and AR(v567es) in Patients with Prostate Cancer. J. Urol. 2016, 196, 1758–1763. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Sprenger, C.; Sun, S.; Epilepsia, K.S.; Haugk, K.; Zhang, X.; Coleman, I.; Nelson, P.S.; Plymate, S. AR variant ARv567es induces carcinogenesis in a novel transgenic mouse model of prostate cancer. Neoplasia 2013, 15, 1009–1017. [Google Scholar] [CrossRef] [Green Version]
- Kumar, R. Emerging role of glucocorticoid receptor in castration resistant prostate cancer: A potential therapeutic target. J. Cancer 2020, 11, 696–701. [Google Scholar] [CrossRef] [Green Version]
- Szmulewitz, R.Z.; Chung, E.; Al-Ahmadie, H.; Daniel, S.; Kocherginsky, M.; Razmaria, A.; Zagaja, G.P.; Brendler, C.B.; Stadler, W.M.; Conzen, S.D. Serum/glucocorticoid-regulated kinase 1 expression in primary human prostate cancers. Prostate 2012, 72, 157–164. [Google Scholar] [CrossRef]
- Sahu, B.; Laakso, M.; Pihlajamaa, P.; Ovaska, K.; Sinielnikov, I.; Hautaniemi, S.; Jänne, O.A. FoxA1 specifies unique androgen and glucocorticoid receptor binding events in prostate cancer cells. Cancer Res. 2013, 73, 1570–1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kach, J.; Conzen, S.D.; Szmulewitz, R.Z. Targeting the glucocorticoid receptor in breast and prostate cancers. Sci. Transl. Med. 2015, 7, 305ps19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, T.Z.; Jin, F.S.; Xie, L.P.; Li, L.C. Relationship between glucocorticoid receptor signal pathway and androgen-independent prostate cancer. Urol. Int. 2008, 81, 228–233. [Google Scholar] [CrossRef]
- Hirayama, Y.; Sadar, M.D. Does increased expression of glucocorticoid receptor support application of antagonists to this receptor for the treatment of castration resistant prostate cancer? AME Med. J. 2018, 3, 66. [Google Scholar] [CrossRef]
- Arora, V.K.; Schenkein, E.; Murali, R.; Subudhi, S.K.; Wongvipat, J.; Balbas, M.D.; Shah, N.; Cai, L.; Efstathiou, E.; Logothetis, C.; et al. Glucocorticoid receptor confers resistance to antiandrogens by bypassing androgen receptor blockade. Cell 2013, 155, 1309–1322. [Google Scholar] [CrossRef] [Green Version]
- Arora, V.K.; Schenkein, E.; Murali, R.; Subudhi, S.K.; Wongvipat, J.; Balbas, M.D.; Shah, N.; Cai, L.; Efstathiou, E.; Logothetis, C.; et al. Upregulation of glucocorticoid receptor-mediated glucose transporter 4 in enzalutamide-resistant prostate cancer. Cancer Sci. 2021, 112, 1899–1910. [Google Scholar] [CrossRef]
- Purayil, H.T.; Daaka, Y. βArrestin1 regulates glucocorticoid receptor mitogenic signaling in castration-resistant prostate cancer. Prostate 2022, 82, 816–825. [Google Scholar] [CrossRef] [PubMed]
- Jasin, M.; Haber, J.E. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair 2016, 44, 6–16. [Google Scholar] [CrossRef] [Green Version]
- Bennett, E.P.; Petersen, B.L.; Johansen, I.E.; Niu, Y.; Yang, Z.; Chamberlain, C.A.; Met, Ö.; Wandall, H.H.; Frödin, M. INDEL detection, the ‘Achilles heel’ of precise genome editing: A survey of methods for accurate profiling of gene editing induced indels. Nucleic Acids Res. 2020, 48, 11958–11981. [Google Scholar] [CrossRef]
- Matveev, V.B.; Kirichek, A.A.; Filippova, M.G.; Savinkova, A.V.; Khalmurzaev, O.A.; Lyubchenko, L.N. Impact of germline BRCA2 and CHEK2 mutations on time to castration resistance in patients with metastatic hormone-nave prostate cancer. Urologiia 2019, 5, 79–85. (In Russian) [Google Scholar] [CrossRef] [PubMed]
- Gallagher, D.J.; Gaudet, M.M.; Pal, P.; Kirchhoff, T.; Balistreri, L.; Vora, K.; Bhatia, J.; Stadler, Z.; Fine, S.W.; Reuter, V.; et al. Germline BRCA mutations denote a clinicopathologic subset of prostate cancer. Clin. Cancer Res. 2010, 16, 2115–2121. [Google Scholar] [CrossRef] [Green Version]
- Lang, S.H.; Swift, S.L.; White, H.; Misso, K.; Kleijnen, J.; Quek, R. A systematic review of the prevalence of DNA damage response gene mutations in prostate cancer. Int. J. Oncol. 2019, 55, 597–616. [Google Scholar] [CrossRef] [Green Version]
- Schiewer, M.J.; Knudsen, K.E. DNA Damage Response in Prostate Cancer. Cold Spring Harb. Perspect. Med. 2019, 9, a030486. [Google Scholar] [CrossRef]
- Wang, M.; Li, Q.; Gu, C.; Zhu, Y.; Yang, Y.; Wang, J.; Jin, L.; He, J.; Ye, D.; Wei, Q. Polymorphisms in nucleotide excision repair genes and risk of primary prostate cancer in Chinese Han populations. Oncotarget 2016, 8, 24362–24371. [Google Scholar] [CrossRef]
- Kote-Jarai, Z.; Jugurnauth, S.; Mulholland, S.; Leongamornlert, D.A.; Guy, M.; Edwards, S.; Tymrakiewitcz, M.; O’Brien, L.; Hall, A.; Wilkinson, R.; et al. A recurrent truncating germline mutation in the BRIP1/FANCJ gene and susceptibility to prostate cancer. Br. J. Cancer 2009, 100, 426–430. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, G.; Armenia, J.; Mazzu, Y.Z.; Nandakumar, S.; Stopsack, K.H.; Atiq, M.O.; Komura, K.; Jehane, L.; Hirani, R.; Chadalavada, K.; et al. Significance of BRCA2 and RB1 Co-loss in Aggressive Prostate Cancer Progression. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2020, 26, 2047–2064. [Google Scholar] [CrossRef]
- Chakraborty, G.; Patail, N.K.; Hirani, R.; Nandakumar, S.; Mazzu, Y.Z.; Yoshikawa, Y.; Atiq, M.; Jehane, L.E.; Stopsack, K.H.; Lee, G.M.; et al. Attenuation of SRC Kinase Activity Augments PARP Inhibitor-mediated Synthetic Lethality in BRCA2-altered Prostate Tumors. Clin. Cancer Res. 2021, 27, 1792–1806. [Google Scholar] [CrossRef]
- Li, L.; Karanika, S.; Yang, G.; Wang, J.; Park, S.; Broom, B.M.; Manyam, G.C.; Wu, W.; Luo, Y.; Basourakos, S.; et al. Androgen receptor inhibitor-induced “BRCAness” and PARP inhibition are synthetically lethal for castration-resistant prostate cancer. Sci. Signal. 2017, 10, eaam7479. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, D.J.; Cronin, A.M.; Milowsky, M.I.; Morris, M.J.; Bhatia, J.; Scardino, P.T.; Eastham, J.A.; Offit, K.; Robson, M.E. Germline BRCA mutation does not prevent response to taxane-based therapy for the treatment of castration-resistant prostate cancer. BJU Int. 2012, 109, 713–719. [Google Scholar] [CrossRef] [Green Version]
- Mateo, J.; Seed, G.; Bertan, C. Genomics of lethal prostate cancer at diagnosis and castration resistance. J. Clin. Investig. 2020, 130, 1743–1751. [Google Scholar] [CrossRef]
- Walker, S.R.; Abdelsalam, R.; Ghosh, S.; Livingstone, J.; Palanisamy, N.; Boutros, P.C.; Yip, S.M.; Lees-Miller, S.P.; Bismar, T.A. Decreased ATM Protein Expression Is Substantiated with PTEN Loss in Defining Aggressive Phenotype of Prostate Cancer Associated with Lethal Disease. Eur. Urol. Open Sci. 2021, 29, 93–101. [Google Scholar] [CrossRef]
- Kwon, D.H.; Chou, J.; Yip, S.M.; Reimers, M.A.; Zhang, L.; Wright, F.; Dhawan, M.S.; Borno, H.T.; Desai, A.; Aggarwal, R.R.; et al. Differential treatment outcomes in BRCA1/2-, CDK12-, and ATM-mutated metastatic castration-resistant prostate cancer. Cancer 2021, 127, 1965–1973. [Google Scholar] [CrossRef] [PubMed]
- Gonnissen, A.; Isebaert, S.; Haustermans, K. Hedgehog signaling in prostate cancer and its therapeutic implication. Int. J. Mol. Sci. 2013, 14, 13979–14007. [Google Scholar] [CrossRef]
- Mimeault, M.; Rachagani, S.; Muniyan, S.; Seshacharyulu, P.; Johansson, S.L.; Datta, K.; Lin, M.-F.; Batra, S.K. Inhibition of hedgehog signaling improves the anti-carcinogenic effects of docetaxel in prostate cancer. Oncotarget 2015, 6, 3887–3903. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Alsaidan, O.A.; Rai, S.; Wu, M.; Shen, H.; Beharry, Z.; Almada, L.L.; Fernandez-Zapico, M.E.; Wang, L.; Cai, H. Stromal Gli signaling regulates the activity and differentiation of prostate stem and progenitor cells. J. Biol. Chem. 2018, 293, 10547–10560. [Google Scholar] [CrossRef] [Green Version]
- Chang, H.-H.; Chen, B.-Y.; Wu, C.-Y.; Tsao, Z.-J.; Chen, Y.-Y.; Chang, C.-P.; Yang, C.-R.; Lin, D.P.-C. Hedgehog overexpression leads to the formation of prostate cancer stem cells with metastatic property irrespective of androgen receptor expression in the mouse model. J. Biomed. Sci. 2011, 18, 6. [Google Scholar] [CrossRef] [Green Version]
- Kasper, M.; Regl, G.; Frischauf, A.-M.; Aberger, F. GLI transcription factors: Mediators of oncogenic Hedgehog signalling. Eur. J. Cancer 2006, 42, 437–445. [Google Scholar] [CrossRef] [PubMed]
- Verma, S.; Shankar, E.; Kalayci, F.N.C.; Mukunda, A.; Alassfar, M.; Singh, V.; Chan, E.R.; MacLennan, G.T.; Gupta, S. Androgen Deprivation Induces Transcriptional Reprogramming in Prostate Cancer Cells to Develop Stem Cell-Like Characteristics. Int. J. Mol. Sci. 2020, 21, 9568. [Google Scholar] [CrossRef]
- Yang, R.; Chen, H.; Guo, D.; Dong, Y.; Miller, D.D.; Li, W.; Mahato, R.I. Polymeric Micellar Delivery of Novel Microtubule Destabilizer and Hedgehog Signaling Inhibitor for Treating Chemoresistant Prostate Cancer. J. Pharmacol. Exp. Ther. 2019, 370, 864–875. [Google Scholar] [CrossRef]
- Clement, V.; Sanchez, P.; de Tribolet, N.; Radovanovic, I.; Ruiz i Altaba, A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr. Biol. 2007, 17, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.-A.; Pan, S.-C.; Chu, I.; Lai, R.-Y.; Wei, Y.-H. Targeting cancer stem cells from a metabolic perspective. Exp. Biol. Med. 2020, 245, 465–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abad, E.; García-Mayea, Y.; Mir, C.; Sebastian, D.; Zorzano, A.; Potesil, D.; Zdrahal, Z.; Lyakhovich, A.; Lleonart, M.E. Common Metabolic Pathways Implicated in Resistance to Chemotherapy Point to a Key Mitochondrial Role in Breast Cancer. Mol. Cell Proteom. 2019, 18, 231–244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gowda, P.S.; Deng, J.D.; Mishra, S.; Bandyopadhyay, A.; Liang, S.; Lin, S.; Mahalingam, D.; Sun, L.-Z. Inhibition of hedgehog and androgen receptor signaling pathways produced synergistic suppression of castration-resistant prostate cancer progression. Mol. Cancer Res. 2013, 11, 1448–1461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Feuerstein, M.A.; Levina, E.; Baghel, P.S.; Carkner, R.D.; Tanner, M.J.; Shtutman, M.; Vacherot, F.; Terry, S.; de la Taille, A.; et al. Hedgehog/Gli supports androgen signaling in androgen deprived and androgen independent prostate cancer cells. Mol. Cancer 2010, 9, 89. [Google Scholar] [CrossRef] [Green Version]
- Li, N.; Chen, M.; Truong, S.; Yan, C.; Buttyan, R. Determinants of Gli2 co-activation of wildtype and naturally truncated androgen receptors. Prostate 2014, 74, 1400–1410. [Google Scholar] [CrossRef]
- Xia, L.; Bouamar, H.; Gu, X.; Zeballos, C.; Qin, T.; Wang, B.; Zhou, Y.; Wang, Y.; Yang, J.; Zhu, H.; et al. Gli2 mediates the development of castration-resistant prostate cancer. Int. J. Oncol. 2020, 57, 100–112. [Google Scholar] [CrossRef]
- Burleson, M.; Deng, J.J.; Qin, T.; Duong, T.M.; Yan, Y.; Gu, X.; Das, D.; Easley, A.; Liss, M.A.; Yew, P.R.; et al. GLI3 Is Stabilized by SPOP Mutations and Promotes Castration Resistance via Functional Cooperation with Androgen Receptor in Prostate Cancer. Mol. Cancer Res. 2021, 20, 62–76. [Google Scholar] [CrossRef]
- Kolinsky, M.P.; Rescigno, P.; Bianchini, D.; Zafeiriou, Z.; Mehra, N.; Mateo, J.; Michalarea, V.; Riisnaes, R.; Crespo, M.; Figueiredo, I.; et al. A phase I dose-escalation study of enzalutamide in combination with the AKT inhibitor AZD5363 (capivasertib) in patients with metastatic castration-resistant prostate cancer. Ann. Oncol. 2020, 31, 619–625. [Google Scholar] [CrossRef] [Green Version]
- Sarker, D.; Reid, A.H.M.; Yap, T.A.; de Bono, J.S. Targeting the PI3K/AKT Pathway for the Treatment of Prostate Cancer. Clin. Cancer Res. 2009, 15, 4799–4805. [Google Scholar] [CrossRef] [Green Version]
- Gasmi, A.; Roubaud, G.; Dariane, C.; Barret, E.; Beauval, J.-B.; Brureau, L.; Créhange, G.; Fiard, G.; Fromont, G.; Gauthé, M.; et al. Overview of the Development and Use of Akt Inhibitors in Prostate Cancer. J. Clin. Med. 2021, 11, 160. [Google Scholar] [CrossRef]
- Kato, M.; Banuelos, C.A.; Imamura, Y.; Leung, J.K.; Caley, D.P.; Wang, J.; Mawji, N.R.; Sadar, M.D. Cotargeting Androgen Receptor Splice Variants and mTOR Signaling Pathway for the Treatment of Castration-Resistant Prostate Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 2744–2754. [Google Scholar] [CrossRef] [Green Version]
- Kaarbø, M.; Mikkelsen, Ø.L.; Malerød, L.; Qu, S.; Lobert, V.H.; Akgul, G.; Halvorsen, T.; Mælandsmo, G.M.; Saatcioglu, F. PI3K-AKT-mTOR Pathway is Dominant over Androgen Receptor Signaling in Prostate Cancer Cells. Anal. Cell. Pathol. 2010, 32, 11–27. [Google Scholar] [CrossRef]
- Manning, B.D.; Cantley, L.C. AKT/PKB signaling: Navigating downstream. Cell 2007, 129, 1261–1274. [Google Scholar] [CrossRef] [Green Version]
- Engelman, J.A.; Luo, J.; Cantley, L.C. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 2006, 7, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwan, E.M.; Dai, C.; Fettke, H.; Hauser, C.; Docanto, M.M.; Bukczynska, P.; Ng, N.; Foroughi, S.; Graham, L.-J.K.; Mahon, K.; et al. Plasma Cell-Free DNA Profiling of PTEN-PI3K-AKT Pathway Aberrations in Metastatic Castration-Resistant Prostate Cancer. JCO Precis. Oncol. 2021, 5, PO.20.00424. [Google Scholar] [CrossRef]
- Pungsrinont, T.; Kallenbach, J.; Baniahmad, A. Role of PI3K-AKT-mTOR Pathway as a Pro-Survival Signaling and Resistance-Mediating Mechanism to Therapy of Prostate Cancer. Int. J. Mol. Sci. 2021, 22, 11088. [Google Scholar] [CrossRef]
- Datta, S.R.; Dudek, H.; Tao, X.; Masters, S.; Fu, H.; Gotoh, Y.; Greenberg, M.E. Akt Phosphorylation of BAD Couples Survival Signals to the Cell-Intrinsic Death Machinery. Cell 1997, 91, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Pilling, A.B.; Hwang, C. Targeting prosurvival BCL2 signaling through Akt blockade sensitizes castration-resistant prostate cancer cells to enzalutamide. Prostate 2019, 79, 1347–1359. [Google Scholar] [CrossRef]
- Carver, B.S.; Chapinski, C.; Wongvipat, J.; Hieronymus, H.; Chen, Y.; Chandarlapaty, S.; Arora, V.K.; Le, C.; Koutcher, J.; Scher, H.; et al. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell 2011, 19, 575–586. [Google Scholar] [CrossRef] [Green Version]
- Qi, W.; Morales, C.; Cooke, L.S.; Johnson, B.; Somer, B.; Mahadevan, D. Reciprocal feedback inhibition of the androgen receptor and PI3K as a novel therapy for castrate-sensitive and -resistant prostate cancer. Oncotarget 2015, 6, 41976–41987. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Lamoureux, F.; Crafter, C.; Davies, B.R.; Beraldi, E.; Fazli, L.; Kim, S.; Thaper, D.; Gleave, M.E.; Zoubeidi, A. Synergistic Targeting of PI3K/AKT Pathway and Androgen Receptor Axis Significantly Delays Castration-Resistant Prostate Cancer Progression In Vivo. Mol. Cancer Ther. 2013, 12, 2342–2355. [Google Scholar] [CrossRef] [Green Version]
- Mulholland, D.J.; Tran, L.M.; Li, Y.; Cai, H.; Morim, A.; Wang, S.; Plaisier, S.; Garraway, I.P.; Huang, J.; Graeber, T.G.; et al. Cell autonomous role of PTEN in regulating castration-resistant prostate cancer growth. Cancer Cell 2011, 19, 792–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edlind, M.P.; Hsieh, A.C. PI3K-AKT-mTOR signaling in prostate cancer progression and androgen deprivation therapy resistance. Asian J. 2014, 16, 378–386. [Google Scholar] [CrossRef]
- Shorning, B.Y.; Dass, M.S.; Smalley, M.J.; Pearson, H.B. The PI3K-AKT-mTOR Pathway and Prostate Cancer: At the Crossroads of AR, MAPK, and WNT Signaling. Int. J. Mol. Sci. 2020, 21, 4507. [Google Scholar] [CrossRef]
- Robinson, D.; van Allen, E.M.; Wu, Y.-M.; Schultz, N.; Lonigro, R.J.; Mosquera, J.-M.; Montgomery, B.; Taplin, M.-E.; Pritchard, C.C.; Attard, G.; et al. Integrative clinical genomics of advanced prostate cancer. Cell 2015, 161, 1215–1228. [Google Scholar] [CrossRef] [Green Version]
- Koh, C.M.; Bieberich, C.J.; Dang, C.V.; Nelson, W.G.; Yegnasubramanian, S.; de Marzo, A.M. MYC and Prostate Cancer. Genes Cancer 2010, 1, 617–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansom, O.J.; Meniel, V.S.; Muncan, V.; Phesse, T.J.; Wilkins, J.A.; Reed, K.R.; Vass, J.K.; Athineos, D.; Clevers, H.; Clarke, A.R. Myc deletion rescues Apc deficiency in the small intestine. Nature 2007, 446, 676–679. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Boutros, M. Wnt signaling in cancer. Oncogene 2017, 36, 1461–1473. [Google Scholar] [CrossRef]
- Stewart, D.J. Wnt Signaling Pathway in Non-Small Cell Lung Cancer. JNCI J. Natl. Cancer Inst. 2013, 106, djt356. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Chen, M.W.; Terry, S.; Vacherot, F.; Bemis, D.L.; Capodice, J.; Kitajewski, J.; de la Taille, A.; Benson, M.C.; Guo, Y.; et al. Complex regulation of human androgen receptor expression by Wnt signaling in prostate cancer cells. Oncogene 2006, 25, 3436–3444. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.A.; Logan, S.K. Revisiting the role of Wnt/β-catenin signaling in prostate cancer. Mol. Cell. Endocrinol. 2018, 462, 3–8. [Google Scholar] [CrossRef] [PubMed]
- Koushyar, S.; Meniel, V.S.; Phesse, T.J.; Pearson, H.B. Exploring the Wnt Pathway as a Therapeutic Target for Prostate Cancer. Biomolecules 2022, 12, 309. [Google Scholar] [CrossRef] [PubMed]
- Zi, X.; Guo, Y.; Simoneau, A.R.; Hope, C.; Xie, J.; Holcombe, R.F.; Hoang, B.H. Expression of Frzb/Secreted Frizzled-Related Protein 3, a Secreted Wnt Antagonist, in Human Androgen-Independent Prostate Cancer PC-3 Cells Suppresses Tumor Growth and Cellular Invasiveness. Cancer Res. 2005, 65, 9762–9770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khurana, N.; Sikka, S.C. Interplay Between SOX9, Wnt/β-Catenin and Androgen Receptor Signaling in Castration-Resistant Prostate Cancer. Int. J. Mol. Sci. 2019, 20, 2066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Wang, L.; Zhang, M.; Melamed, J.; Liu, X.; Reiter, R.; Wei, J.; Peng, Y.; Zou, X.; Pellicer, A.; et al. LEF1 in androgen-independent prostate cancer: Regulation of androgen receptor expression, prostate cancer growth, and invasion. Cancer Res. 2009, 69, 3332–3338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Wang, D.; Wan, X.; Xu, Y.; Lu, Y.; Kong, Z.; Li, D.; Gu, W.; Wang, C.; Li, Y.; et al. Crosstalk Between AR and Wnt Signaling Promotes Castration-Resistant Prostate Cancer Growth. Onco Targets Ther. 2020, 13, 9257–9267. [Google Scholar] [CrossRef]
- Lee, E.; Ha, S.; Logan, S.K. Divergent Androgen Receptor and Beta-Catenin Signaling in Prostate Cancer Cells. PLoS ONE 2015, 10, e0141589. [Google Scholar] [CrossRef] [Green Version]
- Pawlowski, J.E.; Ertel, J.R.; Allen, M.P.; Xu, M.; Butler, C.; Wilson, E.M.; Wierman, M.E. Liganded Androgen Receptor Interaction with β-Catenin. J. Biol. Chem. 2002, 277, 20702–20710. [Google Scholar] [CrossRef] [Green Version]
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive Transcriptional Activation by a β-Catenin-Tcf Complex in APC −/− Colon Carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandsmark, E.; Hansen, A.F.; Selnæs, K.M.; Bertilsson, H.; Bofin, A.M.; Wright, A.J.; Viset, T.; Richardsen, E.; Drabløs, F.; Bathen, T.F.; et al. A novel non-canonical Wnt signature for prostate cancer aggressiveness. Oncotarget 2017, 8, 9572–9586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minami, Y.; Oishi, I.; Endo, M.; Nishita, M. Ror-family receptor tyrosine kinases in noncanonical Wnt signaling: Their implications in developmental morphogenesis and human diseases. Dev. Dyn. 2009, 239, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.T.; Kwon, S.J.; Kim, J.; Kwon, Y.S.; Lee, N.; Hong, J.H.; Jamieson, C.; Kim, W.-J.; Kim, I.Y. WNT5A induces castration-resistant prostate cancer via CCL2 and tumour-infiltrating macrophages. Br. J. Cancer 2018, 118, 670–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, C.-Y.; Wu, K.-Y.; Lin, T.-Y.; Chen, C.-C. The Crosstalk of Long Non-Coding RNA and MicroRNA in Castration-Resistant and Neuroendocrine Prostate Cancer: Their Interaction and Clinical Importance. Int. J. Mol. Sci. 2021, 23, 392. [Google Scholar] [CrossRef]
- Lim, M.C.J.; Baird, A.-M.; Aird, J.; Greene, J.; Kapoor, D.; Gray, S.G.; McDermott, R.; Finn, S.P. RNAs as Candidate Diagnostic and Prognostic Markers of Prostate Cancer—From Cell Line Models to Liquid Biopsies. Diagnostics 2018, 8, 60. [Google Scholar] [CrossRef] [Green Version]
- Xie, G.; Jiang, R. Non-coding RNAs in castration-resistant prostate cancer. Zhonghua Nan Ke Xue 2015, 21, 1014–1019. [Google Scholar]
- Rönnau, C.G.H.; Fussek, S.; Smit, F.P.; Aalders, T.W.; van Hooij, O.; Pinto, P.M.C.; Burchardt, M.; Schalken, J.A.; Verhaegh, G.W. Upregulation of miR-3195, miR-3687 and miR-4417 is associated with castration-resistant prostate cancer. World J. Urol. 2021, 39, 3789–3797. [Google Scholar] [CrossRef]
- Goto, Y.; Kojima, S.; Nishikawa, R.; Kurozumi, A.; Kato, M.; Enokida, H.; Matsushita, R.; Yamazaki, K.; Ishida, Y.; Nakagawa, M.; et al. MicroRNA expression signature of castration-resistant prostate cancer: The microRNA-221/222 cluster functions as a tumour suppressor and disease progression marker. Br. J. Cancer 2015, 113, 1055–1065. [Google Scholar] [CrossRef]
- Doldi, V.; El Bezawy, R.; Zaffaroni, N. MicroRNAs as Epigenetic Determinants of Treatment Response and Potential Therapeutic Targets in Prostate Cancer. Cancers 2021, 13, 2380. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, K.Y.; Liu, Q.; Cao, Q. Androgen Receptor-Related Non-coding RNAs in Prostate Cancer. Front. Cell. Dev. Biol. 2021, 9, 660853. [Google Scholar] [CrossRef] [PubMed]
- Arrighetti, N.; Beretta, G.L. miRNAs as Therapeutic Tools and Biomarkers for Prostate Cancer. Pharmaceutics 2021, 13, 380. [Google Scholar] [CrossRef] [PubMed]
- Gui, B.; Hsieh, C.-L.; Kantoff, P.W.; Kibel, A.S.; Jia, L. Androgen receptor-mediated downregulation of microRNA-221 and -222 in castration-resistant prostate cancer. PLoS ONE 2017, 12, e0184166. [Google Scholar] [CrossRef]
- Guo, T.; Wang, Y.; Jia, J.; Mao, X.; Stankiewicz, E.; Scandura, G.; Burke, E.; Xu, L.; Marzec, J.; Davies, C.R.; et al. The Identification of Plasma Exosomal miR-423-3p as a Potential Predictive Biomarker for Prostate Cancer Castration-Resistance Development by Plasma Exosomal miRNA Sequencing. Front. Cell Dev. Biol. 2021, 8, 602493. [Google Scholar] [CrossRef] [PubMed]
- Jalava, S.E.; Urbanucci, A.; Latonen, L.; Waltering, K.K.; Sahu, B.; Jänne, O.A.; Seppälä, J.; Lähdesmäki, H.; Tammela, T.L.J.; Visakorpi, T. Androgen-regulated miR-32 targets BTG2 and is overexpressed in castration-resistant prostate cancer. Oncogene 2012, 31, 4460–4471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakamoto, S. Editorial Comment to Functional significance of aberrantly expressed microRNAs in prostate cancer. Int. J. Urol. 2015, 22, 252–253. [Google Scholar] [CrossRef] [PubMed]
- Lo, U.G.; Lee, C.-F.; Lee, M.-S.; Hsieh, J.-T. The Role and Mechanism of Epithelial-to-Mesenchymal Transition in Prostate Cancer Progression. Int. J. Mol. Sci. 2017, 18, 2079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Jia, D.; Kim, H.; Abd Elmageed, Z.Y.; Datta, A.; Davis, R.; Srivastav, S.; Moroz, K.; Crawford, B.E.; Moparty, K.; et al. Dysregulation of miR-212 Promotes Castration Resistance through hnRNPH1-Mediated Regulation of AR and AR-V7: Implications for Racial Disparity of Prostate Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 1744–1756. [Google Scholar] [CrossRef] [Green Version]
- Kroiss, A.; Vincent, S.; Decaussin-Petrucci, M.; Meugnier, E.; Viallet, J.; Ruffion, A.; Chalmel, F.; Samarut, J.; Allioli, N. Androgen-regulated microRNA-135a decreases prostate cancer cell migration and invasion through downregulating ROCK1 and ROCK2. Oncogene 2014, 34, 2846–2855. [Google Scholar] [CrossRef]
- Sikand, K.; Slaibi, J.E.; Singh, R.; Slane, S.D.; Shukla, G.C. miR 488* inhibits androgen receptor expression in prostate carcinoma cells. Int. J. Cancer 2011, 129, 810–819. [Google Scholar] [CrossRef]
- Das, D.K.; Persaud, L.; Sauane, M. MicroRNA-4719 and microRNA-6756-5p Correlate with Castration-Resistant Prostate Cancer Progression through Interleukin-24 Regulation. Noncoding RNA 2019, 5, 10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, M.K.; Niknafs, Y.S.; Malik, R.; Singhal, U.; Sahu, A.; Hosono, Y.; Barrette, T.R.; Prensner, J.R.; Evans, J.R.; Zhao, S.; et al. The landscape of long noncoding RNAs in the human transcriptome. Nat. Genet. 2015, 47, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef] [PubMed]
- Hangauer, M.J.; Vaughn, I.W.; McManus, M.T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 2013, 9, e1003569. [Google Scholar] [CrossRef] [PubMed]
- Ballarino, M.; Morlando, M.; Fatica, A.; Bozzoni, I. Non-coding RNAs in muscle differentiation and musculoskeletal disease. J. Clin. Investig. 2016, 126, 2021–2030. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Liao, Q.; Li, W.; Deng, G.; Jia, M.; Fang, Q.; Ji, H.; Meng, M. The lncRNA PTTG3P promotes the progression of CRPC via upregulating PTTG1. Bull. Cancer 2021, 108, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Ghildiyal, R.; Sawant, M.; Renganathan, A.; Mahajan, K.; Kim, E.H.; Luo, J.; Dang, H.X.; Maher, C.A.; Feng, F.Y.; Mahajan, N.P. Loss of Long Noncoding RNA NXTAR in Prostate Cancer Augments Androgen Receptor Expression and Enzalutamide Resistance. Cancer Res. 2022, 82, 155–168. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, M.; Yang, Y.; Li, Q.; Yu, J.; Zhu, S.; Niu, Y.; Shang, Z. Targeting KDM4A-AS1 represses AR/AR-Vs deubiquitination and enhances enzalutamide response in CRPC. Oncogene 2022, 41, 387–399. [Google Scholar] [CrossRef]
- Zhang, B.; Zhang, M.; Shen, C.; Liu, G.; Zhang, F.; Hou, J.; Yao, W. LncRNA PCBP1-AS1-mediated AR/AR-V7 deubiquitination enhances prostate cancer enzalutamide resistance. Cell Death Dis. 2021, 12, 856. [Google Scholar] [CrossRef]
- Zhang, A.; Zhao, J.C.; Kim, J.; Fong, K.-W.; Yang, Y.A.; Chakravarti, D.; Mo, Y.-Y.; Yu, J. LncRNA HOTAIR Enhances the Androgen-Receptor-Mediated Transcriptional Program and Drives Castration-Resistant Prostate Cancer. Cell Rep. 2015, 13, 209–221. [Google Scholar] [CrossRef] [Green Version]
- Ostano, P.; Mello-Grand, M.; Sesia, D.; Gregnanin, I.; Peraldo-Neia, C.; Guana, F.; Jachetti, E.; Farsetti, A.; Chiorino, G. Gene Expression Signature Predictive of Neuroendocrine Transformation in Prostate Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, Y.-T.; Lin, T.-P.; Tang, J.-T.; Campbell, M.; Luo, Y.-L.; Lu, S.-Y.; Yang, C.-P.; Cheng, T.-Y.; Chang, C.-H.; Liu, T.-T.; et al. HOTAIR is a REST-regulated lncRNA that promotes neuroendocrine differentiation in castration resistant prostate cancer. Cancer Lett. 2018, 433, 43–52. [Google Scholar] [CrossRef] [PubMed]
- You, Z.; Liu, C.; Wang, C.; Ling, Z.; Wang, Y.; Wang, Y.; Zhang, M.; Chen, S.; Xu, B.; Guan, H.; et al. LncRNA CCAT1 Promotes Prostate Cancer Cell Proliferation by Interacting with DDX5 and MIR-28-5P. Mol. Cancer Ther. 2019, 18, 2469–2479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mao, Y.; Yang, G.; Li, Y.; Liang, G.; Xu, W.; Hu, M. Advances in the Current Understanding of the Mechanisms Governing the Acquisition of Castration-Resistant Prostate Cancer. Cancers 2022, 14, 3744. https://doi.org/10.3390/cancers14153744
Mao Y, Yang G, Li Y, Liang G, Xu W, Hu M. Advances in the Current Understanding of the Mechanisms Governing the Acquisition of Castration-Resistant Prostate Cancer. Cancers. 2022; 14(15):3744. https://doi.org/10.3390/cancers14153744
Chicago/Turabian StyleMao, Yifeng, Gaowei Yang, Yingbang Li, Guowu Liang, Wangwang Xu, and Mingqiu Hu. 2022. "Advances in the Current Understanding of the Mechanisms Governing the Acquisition of Castration-Resistant Prostate Cancer" Cancers 14, no. 15: 3744. https://doi.org/10.3390/cancers14153744
APA StyleMao, Y., Yang, G., Li, Y., Liang, G., Xu, W., & Hu, M. (2022). Advances in the Current Understanding of the Mechanisms Governing the Acquisition of Castration-Resistant Prostate Cancer. Cancers, 14(15), 3744. https://doi.org/10.3390/cancers14153744






