3D Disease Modelling of Hard and Soft Cancer Using PHA-Based Scaffolds
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Chemicals Used
2.2. Production and Extraction of P(3HB) and P(3HO-co-3HD)
2.3. Gas Chromatography Mass Spectrometry (GC-MS)
2.4. Production of the 3D PHA Scaffolds
2.5. Cell Culture on 3D PHA Scaffolds
2.6. Cell Proliferation Studies
2.7. Scanning Electron Microscopy (SEM)
2.7.1. SEM Characterisation of the 3D PHA Scaffolds
2.7.2. SEM Characterisation of the Cell Laden 3D PHA Scaffolds
2.8. Confocal Microscopy
2.9. Total RNA Extraction, cDNA Synthesis and qRT-PCR
2.10. Data Analysis
3. Results
3.1. Production and Chemical Characterisation of the Polymer to Make 3D PHA Scaffolds
3.2. Production and Characterisation of the 3D PHA Scaffold
3.3. Cell Culture on the 3D PHA Scaffolds to Create the Disease Models
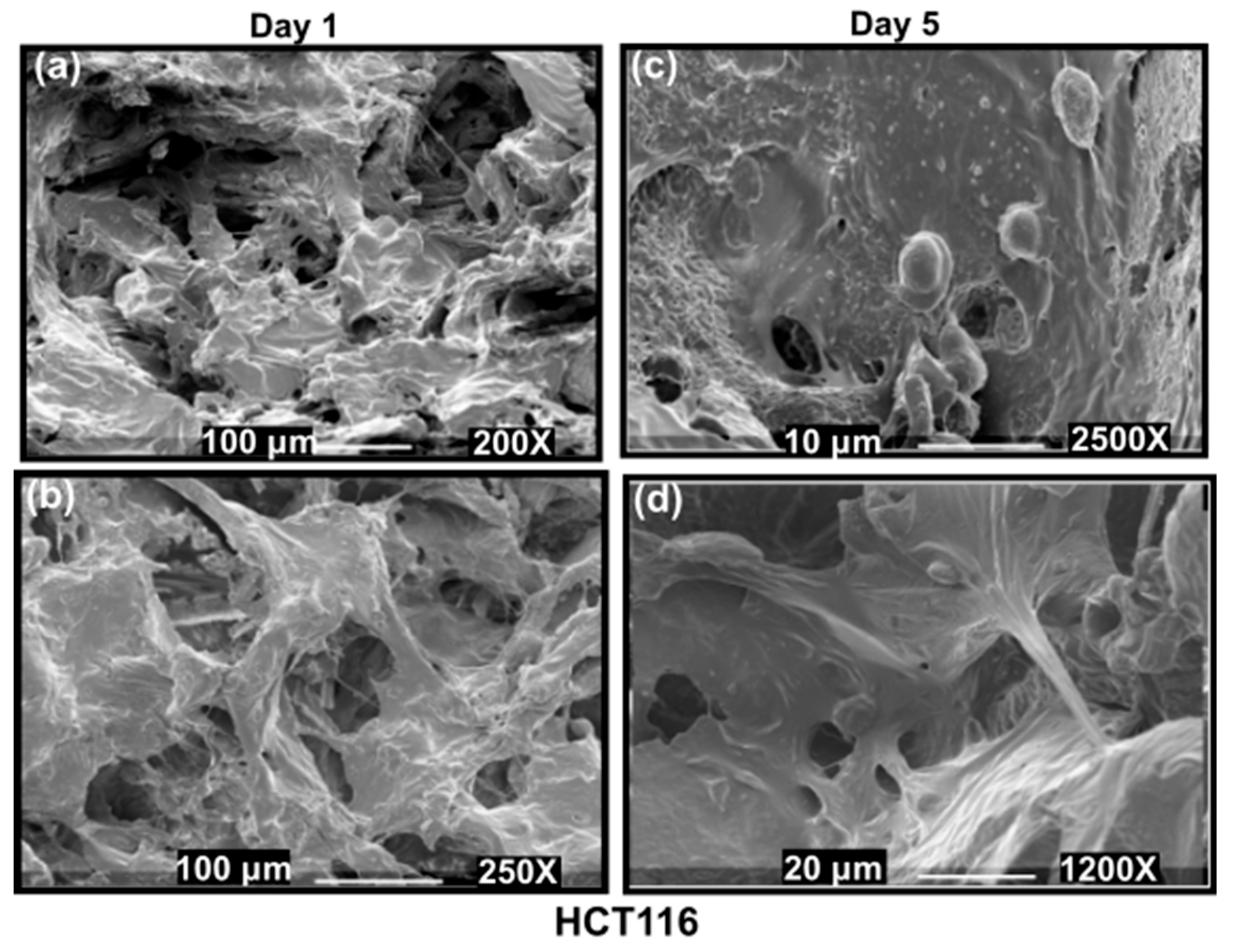
3.3.1. Hard Tumour Disease Models
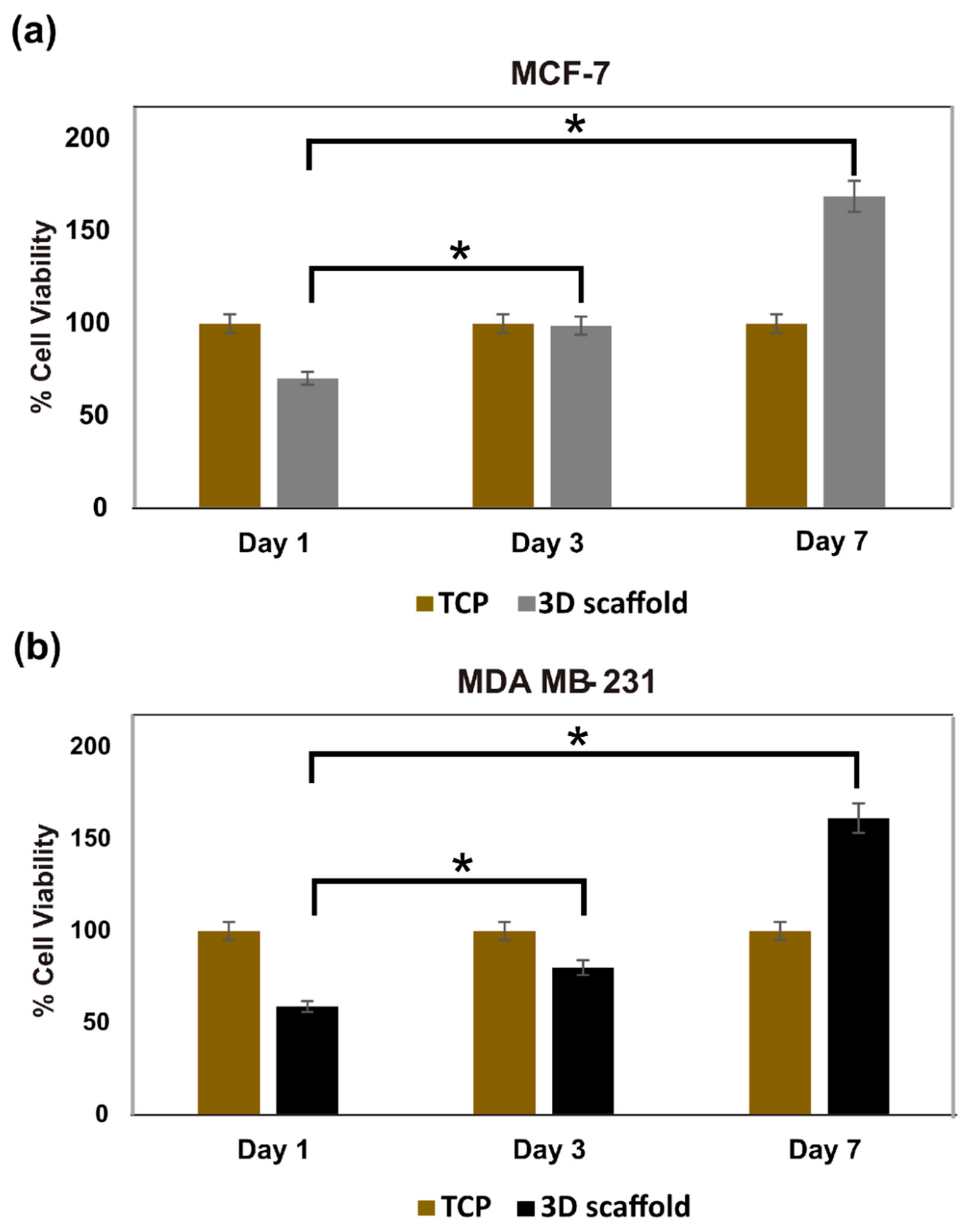
3.3.2. Cell Viability of Hard Cancer 3D Models
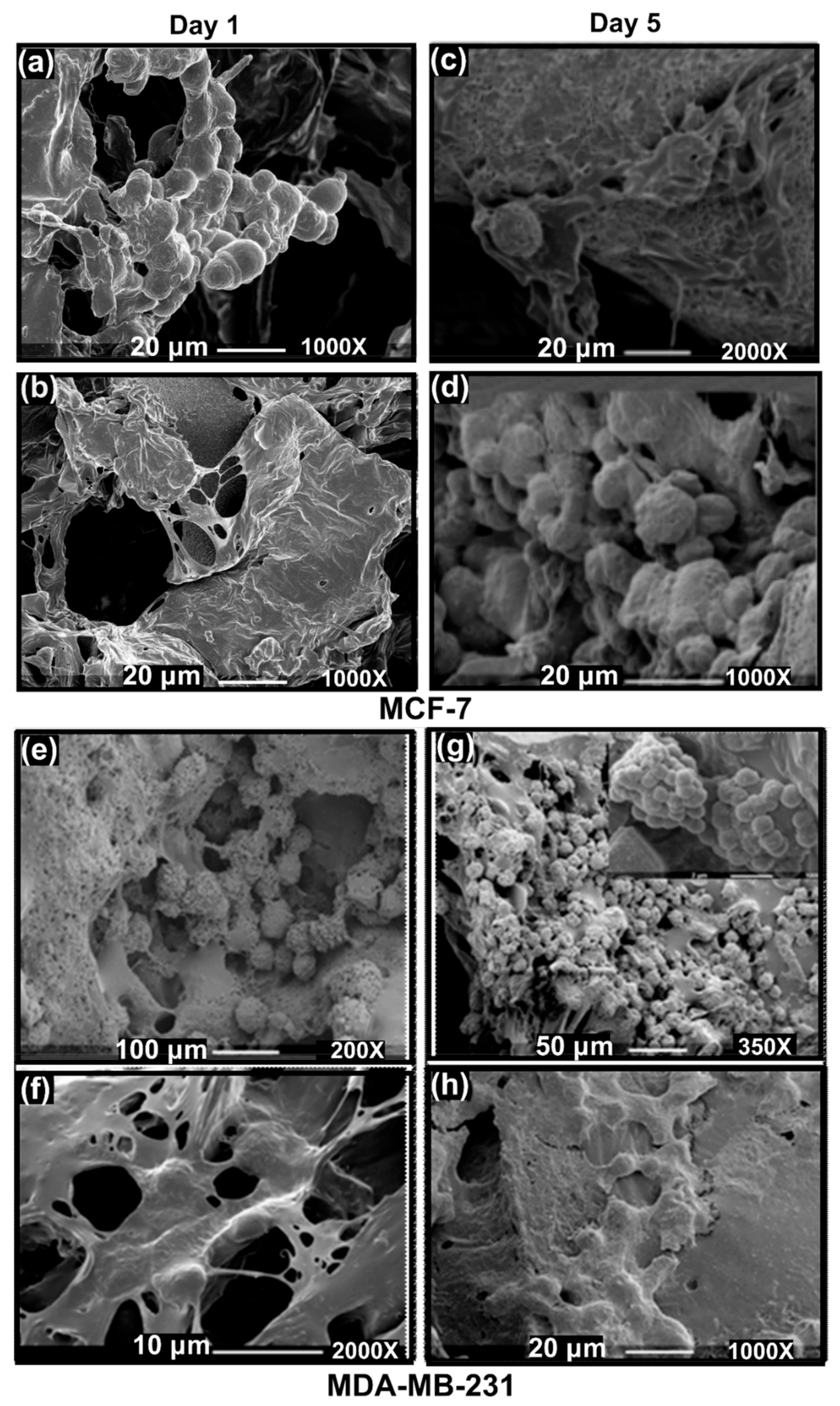
3.3.3. SEM Imaging of the Disease Models: Formation of Hard Cancer 3D Models
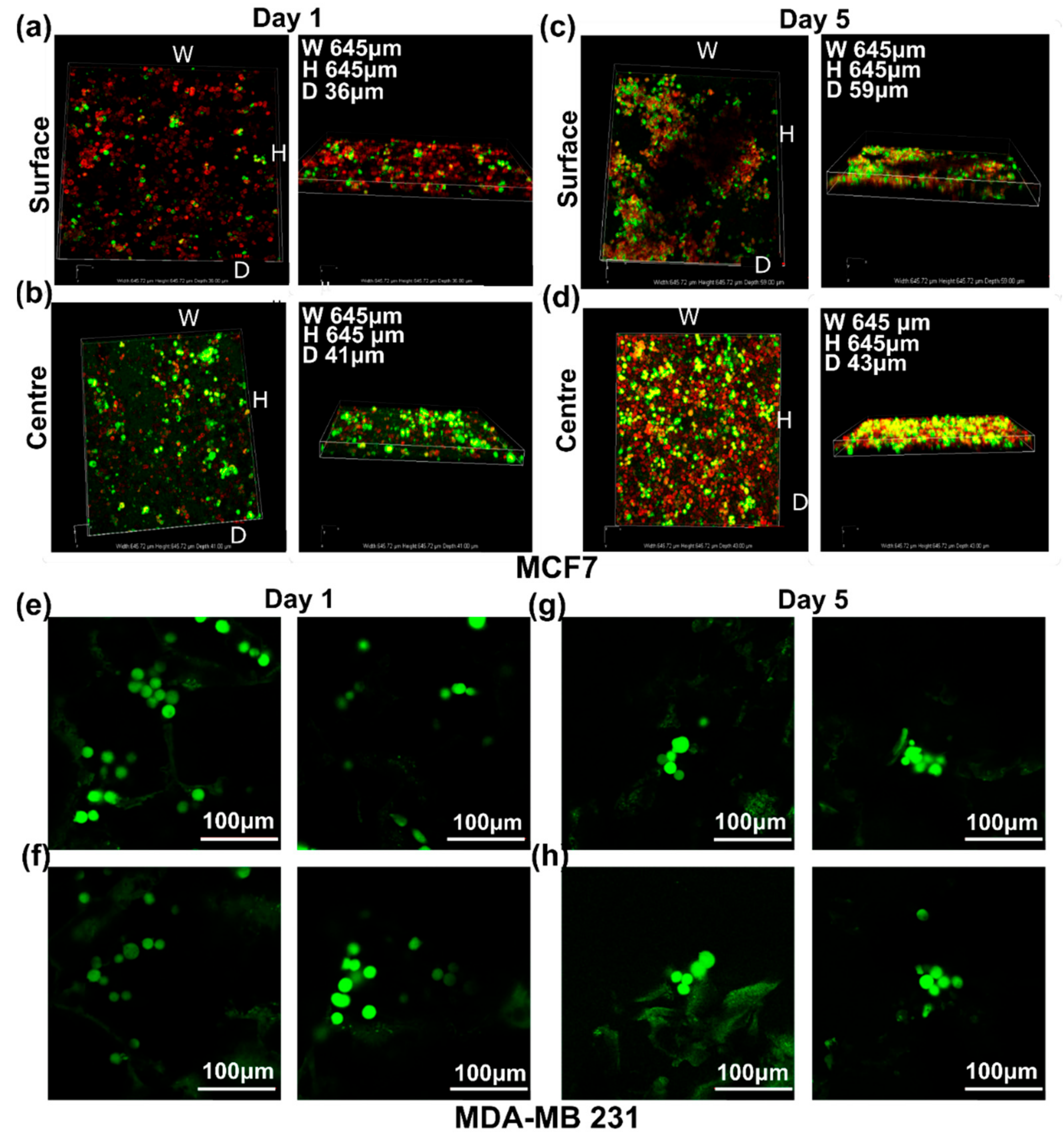
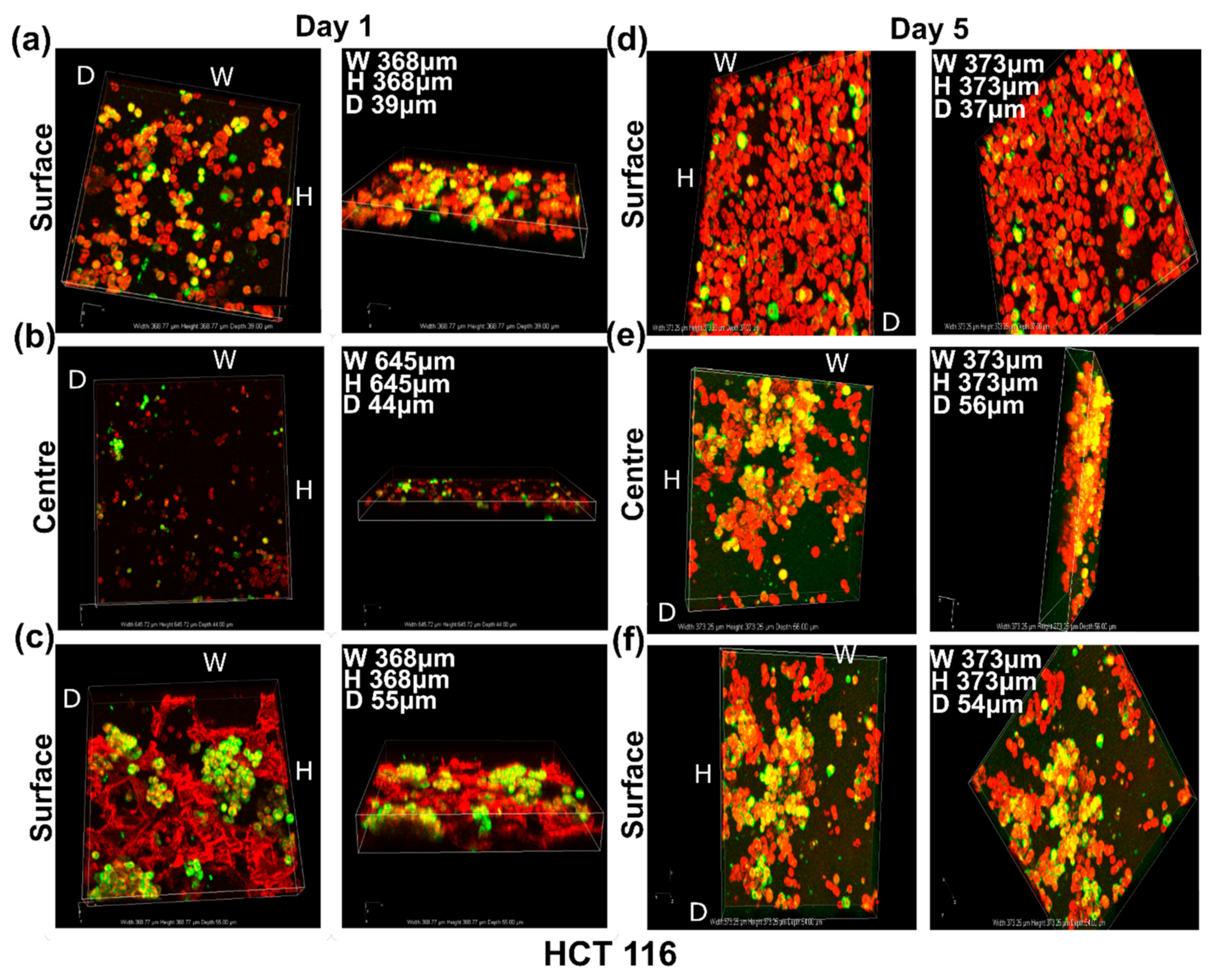
3.3.4. Live Cell Assessment on the Hard Cancer Disease Models
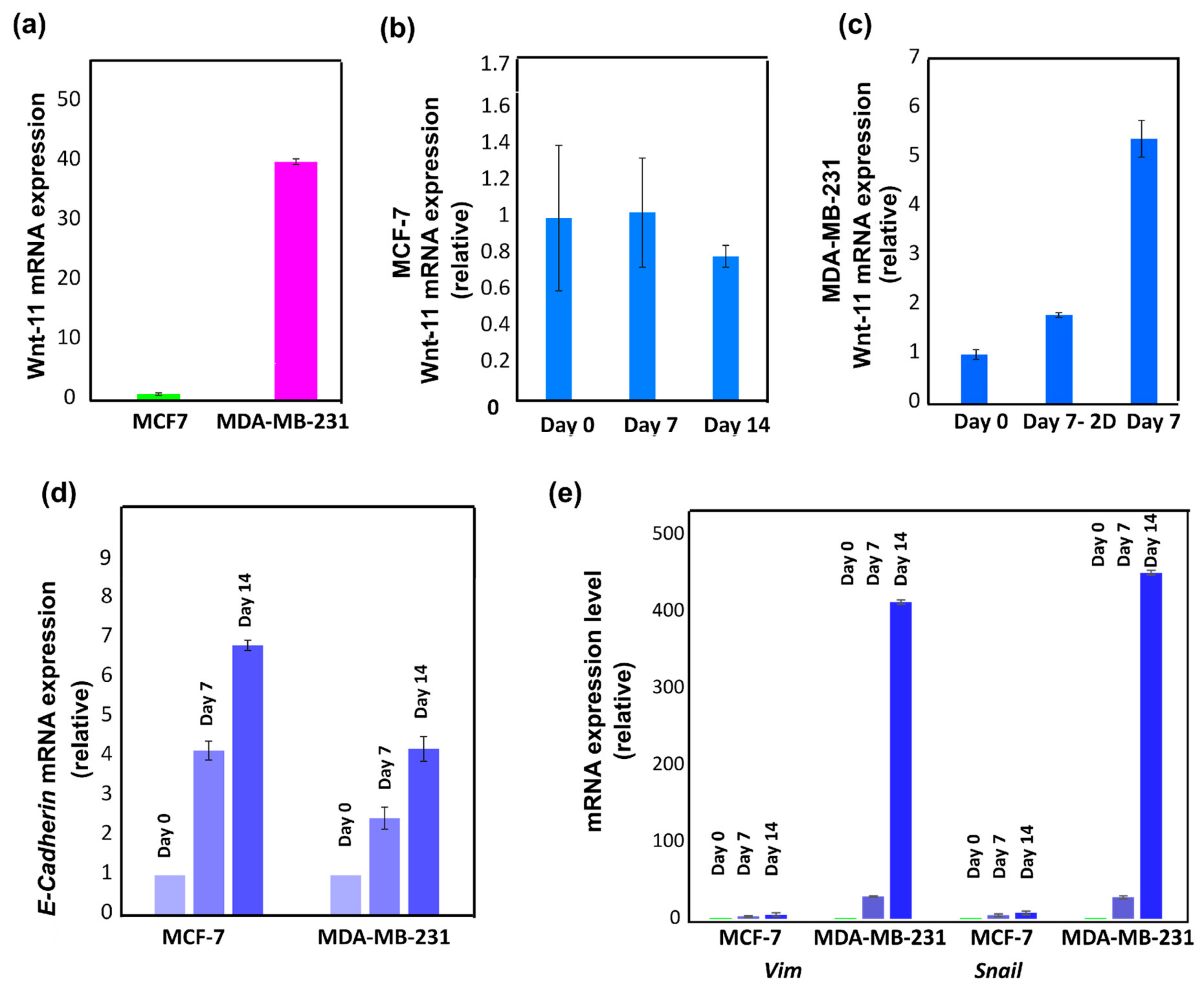
3.3.5. Wnt-11 and E-Cadherin mRNA Expression Profiling
3.4. Cancer Disease Modelling for Soft Tumours
3.4.1. SEM Imaging of the Disease Models: Formation of Soft Cancer 3D Models
3.4.2. Live Cell Assessment on the Soft Cancer Disease Models
3.5. Comparison of Growth Patterns in Soft and Hard Cancer Disease Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Liverani, C.; De Vita, A.; Minardi, S.; Kang, Y.; Mercatali, L.; Amadori, D.; Bongiovanni, A.; La Manna, F.; Ibrahim, T.; Tasciotti, E. A biomimetic 3D model of hypoxia-driven cancer progression. Sci. Rep. 2019, 9, 12263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asghar, W.; El Assal, R.; Shafiee, H.; Pitteri, S.; Paulmurugan, R.; Demirci, U. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today 2015, 18, 539–553. [Google Scholar] [CrossRef]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S. Beyond the Petri dish. Nat. Biotechnol. 2004, 22, 151–152. [Google Scholar] [CrossRef]
- Benton, G.; George, J.; Kleinman, H.K.; Arnaoutova, I.P. Advancing science and technology via 3D culture on basement membrane matrix. J. Cell. Physiol. 2009, 221, 18–25. [Google Scholar] [CrossRef]
- Yamada, K.M.; Cukierman, E. Modeling Tissue Morphogenesis and Cancer in 3D. Cell 2007, 130, 601–610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lutolf, M.P.; Gilbert, P.M.; Blau, H.M. Designing materials to direct stem-cell fate. Nature 2009, 462, 433–441. [Google Scholar] [CrossRef] [Green Version]
- Hutmacher, D.W.; Loessner, D.; Rizzi, S.; Kaplan, D.L.; Mooney, D.J.; Clements, J.A. Can tissue engineering concepts advance tumor biology research? Trends Biotechnol. 2010, 28, 125–133. [Google Scholar] [CrossRef]
- Duval, K.; Grover, H.; Han, L.H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling physiological events in 2D vs. 3D cell culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919. [Google Scholar] [CrossRef]
- Siolas, D.; Hannon, G.J. Patient-derived tumor xenografts: Transforming clinical samples into mouse models. Cancer Res. 2013, 73, 5315–5319. [Google Scholar] [CrossRef] [Green Version]
- Wilding, J.L.; Bodmer, W.F. Cancer cell lines for drug discovery and development. Cancer Res. 2014, 74, 2377–2384. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Chang, H.; Giricz, O.; Lee, G.Y.; Baehner, F.L.; Gray, J.W.; Bissell, M.J.; Kenny, P.A.; Parvin, B. Molecular predictors of 3D morphogenesis by breast cancer cell lines in 3D culture. PLoS Comput. Biol. 2010, 6, e1000684. [Google Scholar] [CrossRef] [Green Version]
- Fischbach, C.; Chen, R.; Matsumoto, T.; Schmelzle, T.; Brugge, J.S.; Polverini, P.J.; Mooney, D.J. Engineering tumors with 3D scaffolds. Nat. Methods 2007, 4, 855–860. [Google Scholar] [CrossRef]
- Fischbach, C.; Hyun, J.K.; Hsiong, S.X.; Evangelista, M.B.; Yuen, W.; Mooney, D.J. Cancer cell angiogenic capability is regulated by 3D culture and integrin engagement. Proc. Natl. Acad. Sci. USA 2009, 106, 399–404. [Google Scholar] [CrossRef] [Green Version]
- Kenny, P.A.; Bissell, M.J. Tumor reversion: Correction of malignant behavior by microenvironmental cues. Int. J. Cancer 2003, 107, 688–695. [Google Scholar] [CrossRef] [Green Version]
- Kenny, H.A.; Krausz, T.; Yamada, S.D.; Lengyel, E. Use of a novel 3D culture model to elucidate the role of mesothelial cells, fibroblasts and extra-cellular matrices on adhesion and invasion of ovarian cancer cells to the omentum. Int. J. Cancer 2007, 121, 1463–1472. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Jeong, J.; DeVolder, R.J.; Cha, C.; Wang, F.; Tong, Y.W.; Kong, H. A cell-instructive hydrogel to regulate malignancy of 3D tumor spheroids with matrix rigidity. Biomaterials 2011, 32, 9308–9315. [Google Scholar] [CrossRef] [PubMed]
- Loessner, D.; Stok, K.S.; Lutolf, M.P.; Hutmacher, D.W.; Clements, J.A.; Rizzi, S.C. Bioengineered 3D platform to explore cell-ECM interactions and drug resistance of epithelial ovarian cancer cells. Biomaterials 2010, 31, 8494–8506. [Google Scholar] [CrossRef] [Green Version]
- Eke, I.; Cordes, N. Radiobiology goes 3D: How ECM and cell morphology impact on cell survival after irradiation. Radiother. Oncol. 2011, 99, 271–278. [Google Scholar] [CrossRef] [PubMed]
- Weiss, M.S.; Bernabé, B.P.; Shikanov, A.; Bluver, D.A.; Mui, M.D.; Shin, S.; Broadbelt, L.J.; Shea, L.D. The impact of adhesion peptides within hydrogels on the phenotype and signaling of normal and cancerous mammary epithelial cells. Biomaterials 2012, 33, 3548–3559. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scanlon, S.E.; Glazer, P.M. Multifaceted control of DNA repair pathways by the hypoxic tumor microenvironment. DNA Repair 2015, 32, 180–189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hammond, E.M.; Asselin, M.C.; Forster, D.; O’Connor, J.P.B.; Senra, J.M.; Williams, K.J. The Meaning, Measurement and Modification of Hypoxia in the Laboratory and the Clinic. Clin. Oncol. 2014, 26, 277–288. [Google Scholar] [CrossRef]
- Wilson, W.R.; Hay, M.P. Targeting hypoxia in cancer therapy. Nat. Rev. Cancer 2011, 11, 393–410. [Google Scholar] [CrossRef]
- Bristow, R.G. Hypoxia, DNA Repair, and Genetic Instability. AACR Educ. Book 2008, 2008, 287–291. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Zhao, J.; Zhang, L.; Wei, F.; Lian, Y.; Wu, Y.; Gong, Z.; Zhang, S.; Zhou, J.; Cao, K.; et al. Role of tumor microenvironment in tumorigenesis. J. Cancer 2017, 8, 761–773. [Google Scholar] [CrossRef]
- Höckel, M.; Vaupel, P. Tumor hypoxia: Definitions and current clinical, biologic, and molecular aspects. J. Natl. Cancer Inst. 2001, 93, 266–276. [Google Scholar] [CrossRef] [Green Version]
- Ricci, C.; Moroni, L.; Danti, S. Cancer tissue engineering new perspectives in understanding the biology of solid tumours a critical review. OA Tissue Eng. 2013, 1, 4. [Google Scholar] [CrossRef] [Green Version]
- Hirt, C.; Papadimitropoulos, A.; Muraro, M.G.; Mele, V.; Panopoulos, E.; Cremonesi, E.; Ivanek, R.; Schultz-Thater, E.; Droeser, R.A.; Mengus, C.; et al. Bioreactor-engineered cancer tissue-like structures mimic phenotypes, gene expression profiles and drug resistance patterns observed “in vivo”. Biomaterials 2015, 62, 138–146. [Google Scholar] [CrossRef]
- Riedl, A.; Schlederer, M.; Pudelko, K.; Stadler, M.; Walter, S.; Unterleuthner, D.; Unger, C.; Kramer, N.; Hengstschläger, M.; Kenner, L.; et al. Comparison of cancer cells in 2D vs 3D culture reveals differences in AKT-mTOR-S6K signaling and drug responses. J. Cell Sci. 2017, 130, 203–218. [Google Scholar]
- Cannon, T.M.; Shah, A.T.; Skala, M.C. Autofluorescence imaging captures heterogeneous drug response differences between 2D and 3D breast cancer cultures. Biomed. Opt. Express 2017, 8, 1911–1925. [Google Scholar] [CrossRef] [Green Version]
- Hashemzadeh, A.H. Comparison between Three-Dimensional Spheroid and Two-Dimensional MonolayerA549 Lung Cancer and PC9 Normal Cell Lines under Treatment of Silver Nanoparticles. J. Biotechnol. 2019, 10, 573–580. [Google Scholar]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [Green Version]
- Kenny, P.A.; Lee, G.Y.; Myers, C.A.; Neve, R.M.; Semeiks, J.R.; Spellman, P.T.; Lorenz, K.; Lee, E.H.; Barcellos-Hoff, M.H.; Petersen, O.W.; et al. The morphologies of breast cancer cell lines in three-dimensional assays correlate with their profiles of gene expression. Mol. Oncol. 2007, 1, 84–96. [Google Scholar] [CrossRef]
- Rahmanzadeh, R.; Rai, P.; Celli, J.P.; Rizvi, I.; Baron-Lühr, B.; Gerdes, J.; Hasan, T. Ki-67 as a molecular target for therapy in an in vitro three-dimensional model for ovarian cancer. Cancer Res. 2010, 70, 9234–9242. [Google Scholar] [CrossRef] [Green Version]
- Rizvi, I.; Celli, J.P.; Evans, C.L.; Abu-Yousif, A.O.; Muzikansky, A.; Pogue, B.W.; Finkelstein, D.; Hasan, T. Synergistic enhancement of carboplatin efficacy with photodynamic therapy in a three-dimensional model for micrometastatic ovarian cancer. Cancer Res. 2010, 70, 9319–9328. [Google Scholar] [CrossRef] [Green Version]
- Lv, D.; Hu, Z.; Lu, L.; Lu, H.; Xu, X. Three-dimensional cell culture: A powerful tool in tumor research and drug discovery. Oncol. Lett. 2017, 14, 6999–7010. [Google Scholar] [CrossRef] [Green Version]
- Nii, T.; Makino, K.; Tabata, Y. Three-dimensional culture system of cancer cells combined with biomaterials for drug screening. Cancers 2020, 12, 2754. [Google Scholar] [CrossRef]
- Tamayol, A.; Najafabadi, A.H.; Aliakbarian, B.; Arab-Tehrany, E.; Akbari, M.; Annabi, N.; Juncker, D.; Khademhosseini, A. Hydrogel Templates for Rapid Manufacturing of Bioactive Fibers and 3D Constructs. Adv. Healthc. Mater. 2015, 4, 2146–2153. [Google Scholar] [CrossRef]
- Liu, C.; Liu, Y.; Xu, X.X.; Wu, H.; Xie, H.G.; Chen, L.; Lu, T.; Yang, L.; Guo, X.; Sun, G.W.; et al. Potential effect of matrix stiffness on the enrichment of tumor initiating cells under three-dimensional culture conditions. Exp. Cell Res. 2015, 330, 123–134. [Google Scholar] [CrossRef] [PubMed]
- DelNero, P.; Lane, M.; Verbridge, S.S.; Kwee, B.; Kermani, P.; Hempstead, B.; Stroock, A.; Fischbach, C. 3D culture broadly regulates tumor cell hypoxia response and angiogenesis via pro-inflammatory pathways. Biomaterials 2015, 55, 110–118. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hayashi, K.; Tabata, Y. Preparation of stem cell aggregates with gelatin microspheres to enhance biological functions. Acta Biomater. 2011, 7, 2797–2803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nii, T.; Makino, K.; Tabata, Y. A Cancer Invasion Model Combined with Cancer-Associated Fibroblasts Aggregates Incorporating Gelatin Hydrogel Microspheres Containing a p53 Inhibitor. Tissue Eng. Part C Methods 2019, 25, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Dhiman, H.K.; Ray, A.R.; Panda, A.K. Characterization and evaluation of chitosan matrix for in vitro growth of MCF-7 breast cancer cell lines. Biomaterials 2004, 25, 5147–5154. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Ramay, H.R.; Hauch, K.D.; Xiao, D.; Zhang, M. Chitosan-alginate hybrid scaffolds for bone tissue engineering. Biomaterials 2005, 26, 3919–3928. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.S.; Dai, L.G.; Yen, B.L.; Hsu, S.H. Spheroid formation of mesenchymal stem cells on chitosan and chitosan-hyaluronan membranes. Biomaterials 2011, 32, 6929–6945. [Google Scholar] [CrossRef]
- Stylianopoulos, T. The Solid Mechanics of Cancer and Strategies for Improved Therapy. J. Biomech. Eng. 2017, 139, 21004. [Google Scholar] [CrossRef]
- Wei, S.C.; Fattet, L.; Tsai, J.H.; Guo, Y.; Pai, V.H.; Majeski, H.E.; Chen, A.C.; Sah, R.L.; Taylor, S.S.; Engler, A.J.; et al. Matrix stiffness drives epithelial-mesenchymal transition and tumour metastasis through a TWIST1-G3BP2 mechanotransduction pathway. Nat. Cell Biol. 2015, 17, 678–688. [Google Scholar] [CrossRef]
- Samani, A.; Zubovits, J.; Plewes, D. Elastic moduli of normal and pathological human breast tissues: An inversion-technique-based investigation of 169 samples. Phys. Med. Biol. 2007, 52, 1565–1576. [Google Scholar] [CrossRef]
- Kawano, S.; Kojima, M.; Higuchi, Y.; Sugimoto, M.; Ikeda, K.; Sakuyama, N.; Takahashi, S.; Hayashi, R.; Ochiai, A.; Saito, N. Assessment of elasticity of colorectal cancer tissue, clinical utility, pathological and phenotypical relevance. Cancer Sci. 2015, 106, 1232–1239. [Google Scholar] [CrossRef]
- Muneer, F.; Rasul, I.; Azeem, F.; Siddique, M.H.; Zubair, M.; Nadeem, H. Microbial Polyhydroxyalkanoates (PHAs): Efficient Replacement of Synthetic Polymers. J. Polym. Environ. 2020, 28, 2301–2323. [Google Scholar] [CrossRef]
- Shishatskaya, E.I.; Volova, T.G. A comparative investigation of biodegradable polyhydroxyalkanoate films as matrices for in vitro cell cultures. J. Mater. Sci. Mater. Med. 2004, 15, 915–923. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, Y.; Chen, G.Q. Medical application of microbial biopolyesters polyhydroxyalkanoates. Artif. Cells Blood Substit. Biotechnol. 2009, 37, 1–12. [Google Scholar] [CrossRef]
- Misra, S.K.; Valappil, S.P.; Roy, I.; Boccaccini, A.R. Polyhydroxyalkanoate (PHA)/inorganic phase composites for tissue engineering applications. Biomacromolecules 2006, 7, 2249–2258. [Google Scholar] [CrossRef]
- Basnett, P.; Marcello, E.; Lukasiewicz, B.; Nigmatullin, R.; Paxinou, A.; Ahmad, M.H.; Gurumayum, B.; Roy, I. Antimicrobial materials with lime oil and a poly(3-hydroxyalkanoate) produced via valorisation of sugar cane molasses. J. Funct. Biomater. 2020, 11, 24. [Google Scholar] [CrossRef]
- Deng, Y.; Lin, X.S.; Zheng, Z.; Deng, J.G.; Chen, J.C.; Ma, H.; Chen, G.Q. Poly (hydroxybutyrate-co-hydroxyhexanoate) promoted production of extracellular matrix of articular cartilage chondrocytes in vitro. Biomaterials 2003, 24, 4273–4281. [Google Scholar] [CrossRef]
- Wang, Y.; Bian, Y.Z.; Wu, Q.; Chen, G.Q. Evaluation of three-dimensional scaffolds prepared from poly (3-hydroxybutyrate-co-3-hydroxyhexanoate) for growth of allogeneic chondrocytes for cartilage repair in rabbits. Biomaterials 2008, 29, 2858–2868. [Google Scholar] [CrossRef]
- Pohl, S.G.; Brook, N.; Agostino, M.; Arfuso, F.; Kumar, A.P.; Dharmarajan, A. Wnt signaling in triple-negative breast cancer. Oncogenesis 2017, 6, e310. [Google Scholar] [CrossRef] [Green Version]
- Dwyer, M.A.; Joseph, J.D.; Wade, H.E.; Eaton, M.L.; Kunder, R.S.; Kazmin, D.; Chang, C.Y.; McDonnell, D.P. WNT11 expression is induced by estrogen-related receptor α and β-catenin and acts in an autocrine manner to increase cancer cell migration. Cancer Res. 2010, 70, 9298–9308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murillo-Garzón, V.; Gorroño-Etxebarria, I.; Åkerfelt, M.; Puustinen, M.C.; Sistonen, L.; Nees, M.; Carton, J.; Waxman, J.; Kypta, R.M. Frizzled-8 integrates Wnt-11 and transforming growth factor-β signaling in prostate cancer. Nat. Commun. 2018, 9, 554–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Xiao, Z.; Meng, Y.; Zhao, Y.; Han, J.; Su, G.; Chen, B.; Dai, J. The enhancement of cancer stem cell properties of MCF-7 cells in 3D collagen scaffolds for modeling of cancer and anti-cancer drugs. Biomaterials 2012, 33, 1437–1444. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Lim, C.T. Tumor dissemination: An EMT affair. Cancer Cell 2013, 23, 272–273. [Google Scholar] [CrossRef] [Green Version]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-Mesenchymal Transition Generates Cells with Properties of Stem Cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Bueno, G.; Cubillo, E.; Sarrió, D.; Peinado, H.; Rodríguez-Pinilla, S.M.; Villa, S.; Bolós, V.; Jordá, M.; Fabra, A.; Portillo, F.; et al. Genetic profiling of epithelial cells expressing E-cadherin repressors reveals a distinct role for snail, Slug, and E47 Factors in epithelial- mesenchymal transition. Cancer Res. 2006, 66, 9543–9556. [Google Scholar] [CrossRef] [Green Version]
- Wachsmannova, L.; Stevurkova, V.; Ciernikova, S. Changes in SNAI1 and VIM gene expression in Caco2 cells after co-cultivation with bacteria from colorectal cancer biopsies. Neoplasma 2019, 66, 271–275. [Google Scholar] [CrossRef]
- Berx, G.; Cleton-Jansen, A.M.; Nollet, F.; De Leeuw, W.J.F.; Van De Vijver, M.J.; Cornelisse, C.; Van Roy, F. E-cadherin is a tumour/invasion suppressor gene mutated in human lobular breast cancers. EMBO J. 1995, 14, 6107–6115. [Google Scholar] [CrossRef]
- Prat, A.; Perou, C.M. Deconstructing the molecular portraits of breast cancer. Mol. Oncol. 2011, 5, 5–23. [Google Scholar] [CrossRef]
- Basnett, P.; Lukasiewicz, B.; Marcello, E.; Gura, H.K.; Knowles, J.C.; Roy, I. Production of a novel medium chain length poly (3-hydroxyalkanoate) using unprocessed biodiesel waste and its evaluation as a tissue engineering scaffold. Microb. Biotechnol. 2017, 10, 1384–1399. [Google Scholar] [CrossRef]
- Basnett, P.; Marcello, E.; Lukasiewicz, B.; Panchal, B.; Nigmatullin, R.; Knowles, J.C.; Roy, I. Biosynthesis and characterization of a novel, biocompatible medium chain length polyhydroxyalkanoate by Pseudomonas mendocina CH50 using coconut oil as the carbon source. J. Mater. Sci. Mater. Med. 2018, 29, 179. [Google Scholar] [CrossRef]
- Taylor, C.S.; Illangakoon, U.; Dawson, J.I.; Kanczler, J.M.; Behbehani, M.; Humphrey, E.; Majid, Q.; Lukasiewicz, B.; Nigmatullin, R.; Heseltine, P.; et al. Harnessing Polyhydroxyalkanoates and Pressurized Gyration for Hard and Soft Tissue Engineering. ACS Appl. Mater. Interfaces 2021, 13, 32624–32639. [Google Scholar]
- Theodoridis, K.; Aggelidou, E.; Manthou, M.E.; Keklikoglou, K.; Tsimponis, A.; Demiri, E.; Bakopoulou, A.; Mihailidis, A.; Kritis, A. An effective device and method for enhanced cell growth in 3D scaffolds: Investigation of cell seeding and proliferation under static and dynamic conditions. Mater. Sci. Eng. C 2020, 114, 111060. [Google Scholar] [CrossRef]
- Uysal-Onganer, P.; Kawano, Y.; Caro, M.; Walker, M.M.; Diez, S.; Darrington, R.S.; Waxman, J.; Kypta, R.M. Wnt-11 promotes neuroendocrine-like differentiation, survival and migration of prostate cancer cells. Mol. Cancer 2010, 9, 55. [Google Scholar] [CrossRef] [Green Version]
- Nozato, M.; Kaneko, S.; Nakagawara, A.; Komuro, H. Epithelial-mesenchymal transition-related gene expression as a new prognostic marker for neuroblastoma. Int. J. Oncol. 2013, 42, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.S.; Liu, L.Q.; Xu, C.; Zhao, Y.H.; Zhu, X.D.; Huang, S.Z. Reference gene selection for quantitative real-time RT-PCR normalization in Iris. lactea var. chinensis roots under cadmium, lead, and salt stress conditions. Sci. World J. 2014, 2014, 532713. [Google Scholar]
- Arisan, E.D.; Rencuzogullari, O.; Freitas, I.L.; Radzali, S.; Keskin, B.; Kothari, A.; Warford, A.; Uysal-Onganer, P. Upregulated wnt-11 and mir-21 expression trigger epithelial mesenchymal transition in aggressive prostate cancer cells. Biology 2020, 9, 52. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Mezencev, R.; Kim, B.; Wang, L.; McDonald, J.; Sulchek, T. Cell Stiffness Is a Biomarker of the Metastatic Potential of Ovarian Cancer Cells. PLoS ONE 2012, 7, e46609. [Google Scholar] [CrossRef] [Green Version]
- Kraning-Rush, C.M.; Califano, J.P.; Reinhart-King, C.A. Cellular traction stresses increase with increasing metastatic potential. PLoS ONE 2012, 7, e32572. [Google Scholar] [CrossRef] [Green Version]
- Yousafzai, M.S.; Coceano, G.; Bonin, S.; Niemela, J.; Scoles, G.; Cojoc, D. Investigating the effect of cell substrate on cancer cell stiffness by optical tweezers. J. Biomech. 2017, 60, 266–269. [Google Scholar]
- Jaganathan, H.; Gage, J.; Leonard, F.; Srinivasan, S.; Souza, G.R.; Dave, B.; Godin, B. Three-dimensional in vitro co-culture model of breast tumor using magnetic levitation. Sci. Rep. 2014, 4, 6468. [Google Scholar] [CrossRef] [Green Version]
- Tryndyak, V.P.; Beland, F.A.; Pogribny, I.P. E-cadherin transcriptional down-regulation by epigenetic and microRNA-200 family alterations is related to mesenchymal and drug-resistant phenotypes in human breast cancer cells. Int. J. Cancer 2010, 126, 2575–2583. [Google Scholar] [CrossRef]
- Narod, S.A.; Iqbal, J.; Jakubowska, A.; Huzarski, T.; Sun, P.; Cybulski, C.; Gronwald, J.; Byrski, T.; Lubinski, J. Are two-centimeter breast cancers large or small? Curr. Oncol. 2013, 20, 205–211. [Google Scholar] [CrossRef] [Green Version]
- Iyer, J.G.; Storer, B.E.; Paulson, K.G.; Lemos, B.; Phillips, J.L.; Bichakjian, C.K.; Zeitouni, N.; Gershenwald, J.E.; Sondak, V.; Otley, C.C.; et al. Relationships between primary tumor size, number of involved nodes and survival among 8044 cases of Merkel cell carcinoma JG. J. Am. Acad. Dermatol. 2014, 70, 637–643. [Google Scholar] [CrossRef] [Green Version]
- Cavo, M.; Fato, M.; Peñuela, L.; Beltrame, F.; Raiteri, R.; Scaglione, S. Microenvironment complexity and matrix stiffness regulate breast cancer cell activity in a 3D in vitro model. Sci. Rep. 2016, 6, 35367. [Google Scholar] [CrossRef] [Green Version]
- Dwivedi, R.; Pandey, R.; Kumar, S.; Mehrotra, D. Poly hydroxyalkanoates (PHA): Role in bone scaffolds. J. Oral Biol. Craniofacial Res. 2020, 10, 389–392. [Google Scholar] [CrossRef]
- Madl, C.M.; Katz, L.M.; Heilshorn, S.C. Tuning Bulk Hydrogel Degradation by Simultaneous Control of Proteolytic Cleavage Kinetics and Hydrogel Network Architecture. ACS Macro Lett. 2018, 7, 1302–1307. [Google Scholar] [CrossRef]
- Constantinides, C.; Basnett, P.; Lukasiewicz, B.; Carnicer, R.; Swider, E.; Majid, Q.A.; Srinivas, M.; Carr, C.A.; Roy, I. In Vivo Tracking and 1H/19F Magnetic Resonance Imaging of Biodegradable Polyhydroxyalkanoate/Polycaprolactone Blend Scaffolds Seeded with Labeled Cardiac Stem Cells. ACS Appl. Mater. Interfaces 2018, 10, 25056–25068. [Google Scholar] [CrossRef] [Green Version]
- Angelini, S.; National, I.; Cerruti, P.; National, I.; Santagata, G.; National, I.; Immirzi, B.; National, I. Microbial Factories; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar]
- Chen, Y.J.; Huang, Y.C.; Lee, C.Y. Production and characterization of medium-chain-length polyhydroxyalkanoates by Pseudomonas mosselii TO7. J. Biosci. Bioeng. 2014, 118, 145–152. [Google Scholar] [CrossRef]
- Singh, M.; Patel, S.K.S.; Kalia, V.C. Bacillus subtilis as potential producer for polyhydroxyalkanoates. Microb. Cell Factories 2009, 9, 38. [Google Scholar] [CrossRef] [Green Version]
- Sola, A.; Bertacchini, J.; D’Avella, D.; Anselmi, L.; Maraldi, T.; Marmiroli, S.; Messori, M. Development of solvent-casting particulate leaching (SCPL) polymer scaffolds as improved three-dimensional supports to mimic the bone marrow niche. Mater. Sci. Eng. C 2019, 96, 153–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, D.J.; Park, S.J.; Gu, B.K.; Kim, Y.J.; Chung, S.; Kim, C.H. Effect of the pore size in a 3D bioprinted gelatin scaffold on fibroblast proliferation. J. Ind. Eng. Chem. 2018, 67, 388–395. [Google Scholar] [CrossRef]
- Loh, Q.L.; Choong, C. Three-dimensional scaffolds for tissue engineering applications: Role of porosity and pore size. Tissue Eng. Part B Rev. 2013, 19, 485–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurski, L.A.; Petrelli, N.J.; Jia, X.; Farach-Carson, M.C. 3D Matrices for Anti-Cancer Drug Testing and Development. Oncol. Issues 2010, 25, 20–25. [Google Scholar] [CrossRef]
- Florczyk, S.J.; Kievit, F.M.; Wang, K.; Erickson, A.E.; Ellenbogen, R.G.; Zhang, M. 3D porous chitosan-alginate scaffolds promote proliferation and enrichment of cancer stem-like cells. J. Mater. Chem. B 2016, 4, 6326–6334. [Google Scholar] [CrossRef] [PubMed]
- Ivers, L.P.; Cummings, B.; Owolabi, F.; Welzel, K.; Klinger, R.; Saitoh, S.; O’Connor, D.; Fujita, Y.; Scholz, D.; Itasaki, N. Dynamic and influential interaction of cancer cells with normal epithelial cells in 3D culture. Cancer Cell Int. 2014, 14, 38. [Google Scholar] [CrossRef] [Green Version]
- Do Amaral, J.B.; Rezende-Teixeira, P.; Freitas, V.M.; MacHado-Santelli, G.M. MCF-7 cells as a three-dimensional model for the study of human breast cancer. Tissue Eng. Part C Methods 2011, 17, 1097–1107. [Google Scholar] [CrossRef]
- Kievit, F.M.; Florczyk, S.J.; Leung, M.C.; Veiseh, O.; Park, J.O.; Disis, M.L.; Zhang, M. Chitosan-alginate 3D scaffolds as a mimic of the glioma tumor microenvironment. Biomaterials 2010, 31, 5903–5910. [Google Scholar]
- Pauff, S.M.; Miller, S.C. Proliferation and enrichment of CD133 + glioblastoma cancer stem cells on 3D chitosan-alginate scaffolds. Bone 2012, 78, 711–716. [Google Scholar]
- Huang, Y.J.; Hsu, S.H. Acquisition of epithelial-mesenchymal transition and cancer stem-like phenotypes within chitosan-hyaluronan membrane-derived 3D tumor spheroids. Biomaterials 2014, 35, 10070–10079. [Google Scholar] [CrossRef]
- Wang, K.; Kievit, F.M.; Erickson, A.E.; Silber, J.R.; Ellenbogen, R.G.; Zhang, M. Culture on 3D Chitosan-Hyaluronic Acid Scaffolds Enhances Stem Cell Marker Expression and Drug Resistance in Human Glioblastoma Cancer Stem Cells. Adv. Healthc. Mater. 2016, 5, 3173–3181. [Google Scholar] [CrossRef]
- Arisan, E.D.; Rencuzogullari, O.; Cieza-borrella, C.; Miralles Arenas, F.; Dwek, M.; Lange, S.; Uysal-onganer, P. Mir-21 is required for the epithelial–mesenchymal transition in mda-mb-231 breast cancer cells. Int. J. Mol. Sci. 2021, 22, 1557. [Google Scholar] [CrossRef]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef]
- Liu, H.; Wang, H.; Chen, D.; Gu, C.; Huang, J.; Mi, K. Endoplasmic reticulum stress inhibits 3D Matrigel-induced vasculogenic mimicry of breast cancer cells via TGF-β1/Smad2/3 and β-catenin signaling. FEBS Open Bio 2021, 11, 2607–2618. [Google Scholar] [CrossRef]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, ZEB and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef]
- Cano, A.; Pérez-Moreno, M.A.; Rodrigo, I.; Locascio, A.; Blanco, M.J.; Del Barrio, M.G.; Portillo, F.; Nieto, M.A. The transcription factor Snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 2000, 2, 76–83. [Google Scholar] [CrossRef]
- Fabregat, I.; Malfettone, A.; Soukupova, J. New insights into the crossroads between EMT and stemness in the context of cancer. J. Clin. Med. 2016, 5, 37. [Google Scholar] [CrossRef] [Green Version]
- De Craene, B.; Gilbert, B.; Stove, C.; Bruyneel, E.; Van Roy, F.; Berx, G. The transcription factor snail induces tumor cell invasion through modulation of the epithelial cell differentiation program. Cancer Res. 2005, 65, 6237–6244. [Google Scholar] [CrossRef] [Green Version]
- Sarrió, D.; Rodriguez-Pinilla, S.M.; Hardisson, D.; Cano, A.; Moreno-Bueno, G.; Palacios, J. Epithelial-mesenchymal transition in breast cancer relates to the basal-like phenotype. Cancer Res. 2008, 68, 989–997. [Google Scholar] [CrossRef] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomar, A.; Uysal-Onganer, P.; Basnett, P.; Pati, U.; Roy, I. 3D Disease Modelling of Hard and Soft Cancer Using PHA-Based Scaffolds. Cancers 2022, 14, 3549. https://doi.org/10.3390/cancers14143549
Tomar A, Uysal-Onganer P, Basnett P, Pati U, Roy I. 3D Disease Modelling of Hard and Soft Cancer Using PHA-Based Scaffolds. Cancers. 2022; 14(14):3549. https://doi.org/10.3390/cancers14143549
Chicago/Turabian StyleTomar, Akanksha, Pinar Uysal-Onganer, Pooja Basnett, Uttam Pati, and Ipsita Roy. 2022. "3D Disease Modelling of Hard and Soft Cancer Using PHA-Based Scaffolds" Cancers 14, no. 14: 3549. https://doi.org/10.3390/cancers14143549
APA StyleTomar, A., Uysal-Onganer, P., Basnett, P., Pati, U., & Roy, I. (2022). 3D Disease Modelling of Hard and Soft Cancer Using PHA-Based Scaffolds. Cancers, 14(14), 3549. https://doi.org/10.3390/cancers14143549







