MicroRNAs, Tristetraprolin Family Members and HuR: A Complex Interplay Controlling Cancer-Related Processes
Abstract
Simple Summary
Abstract
1. Introduction
2. MiRNAs: From Biogenesis to a Complex Regulatory Network
3. AUBPs Interfering with miRNAs-Dependent Regulation
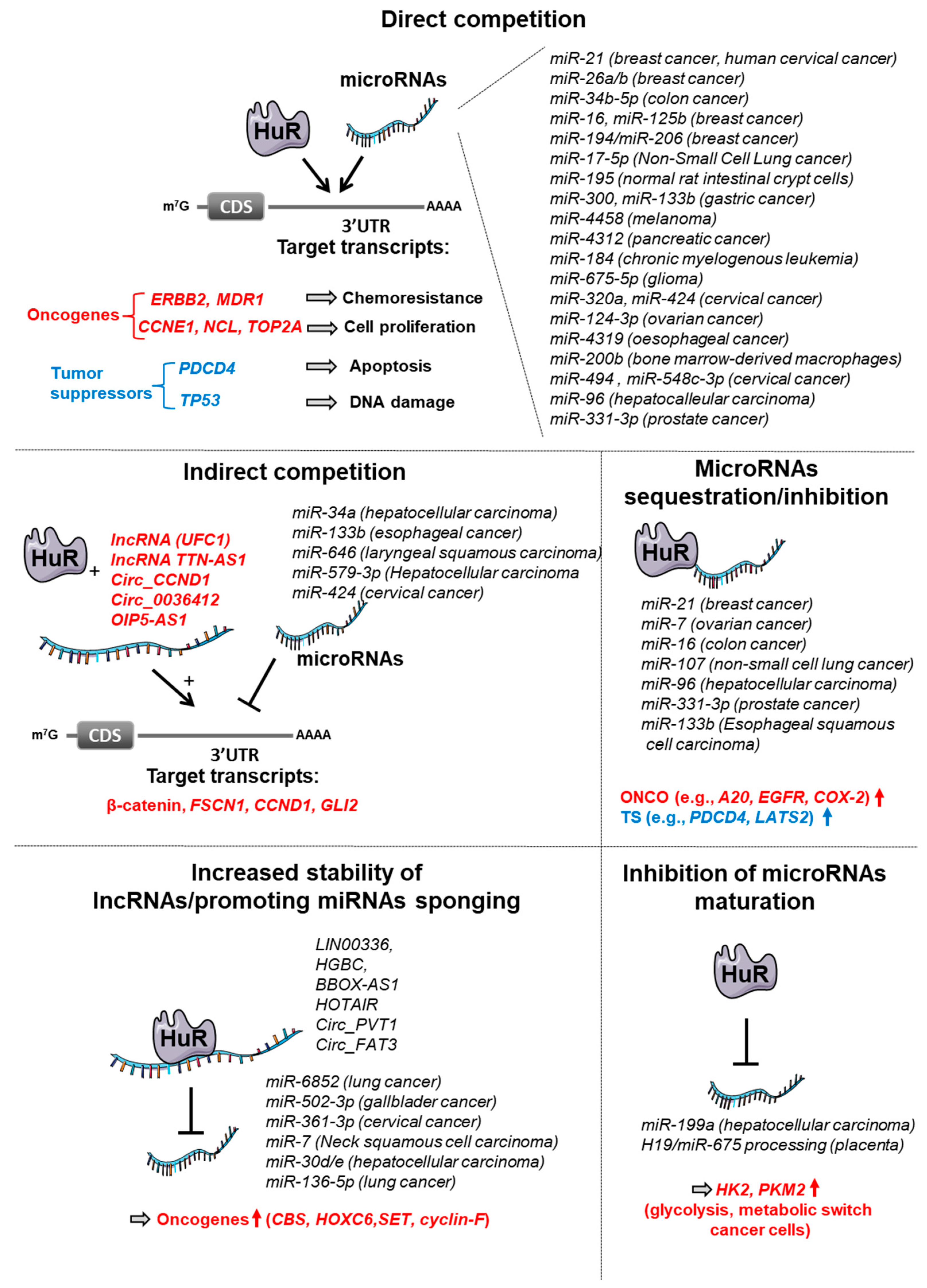
3.1. HuR
3.1.1. Competitive Antagonism between HuR and miRNAs
3.1.2. Other Antagonisms between HuR and miRNAs
3.1.3. Cooperation between HuR and miRNAs
3.2. TTP Family Members (TTP, TIS11, GOS24, NUP475)
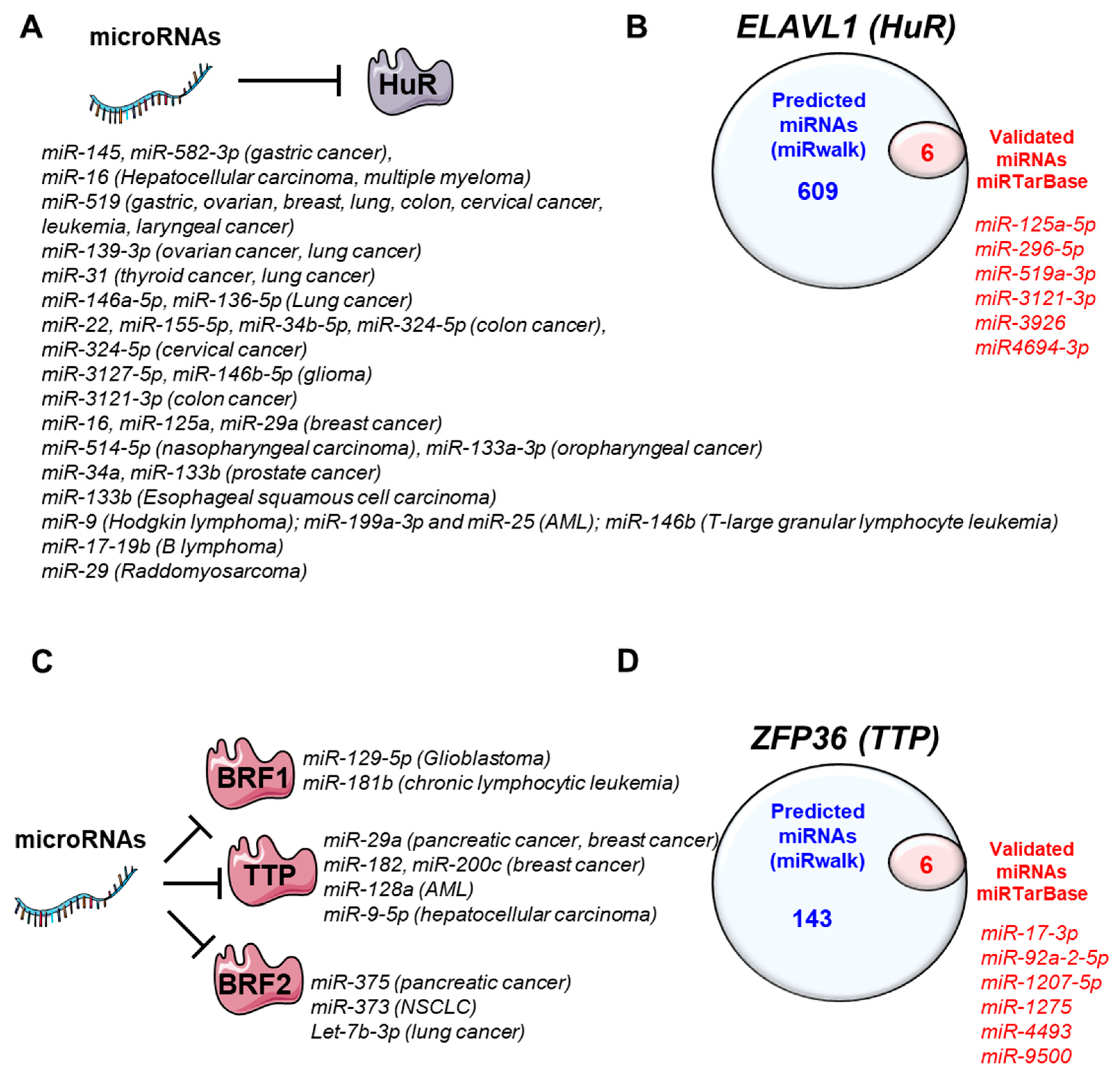
4. MiRNAs-Regulating HuR and TTP Family Members
4.1. HuR
4.2. TTP Family Members
4.2.1. Tristetraprolin (ZFP36)
4.2.2. Butyrate Response Factor 1 (ZFP36L1)
4.2.3. Butyrate Response Factor 2 (ZFP36L2)
| miRNA | Targets (AUBPs) | Models Used | Expression/Activity in Patients | Reference |
|---|---|---|---|---|
| Gastric cancer | ||||
| miR-145 | ELAVL1 | SK-OV-3, A2780 and OVCAR-3 | Down | [161] |
| miR-519 | n.a | Down | [162] | |
| miR-582-3p | HGC27, BGC823 cells | Down | [163] | |
| Ovarian cancer | ||||
| miR-139-3p | ELAVL1 | SK-OV-3, A2780 and OVCAR-3 | Down | [166] |
| miR-519 | Hela, A21780 and HOSE-B cells | Down | [167] | |
| Papillary thyroid carcinoma | ||||
| miR-31 | ELAVL1 | Ovarian carcinoma cells | Down | [169] |
| Non-small cell lung carcinoma | ||||
| miR-139-3p | ELAVL1 | Normal bronchial epithelial cells (BEAS-2B) and NSCLC cells (H1299, H1975, HCC827, H1650 A549) | Down | [172] |
| miR-146a-5p | A549 cells | Down | [173] | |
| miR-31 | n.a | Down | [174] | |
| miR-519 | Hela, A21780 and HOSE-B cells | Down | [167] | |
| miR-373 | ZFP36L2 | A549 cells | Down | [214] |
| Lung adenocarcinoma | ||||
| Let-7b-3p | ZFP36L2 | H1299, A549 cells | Down | [215] |
| Hepatocellular carcinoma | ||||
| miR-16 | ELAVL1 | Hep3B, WRL68 | Down | [165] |
| miR-9-5p | ZFP36 | HuH7, Hep3B | Down | [208] |
| Colorectal cancer | ||||
| miR-22 | ELAVL1 | NCM460, SW480, HT29, HCT15, HCT116, SW620, Caco2, LOVO | Down | [175] |
| miR-324-5p | SW620, SW480, NCM-460 cells | Down | [176] | |
| miR-519 | S1, S1M1 80 (mitoxantrone-resistant), Caco-2, HT-29, SW620 | Down | [168,177] | |
| miR-155-5p | HT-29 | Up | [178,216] | |
| miR-34b-5p | NCM460, SW620, HT-29, HCT116, LoVo, RKO | Down | [60] | |
| miR-3121-3p | DLD-1, HCT116, HT29, SW480 | Down | [179]. | |
| Prostate cancer | ||||
| miR-133b | ELAVL1 | PC3, DU-145 cell lines | Down | [189,217,218] |
| miR-34a | PC3 | Down | [191,219] | |
| Cervical cancer | ||||
| miR-324-5p | ELAVL1 | 33A, ME-180, Hela and Caski | Down | [168,180] |
| miR-519 | Hela cells | Down | [168,180] | |
| Pancreatic cancer | ||||
| miR-29a | ZFP36 | Panc-1, HPDE6c7, BXPC-3 | Up | [204] |
| miR-375 | ZFP36L2 | Pancreatic ductal adenocarcinoma PANC-1 and SW1990 | Down | [213] |
| Glioblastoma | ||||
| miR-3127-5p | ELAVL1 | Down | [183] | |
| miR-146b-5p | Glioma stem cells | Down | [184] | |
| miR-129-5p | ZFP36L1 | LN229, A172, U87, T98G, U251, H4, LN118 and normal astrocytes | Down | [211] |
| Breast cancer | ||||
| miR-16 | ELAVL1 | MDA-MB-231 | Down | [185] |
| miR-125a | MCF-7 | Down | [186,220] | |
| miR-29a | ELAVL1 (indirectly) | MDA-MB-231, MCF-7, MCF12A, MCF10A cells | Up | [188] |
| miR-519 | ELAVL1 | MCF-7 cells | Down | [187] |
| miR-29a | ZFP36 | MDA-MB-231 and MCF-7, MCF12A and MCF10A (normal-like breast cell line), and HEK293 kidney cells | Up | [188,203] |
| miR-182 | ZFP36 | MDA-MB-231, SUM-159, MCF-7, SK-BR-3, MDA-MB-157 | Up | [206] |
| miR-200c | ZFP36 | 4TO7 cells | Up | [209,221] |
| Nasopharyngeal carcinoma | ||||
| miR-514-5p | ELAVL1 | CNE1, CNE2, C666–1 and HNE1 cells | Down | [192] |
| Oropharyngeal squamous cell carcinoma | ||||
| miR-133a-3p | ELAVL1 | UMSCC47 and UMSCC11A cells | Down | [193] |
| Laryngeal squamous cell carcinoma | ||||
| miR-519a | ELAVL1 | laryngeal squamous cell carcinoma human epithelial type 2 cells | Down | [194] |
| Hematologic malignancies | ||||
| miR-519 | ELAVL1 | HL-60 cells (AML) | Up | [196] |
| miR-25 | ELAVL1 (indirectly) | U937 (AML) | Down | [198,222] |
| miR-199a-3p | ELAVL1 | Bone marrow cells | n.a. | [197] |
| miR-25 | ELAVL1 (indirect) | Bone marrow cells | Down | [197,222] |
| miR-146b | ELAVL1 | T-large granular lymphocyte leukemia (T-LGLL), | Down | [200] |
| miR-128a | ZFP36 | AML cells (OCI-AML3 and APL/AML) | Up | [207] |
| miR-181b | ZFP36L1 (not validated) | CLL cells | Down | [212] |
| miR-9 | ELAVL1 | Hodgkin lymphoma L428 cells | Up | [123,223] |
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
References
- Hill, M.; Tran, N. miRNA interplay: Mechanisms and consequences in cancer. Dis. Model. Mech. 2021, 14. [Google Scholar] [CrossRef] [PubMed]
- Monaci, R.; Meoni, S.; Rondine, P.; Leone, G.; Ciuffoletti, L.; Cecchin, A.; Morandini, M. Echotomography in abdominal emergencies. Apropos of 16 controlled clinical cases. (A description of some of them). Minerva Med. 1989, 80, 117–121. [Google Scholar] [PubMed]
- Bernardo, B.C.; Ooi, J.Y.Y.; Lin, R.C.Y.; McMullen, J.R. miRNA therapeutics: A new class of drugs with potential therapeutic applications in the heart. Future Med. Chem. 2015, 7, 1771–1792. [Google Scholar] [CrossRef]
- Yin, X.; Lin, H.; Lin, L.; Miao, L.; He, J.; Zhuo, Z. LncRNAs and CircRNAs in cancer. MedComm 2022, 3, e141. [Google Scholar] [CrossRef] [PubMed]
- Winkler, L.; Dimitrova, N. A mechanistic view of long noncoding RNAs in cancer. Wiley Interdiscip. Rev. RNA 2022, 13, e1699. [Google Scholar] [CrossRef] [PubMed]
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [Google Scholar] [CrossRef] [PubMed]
- Barreau, C.; Paillard, L.; Osborne, H.B. AU-rich elements and associated factors: Are there unifying principles? Nucleic Acids Res. 2005, 33, 7138–7150. [Google Scholar] [CrossRef] [PubMed]
- Lamarre, J.; Gingerich, T.J.; Feige, J.-J. AU-rich elements and the control of gene expression through regulated mRNA stability. Anim. Health Res. Rev. 2004, 5, 49–63. [Google Scholar] [CrossRef]
- Dolicka, D.; Sobolewski, C.; De Sousa, M.C.; Gjorgjieva, M.; Foti, M. mRNA Post-Transcriptional Regulation by AU-Rich Element-Binding Proteins in Liver Inflammation and Cancer. Int. J. Mol. Sci. 2020, 21, 6648. [Google Scholar] [CrossRef]
- Moore, A.E.; Young, L.E.; Dixon, D.A. MicroRNA and AU-rich element regulation of prostaglandin synthesis. Cancer Metastasis Rev. 2011, 30, 419–435. [Google Scholar] [CrossRef]
- Kedde, M.; Agami, R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle 2008, 7, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Huang, Z.; Zhong, S.; Fei, K.; Cao, Y. miR21 inhibits autophagy and promotes malignant development in the bladder cancer T24 cell line. Int. J. Oncol. 2020, 56, 986–998. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Li, C.; Sun, L.; Huang, D.; Li, T.; He, X.; Wu, G.; Yang, Z.; Zhong, X.; Song, L.; et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat. Commun. 2014, 5, 5212. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Guo, M.; Li, Y.; Shen, M.; Kong, D.; Shao, J.; Ding, H.; Tan, S.; Chen, A.; Zhang, F.; et al. RNA-binding protein ZFP36/TTP protects against ferroptosis by regulating autophagy signaling pathway in hepatic stellate cells. Autophagy 2020, 16, 1482–1505. [Google Scholar] [CrossRef]
- Gjorgjieva, M.; Sobolewski, C.; Ay, A.-S.; Abegg, D.; De Sousa, M.C.; Portius, D.; Berthou, F.; Fournier, M.; Maeder, C.; Rantakari, P.; et al. Genetic Ablation of MiR-22 Fosters Diet-Induced Obesity and NAFLD Development. J. Pers. Med. 2020, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Qin, W.; Huo, J.; Zhuo, Q.; Wang, J.; Wang, L. MiR-22 modulates the expression of lipogenesis-related genes and promotes hepatic steatosis in vitro. FEBS Open Bio 2021, 11, 322–332. [Google Scholar] [CrossRef]
- Mohr, A.M.; Mott, J.L. Overview of MicroRNA Biology. Semin. Liver Dis. 2015, 35, 3–11. [Google Scholar] [CrossRef]
- Szczepanek, J.; Skorupa, M.; Tretyn, A. MicroRNA as a Potential Therapeutic Molecule in Cancer. Cells 2022, 11, 1008. [Google Scholar] [CrossRef]
- Winter, J.; Jung, S.; Keller, S.; Gregory, R.I.; Diederichs, S. Many roads to maturity: MicroRNA biogenesis pathways and their regulation. Nat. Cell Biol. 2009, 11, 228–234. [Google Scholar] [CrossRef]
- de Sousa, M.C.; Gjorgjieva, M.; Dolicka, D.; Sobolewski, C.; Foti, M. Deciphering miRNAs’ Action through miRNA Editing. Int. J. Mol. Sci. 2019, 20, 6249. [Google Scholar] [CrossRef]
- Kim, M.; Hur, B.; Kim, S. RDDpred: A condition-specific RNA-editing prediction model from RNA-seq data. BMC Genom. 2016, 17 (Suppl. S1), 85–95. [Google Scholar] [CrossRef] [PubMed]
- Karagkouni, D.; Karavangeli, A.; Paraskevopoulou, M.D.; Hatzigeorgiou, A.G. Characterizing miRNA–lncRNA Interplay. Methods Mol. Biol. 2021, 2372, 243–262. [Google Scholar] [CrossRef] [PubMed]
- Sharma, S.; Verma, S.; Vasudevan, M.; Samanta, S.; Thakur, J.K.; Kulshreshtha, R. The interplay of HuR and miR-3134 in regulation of AU rich transcriptome. RNA Biol. 2013, 10, 1283–1290. [Google Scholar] [CrossRef] [PubMed]
- Baccarini, A.; Brown, B.D. Monitoring MicroRNA Activity and Validating MicroRNA Targets by Reporter-Based Approaches. Methods Mol. Biol. 2010, 667, 215–233. [Google Scholar] [CrossRef]
- Clément, S.; Sobolewski, C.; Gomes, D.; Rojas, A.; Goossens, N.; Conzelmann, S.; Calo, N.; Negro, F.; Foti, M. Activation of the oncogenic miR-21-5p promotes HCV replication and steatosis induced by the viral core 3a protein. Liver Int. 2019, 39, 1226–1236. [Google Scholar] [CrossRef]
- Li, J.-H.; Liu, S.; Zhou, H.; Qu, L.-H.; Yang, J.-H. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein–RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014, 42, D92–D97. [Google Scholar] [CrossRef]
- Nicholson, C.O.; Friedersdorf, M.B.; Keene, J.D. Quantifying RNA binding sites transcriptome-wide using DO-RIP-seq. RNA 2017, 23, 32–46. [Google Scholar] [CrossRef]
- Lebedeva, S.; Jens, M.; Theil, K.; Schwanhäusser, B.; Selbach, M.; Landthaler, M.; Rajewsky, N. Transcriptome-wide Analysis of Regulatory Interactions of the RNA-Binding Protein HuR. Mol. Cell 2011, 43, 340–352. [Google Scholar] [CrossRef]
- Legrand, N.; A Dixon, D.; Sobolewski, C. AU-rich element-binding proteins in colorectal cancer. World J. Gastrointest. Oncol. 2019, 11, 71–90. [Google Scholar] [CrossRef]
- Young, L.E.; Sanduja, S.; Bemis–Standoli, K.; Pena, E.A.; Price, R.L.; Dixon, D.A. The mRNA Binding Proteins HuR and Tristetraprolin Regulate Cyclooxygenase 2 Expression During Colon Carcinogenesis. Gastroenterology 2009, 136, 1669–1679. [Google Scholar] [CrossRef]
- Dixon, D.A.; Tolley, N.D.; King, P.H.; Nabors, L.B.; McIntyre, T.M.; Zimmerman, G.A.; Prescott, S.M. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Investig. 2001, 108, 1657–1665. [Google Scholar] [CrossRef] [PubMed]
- Dixon, D.A.; Balch, G.; Kedersha, N.; Anderson, P.; Zimmerman, G.A.; Beauchamp, R.D.; Prescott, S.M. Regulation of Cyclooxygenase-2 Expression by the Translational Silencer TIA-1. J. Exp. Med. 2003, 198, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Abdelmohsen, K.; Lal, A.; Pullmann, R.; Yang, X.; Galban, S.; Srikantan, S.; Martindale, J.L.; Blethrow, J.; Shokat, K.M.; et al. Nuclear HuR accumulation through phosphorylation by Cdk1. Genes Dev. 2008, 22, 1804–1815. [Google Scholar] [CrossRef]
- Tiedje, C.; Ronkina, N.; Tehrani, M.; Dhamija, S.; Laass, K.; Holtmann, H.; Kotlyarov, A.; Gaestel, M. The p38/MK2-Driven Exchange between Tristetraprolin and HuR Regulates AU–Rich Element–Dependent Translation. PLoS Genet. 2012, 8, e1002977. [Google Scholar] [CrossRef] [PubMed]
- Sandler, H.; Stoecklin, G. Control of mRNA decay by phosphorylation of tristetraprolin. Biochem. Soc. Trans. 2008, 36, 491–496. [Google Scholar] [CrossRef]
- Sengupta, S.; Jang, B.-C.; Wu, M.-T.; Paik, J.-H.; Furneaux, H.; Hla, T. The RNA-binding Protein HuR Regulates the Expression of Cyclooxygenase-2. J. Biol. Chem. 2003, 278, 25227–25233. [Google Scholar] [CrossRef] [PubMed]
- Dean, J.L.E.; Wait, R.; Mahtani, K.R.; Sully, G.; Clark, A.R.; Saklatvala, J. The 3′ Untranslated Region of Tumor Necrosis Factor Alpha mRNA Is a Target of the mRNA-Stabilizing Factor HuR. Mol. Cell. Biol. 2001, 21, 721–730. [Google Scholar] [CrossRef]
- Sawicki, K.T.; Chang, H.-C.; Shapiro, J.S.; Bayeva, M.; De Jesus, A.; Finck, B.N.; Wertheim, J.A.; Blackshear, P.J.; Ardehali, H. Hepatic tristetraprolin promotes insulin resistance through RNA destabilization of FGF21. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Zhang, Z.; Zong, C.; Jiang, M.; Hu, H.; Cheng, X.; Ni, J.; Yi, X.; Jiang, B.; Tian, F.; Chang, M.-W.; et al. Hepatic HuR modulates lipid homeostasis in response to high-fat diet. Nat. Commun. 2020, 11, 3067. [Google Scholar] [CrossRef]
- Rounbehler, R.J.; Fallahi, M.; Yang, C.; Steeves, M.A.; Li, W.; Doherty, J.R.; Schaub, F.X.; Sanduja, S.; Dixon, D.A.; Blackshear, P.J.; et al. Tristetraprolin Impairs Myc-Induced Lymphoma and Abolishes the Malignant State. Cell 2012, 150, 563–574. [Google Scholar] [CrossRef]
- Cao, Y.; Huang, W.; Wu, F.; Shang, J.; Ping, F.; Wang, W.; Li, Y.; Zhao, X.; Zhang, X. ZFP36 protects lungs from intestinal I/R-induced injury and fibrosis through the CREBBP/p53/p21/Bax pathway. Cell Death Dis. 2021, 12, 685. [Google Scholar] [CrossRef] [PubMed]
- Srikantan, S.; Tominaga, K.; Gorospe, M. Functional Interplay between RNA-Binding Protein HuR and microRNAs. Curr. Protein Pept. Sci. 2012, 13, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, N.; Corcoran, D.L.; Nusbaum, J.D.; Reid, D.W.; Georgiev, S.; Hafner, M.; Ascano, M.; Tuschl, T.; Ohler, U.; Keene, J.D. Integrative Regulatory Mapping Indicates that the RNA-Binding Protein HuR Couples Pre-mRNA Processing and mRNA Stability. Mol. Cell 2011, 43, 327–339. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.M.; Steitz, J.A. HuR and mRNA stability. Cell Mol. Life Sci. 2001, 58, 266–277. [Google Scholar] [CrossRef]
- Brody, J.R.; Dixon, D.A. Complex HuR function in pancreatic cancer cells. Wiley Interdiscip. Rev. RNA 2018, 9, e1469. [Google Scholar] [CrossRef] [PubMed]
- Grammatikakis, I.; Abdelmohsen, K.; Gorospe, M. Posttranslational control of HuR function. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Doller, A.; Pfeilschifter, J.; Eberhardt, W. Signalling pathways regulating nucleo-cytoplasmic shuttling of the mRNA-binding protein HuR. Cell. Signal. 2008, 20, 2165–2173. [Google Scholar] [CrossRef]
- Dixon, D.A.; Blanco, F.F.; Bruno, A.; Patrignani, P. Mechanistic aspects of COX-2 expression in colorectal neoplasia. Recent Results Cancer Res. 2013, 191, 7–37. [Google Scholar] [CrossRef]
- A Dixon, D. Regulation of COX-2 Expression in Human Cancers. Prog. Exp. Tumor. Res. 2003, 37, 52–71. [Google Scholar] [CrossRef]
- Wu, X.; Gardashova, G.; Lan, L.; Han, S.; Zhong, C.; Marquez, R.T.; Wei, L.; Wood, S.; Roy, S.; Gowthaman, R.; et al. Targeting the interaction between RNA-binding protein HuR and FOXQ1 suppresses breast cancer invasion and metastasis. Commun. Biol. 2020, 3, 193. [Google Scholar] [CrossRef]
- Blanco, F.F.; Preet, R.; Aguado, A.; Vishwakarma, V.; Stevens, L.E.; Vyas, A.; Padhye, S.; Xu, L.; Weir, S.J.; Anant, S.; et al. Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis. Oncotarget 2016, 7, 74043–74058. [Google Scholar] [CrossRef] [PubMed]
- Lal, P.; Cerofolini, L.; D’Agostino, V.G.; Zucal, C.; Fuccio, C.; Bonomo, I.; Dassi, E.; Giuntini, S.; Di Maio, D.; Vishwakarma, V.; et al. Regulation of HuR structure and function by dihydrotanshinone-I. Nucleic Acids Res. 2017, 45, 9514–9527. [Google Scholar] [CrossRef] [PubMed]
- Pfeffer, S.R.; Yang, C.H.; Pfeffer, L. The Role of miR-21 in Cancer. Drug Dev. Res. 2015, 76, 270–277. [Google Scholar] [CrossRef]
- Arghiani, N.; Matin, M.M. miR-21: A Key Small Molecule with Great Effects in Combination Cancer Therapy. Nucleic Acid Ther. 2021, 31, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Poria, D.K.; Guha, A.; Nandi, I.; Ray, P.S. RNA-binding protein HuR sequesters microRNA-21 to prevent translation repression of proinflammatory tumor suppressor gene programmed cell death 4. Oncogene 2016, 35, 1703–1715. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Poria, D.K.; Ray, P.S. RNA-binding proteins La and HuR cooperatively modulate translation repression of PDCD4 mRNA. J. Biol. Chem. 2021, 296, 100154. [Google Scholar] [CrossRef] [PubMed]
- de Sousa, M.C.; Calo, N.; Sobolewski, C.; Gjorgjieva, M.; Clément, S.; Maeder, C.; Dolicka, D.; Fournier, M.; Vinet, L.; Montet, X.; et al. Mir-21 Suppression Promotes Mouse Hepatocarcinogenesis. Cancers 2021, 13, 4983. [Google Scholar] [CrossRef]
- Liwak-Muir, U.; Dobson, C.C.; Naing, T.; Wylie, Q.; Chehade, L.; Baird, S.D.; Chakraborty, P.K.; Holcik, M. ERK8 is a novel HuR kinase that regulates tumour suppressor PDCD4 through a miR-21 dependent mechanism. Oncotarget 2016, 7, 1439–1450. [Google Scholar] [CrossRef][Green Version]
- Tan, S.; Ding, K.; Chong, Q.-Y.; Zhao, J.; Liu, Y.; Shao, Y.; Zhang, Y.; Yu, Q.; Xiong, Z.; Zhang, W.; et al. Post-transcriptional regulation of ERBB2 by miR26a/b and HuR confers resistance to tamoxifen in estrogen receptor-positive breast cancer cells. J. Biol. Chem. 2017, 292, 13551–13564. [Google Scholar] [CrossRef]
- Wang, Y.; Lin, C.; Liu, Y. Molecular mechanism of miR-34b-5p and RNA binding protein HuR binding to lncRNA OIP5-AS1 in colon cancer cells. Cancer Gene Ther. 2022, 29, 612–624. [Google Scholar] [CrossRef]
- Kim, J.; Abdelmohsen, K.; Yang, X.; De, S.; Grammatikakis, I.; Noh, J.H.; Gorospe, M. LncRNA OIP5-AS1/cyrano sponges RNA-binding protein HuR. Nucleic Acids Res. 2016, 44, 2378–2392. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Connick, M.C.; Vanderhoof, J.; Ishak, M.-A.; Hartley, R.S. MicroRNA-16 Modulates HuR Regulation of Cyclin E1 in Breast Cancer Cells. Int. J. Mol. Sci. 2015, 16, 7112–7132. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, L.S.; Gonçalves, N.; Fonseca, N.A.; Moreira, J.N. Cancer Stem Cells and Nucleolin as Drivers of Carcinogenesis. Pharmaceuticals 2021, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Bose, S.; Tholanikunnel, T.E.; Reuben, A.; Tholanikunnel, B.G.; Spicer, E.K. Regulation of nucleolin expression by miR-194, miR-206, and HuR. Mol. Cell. Biochem. 2016, 417, 141–153. [Google Scholar] [CrossRef] [PubMed]
- Xie, W.; Wang, Y.; Zhang, Y.; Xiang, Y.; Wu, N.; Wu, L.; Li, C.; Cai, T.; Ma, X.; Yu, Z.; et al. Single-nucleotide polymorphism rs4142441 and MYC co-modulated long non-coding RNA OSER1-AS1 suppresses non-small cell lung cancer by sequestering ELAVL1. Cancer Sci. 2021, 112, 2272–2286. [Google Scholar] [CrossRef]
- Pei, Y.-F.; Yin, X.-M.; Liu, X.-Q. TOP2A induces malignant character of pancreatic cancer through activating β-catenin signaling pathway. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 197–207. [Google Scholar] [CrossRef]
- Bouchalova, K.; Cizkova, M.; Cwiertka, K.; Trojanec, R.; Hajduch, M. Triple negative breast cancer-current status and prospective targeted treatment based on HER1 (EGFR), TOP2A and C-MYC gene assessment. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech. Repub. 2009, 153, 13–17. [Google Scholar] [CrossRef]
- Zhang, R.; Xu, J.; Zhao, J.; Bai, J.H. Proliferation and invasion of colon cancer cells are suppressed by knockdown of TOP2A. J. Cell. Biochem. 2018, 119, 7256–7263. [Google Scholar] [CrossRef]
- Srikantan, S.; Abdelmohsen, K.; Lee, E.K.; Tominaga, K.; Subaran, S.S.; Kuwano, Y.; Kulshrestha, R.; Panchakshari, R.; Kim, H.H.; Yang, X.; et al. Translational Control of TOP2A Influences Doxorubicin Efficacy. Mol. Cell. Biol. 2011, 31, 3790–3801. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Pan, T.; Li, L.; Li, J.; Yang, H. STIM1 silencing inhibits the migration and invasion of A549 cells. Mol. Med. Rep. 2017, 16, 3283–3289. [Google Scholar] [CrossRef]
- Zhuang, R.; Rao, J.N.; Zou, T.; Liu, L.; Xiao, L.; Cao, S.; Hansraj, N.Z.; Gorospe, M.; Wang, J.-Y. miR-195 competes with HuR to modulate stim1 mRNA stability and regulate cell migration. Nucleic Acids Res. 2013, 41, 7905–7919. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Cao, N.; Wang, G.; Wang, F.; Yang, B.; Wang, J.; Lv, Y.; Chen, Y.; Li, F. HuR Promotes the Progression of Gastric Cancer through Mediating CDC5L Expression. Dis. Markers 2022, 2022, 5141927. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, H.; Zhi, W. SEMA4D under the posttranscriptional regulation of HuR and miR-4319 boosts cancer progression in esophageal squamous cell carcinoma. Cancer Biol. Ther. 2020, 21, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Wang, M.; Cao, L.; Cong, L.; Gao, Y.; Lu, J.; Feng, J.; Shen, B.; Liu, D. miR-4319 Suppresses the Growth of Esophageal Squamous Cell Carcinoma Via Targeting NLRC5. Curr. Mol. Pharmacol. 2020, 13, 144–149. [Google Scholar] [CrossRef]
- Xu, X.; Song, C.; Chen, Z.; Yu, C.; Wang, Y.; Tang, Y.; Luo, J. Downregulation of HuR Inhibits the Progression of Esophageal Cancer through Interleukin-18. Cancer Res. Treat. 2018, 50, 71–87. [Google Scholar] [CrossRef]
- Hao, Z.; Zhang, H.; Cowell, J. Ubiquitin-conjugating enzyme UBE2C: Molecular biology, role in tumorigenesis, and potential as a biomarker. Tumor Biol. 2012, 33, 723–730. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, F.; Liu, M.; Zhao, Q. UBE2C mRNA expression controlled by miR-300 and HuR determines its oncogenic role in gastric cancer. Biochem. Biophys. Res. Commun. 2021, 534, 597–603. [Google Scholar] [CrossRef]
- Tominaga, K.; Srikantan, S.; Lee, E.K.; Subaran, S.S.; Martindale, J.L.; Abdelmohsen, K.; Gorospe, M. Competitive Regulation of Nucleolin Expression by HuR and miR-494. Mol. Cell. Biol. 2011, 31, 4219–4231. [Google Scholar] [CrossRef]
- Morgan, R.; Pandha, H.S. PBX3 in Cancer. Cancers 2020, 12, 431. [Google Scholar] [CrossRef]
- Zhou, H.; Rao, Y.; Sun, Q.; Liu, Y.; Zhou, X.; Chen, Y.; Chen, J. MiR-4458/human antigen R (HuR) modulates PBX3 mRNA stability in melanoma tumorigenesis. Arch. Dermatol. Res. 2020, 312, 665–673. [Google Scholar] [CrossRef]
- Li, C.; Jiang, J.-Y.; Wang, J.-M.; Sun, J.; An, M.-X.; Li, S.; Yan, J.; Wang, H.-Q. BAG3 regulates stability of IL-8 mRNA via interplay between HuR and miR-4312 in PDACs. Cell Death Dis. 2018, 9, 863. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Ni, H.; Li, X.; Liu, H.; Xi, T.; Zheng, L. LncRNA FENDRR attenuates adriamycin resistance via suppressing MDR1 expression through sponging HuR and miR-184 in chronic myelogenous leukaemia cells. FEBS Lett. 2019, 593, 1993–2007. [Google Scholar] [CrossRef] [PubMed]
- Dico, A.L.; Costa, V.; Martelli, C.; Diceglie, C.; Rajata, F.; Rizzo, A.; Mancone, C.; Tripodi, M.; Ottobrini, L.; Alessandro, R.; et al. MiR675-5p Acts on HIF-1α to Sustain Hypoxic Responses: A New Therapeutic Strategy for Glioma. Theranostics 2016, 6, 1105–1118. [Google Scholar] [CrossRef] [PubMed]
- Alfano, L.; Costa, C.; Caporaso, A.; Antonini, D.; Giordano, A.; Pentimalli, F. HUR protects NONO from degradation by mir320, which is induced by p53 upon UV irradiation. Oncotarget 2016, 7, 78127–78139. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chai, Y.; Liu, J.; Zhang, Z.; Liu, L. HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer. Cancer Med. 2016, 5, 1588–1598. [Google Scholar] [CrossRef]
- Ahuja, D.; Goyal, A.; Ray, P.S. Interplay between RNA-binding protein HuR and microRNA-125b regulates p53 mRNA translation in response to genotoxic stress. RNA Biol. 2016, 13, 1152–1165. [Google Scholar] [CrossRef]
- Chang, S.-H.; Lu, Y.-C.; Li, X.; Hsieh, W.-Y.; Xiong, Y.; Ghosh, M.; Evans, T.; Elemento, O.; Hla, T. Antagonistic Function of the RNA-binding Protein HuR and miR-200b in Post-transcriptional Regulation of Vascular Endothelial Growth Factor-A Expression and Angiogenesis. J. Biol. Chem. 2013, 288, 4908–4921. [Google Scholar] [CrossRef]
- Huang, J.-F.; Jiang, H.; Cai, H.; Liu, Y.; Zhu, Y.-Q.; Lin, S.-S.; Hu, T.-T.; Wang, T.-T.; Yang, W.-J.; Xiao, B.; et al. Genome-wide screening identifies oncofetal lncRNA Ptn-dt promoting the proliferation of hepatocellular carcinoma cells by regulating the Ptn receptor. Oncogene 2019, 38, 3428–3445. [Google Scholar] [CrossRef]
- Epis, M.R.; Barker, A.; Giles, K.M.; Beveridge, D.J.; Leedman, P.J. The RNA-binding Protein HuR Opposes the Repression of ERBB-2 Gene Expression by MicroRNA miR-331-3p in Prostate Cancer Cells. J. Biol. Chem. 2011, 286, 41442–41454. [Google Scholar] [CrossRef]
- Lu, Y.-C.; Chang, S.-H.; Hafner, M.; Li, X.; Tuschl, T.; Elemento, O.; Hla, T. ELAVL1 Modulates Transcriptome-wide miRNA Binding in Murine Macrophages. Cell Rep. 2014, 9, 2330–2343. [Google Scholar] [CrossRef]
- Li, Y.; Estep, J.A.; Karginov, F.V. Transcriptome-wide Identification and Validation of Interactions between the miRNA Machinery and HuR on mRNA Targets. J. Mol. Biol. 2018, 430, 285–296. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.; Ge, J.; Xiu, L.; Zhao, Z.; Duan, X.; Tian, L.; Xie, J.; Yang, L.; Li, L. HuR mediates motility of human bone marrow-derived mesenchymal stem cells triggered by sphingosine 1-phosphate in liver fibrosis. J. Mol. Med. 2017, 95, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, L.; Ling, C.; Heng, W. HuR facilitates cancer stemness of lung cancer cells via regulating miR-873/CDK3 and miR-125a-3p/CDK3 axis. Biotechnol. Lett. 2018, 40, 623–631. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, F.; Fang, E.; Xiao, W.; Mei, H.; Li, H.; Li, D.; Song, H.; Wang, J.; Hong, M.; et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019, 26, 1346–1364. [Google Scholar] [CrossRef] [PubMed]
- Pellecchia, S.; Quintavalle, C.; Pallante, P. The control of tumor progression by circular RNAs: Novel prognostic and therapeutic insights resulting from the analysis of the circAGO2/human antigen R complex. Transl. Cancer Res. 2019, 8, S211–S215. [Google Scholar] [CrossRef]
- Zhu, H.; Gan, X.; Jiang, X.; Diao, S.; Wu, H.; Hu, J. ALKBH5 inhibited autophagy of epithelial ovarian cancer through miR-7 and BCL-2. J. Exp. Clin. Cancer Res. 2019, 38, 163. [Google Scholar] [CrossRef]
- Sheng, J.; Sun, H.; Yu, F.-B.; Li, B.; Zhang, Y.; Zhu, Y.-T. The Role of Cyclooxygenase-2 in Colorectal Cancer. Int. J. Med Sci. 2020, 17, 1095–1101. [Google Scholar] [CrossRef]
- Young, L.E.; Moore, A.E.; Sokol, L.; Meisner-Kober, N.; Dixon, D.A. The mRNA Stability Factor HuR Inhibits MicroRNA-16 Targeting of COX-2. Mol. Cancer Res. 2012, 10, 167–180. [Google Scholar] [CrossRef]
- Jin, D.; Guo, J.; Wu, Y.; Yang, L.; Wang, X.; Du, J.; Dai, J.; Chen, W.; Gong, K.; Miao, S.; et al. m6A demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2–mediated YAP activity in NSCLC. Mol. Cancer 2020, 19, 40. [Google Scholar] [CrossRef]
- Panneerdoss, S.; Eedunuri, V.K.; Yadav, P.; Timilsina, S.; Rajamanickam, S.; Viswanadhapalli, S.; Abdelfattah, N.; Onyeagucha, B.C.; Cui, X.; Lai, Z.; et al. Cross-talk among writers, readers, and erasers of m 6 A regulates cancer growth and progression. Sci. Adv. 2018, 4, eaar8263. [Google Scholar] [CrossRef]
- Cao, C.; Sun, J.; Zhang, D.; Guo, X.; Xie, L.; Li, X.; Wu, D.; Liu, L. The Long Intergenic Noncoding RNA UFC1, a Target of MicroRNA 34a, Interacts With the mRNA Stabilizing Protein HuR to Increase Levels of β-Catenin in HCC Cells. Gastroenterology 2015, 148, 415–426. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Yang, B.; Wang, L.; Zhu, Y.; Zhu, X.; Xia, Z.; Zhao, Z.; Xu, L. LncRNA BBOX1-AS1 upregulates HOXC6 expression through miR-361-3p and HuR to drive cervical cancer progression. Cell Prolif. 2020, 53, e12823. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Mao, C.; Ouyang, L.; Liu, Y.; Lai, W.; Liu, N.; Shi, Y.; Chen, L.; Xiao, D.; Yu, F.; et al. Correction to: Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA. Cell Death Differ. 2020, 27, 1447. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.-P.; Jin, Y.-P.; Wu, X.-S.; Yang, Y.; Li, Y.-S.; Li, H.-F.; Xiang, S.-S.; Song, X.-L.; Jiang, L.; Zhang, Y.-J.; et al. Correction to: LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502-3p/SET/AKT axis. Mol. Cancer 2021, 20, 110. [Google Scholar] [CrossRef]
- Zang, Y.; Li, J.; Wan, B.; Tai, Y. circRNA circ-CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR-646. J. Cell. Mol. Med. 2020, 24, 2423–2433. [Google Scholar] [CrossRef]
- Shi, J.; Lv, X.; Zeng, L.; Li, W.; Zhong, Y.; Yuan, J.; Deng, S.; Liu, B.; Yuan, B.; Chen, Y.; et al. CircPVT1 promotes proliferation of lung squamous cell carcinoma by binding to miR-30d/e. J. Exp. Clin. Cancer Res. 2021, 40, 193. [Google Scholar] [CrossRef]
- Jiang, H.; Tian, Y.; Zhao, X.; Zhang, L.; Wu, Z. A circular RNA derived from FAT atypical cadherin 3 promotes lung cancer progression via forming a regulatory loop with oncogenic ELAV like RNA binding protein 1. J. Biochem. 2022, 171, 519–528. [Google Scholar] [CrossRef]
- Zhang, L.; Lou, J.; Lu, M.; Gao, C.; Zhao, S.; Li, B.; Liang, S.; Li, Y.; Li, D.; Liu, M. Suppression of miR-199a maturation by HuR is crucial for hypoxia-induced glycolytic switch in hepatocellular carcinoma. EMBO J. 2015, 34, 2671–2685. [Google Scholar] [CrossRef]
- Lin, C.; Zhang, S.; Wang, Y.; Wang, Y.; Nice, E.; Guo, C.; Zhang, E.; Yu, L.; Li, M.; Liu, C.; et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin. Cancer Res. 2018, 24, 486–498. [Google Scholar] [CrossRef]
- Wang, L.; Bin Li, B.; Yi, X.; Xiao, X.; Zheng, Q.; Ma, L. Circ_0036412 affects the proliferation and cell cycle of hepatocellular carcinoma via hedgehog signaling pathway. J. Transl. Med. 2022, 20, 154. [Google Scholar] [CrossRef]
- Xu, C.Z.; Jiang, C.; Wu, Q.; Liu, L.; Yan, X.; Shi, R. A Feed-Forward Regulatory Loop between HuR and the Long Noncoding RNA HOTAIR Promotes Head and Neck Squamous Cell Carcinoma Progression and Metastasis. Cell Physiol Biochem 2016, 40, 1039–1051. [Google Scholar] [CrossRef] [PubMed]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Wu, D.; Fan, S.; Zhang, Z.; Chen, G.; Lu, J. Upregulation of miR-675-5p induced by lncRNA H19 was associated with tumor progression and development by targeting tumor suppressor p53 in non–small cell lung cancer. J. Cell. Biochem. 2019, 120, 18724–18735. [Google Scholar] [CrossRef] [PubMed]
- Zhu, M.; Chen, Q.; Liu, X.; Sun, Q.; Zhao, X.; Deng, R.; Wang, Y.; Huang, J.; Xu, M.; Yan, J.; et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014, 281, 3766–3775. [Google Scholar] [CrossRef] [PubMed]
- Gunzburg, M.J.; Sivakumaran, A.; Pendini, N.R.; Yoon, J.-H.; Gorospe, M.; Wilce, M.C.; A Wilce, J. Cooperative interplay of let-7 mimic and HuR with MYC RNA. Cell Cycle 2015, 14, 2729–2733. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.H.; Kuwano, Y.; Srikantan, S.; Lee, E.K.; Martindale, J.L.; Gorospe, M. HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 2009, 23, 1743–1748. [Google Scholar] [CrossRef]
- Dhanasekaran, R.; Deutzmann, A.; Mahauad-Fernandez, W.D.; Hansen, A.S.; Gouw, A.M.; Felsher, D.W. The MYC oncogene —the grand orchestrator of cancer growth and immune evasion. Nat. Rev. Clin. Oncol. 2022, 19, 23–36. [Google Scholar] [CrossRef]
- Wang, Y.; Mo, Y.; Wang, L.; Su, P.; Xie, Y. Let-7b contributes to hepatocellular cancer progression through Wnt/β-catenin signaling. Saudi J. Biol. Sci. 2018, 25, 953–958. [Google Scholar] [CrossRef]
- Han, X.; Zhang, J.-J.; Han, Z.-Q.; Zhang, H.-B.; Wang, Z.-A. Correction: Let-7b attenuates cisplatin resistance and tumor growth in gastric cancer by targeting AURKB. Cancer Gene Ther. 2019, 26, 256. [Google Scholar] [CrossRef]
- Chen, F.; Chen, C.; Yang, S.; Gong, W.; Wang, Y.; Cianflone, K.; Tang, J.; Wang, D.W. Let-7b Inhibits Human Cancer Phenotype by Targeting Cytochrome P450 Epoxygenase 2J2. PLoS ONE 2012, 7, e39197. [Google Scholar] [CrossRef]
- Han, X.; Chen, Y.; Yao, N.; Liu, H.; Wang, Z. MicroRNA let-7b suppresses human gastric cancer malignancy by targeting ING1. Cancer Gene Ther. 2015, 22, 122–129. [Google Scholar] [CrossRef] [PubMed]
- Yoon, J.-H.; Abdelmohsen, K.; Srikantan, S.; Yang, X.; Martindale, J.L.; De, S.; Huarte, M.; Zhan, M.; Becker, K.G.; Gorospe, M. LincRNA-p21 Suppresses Target mRNA Translation. Mol. Cell 2012, 47, 648–655. [Google Scholar] [CrossRef] [PubMed]
- Leucci, E.; Zriwil, A.; Gregersen, L.H.; Jensen, K.T.; Obad, S.; Bellan, C.; Leoncini, L.; Kauppinen, S.; Lund, A.H. Inhibition of miR-9 de-represses HuR and DICER1 and impairs Hodgkin lymphoma tumour outgrowth in vivo. Oncogene 2012, 31, 5081–5089. [Google Scholar] [CrossRef] [PubMed]
- Thorne, J.L.; Battaglia, S.; Baxter, D.E.; Hayes, J.L.; Hutchinson, S.A.; Jana, S.; Millican-Slater, R.A.; Smith, L.; Teske, M.C.; Wastall, L.M.; et al. MiR-19b non-canonical binding is directed by HuR and confers chemosensitivity through regulation of P-glycoprotein in breast cancer. Biochim. Biophys. Acta Gene Regul. Mech. 2018, 1861, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Glorian, V.; Maillot, G.; Polès, S.; Iacovoni, J.S.; Favre, G.; Vagner, S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell Death Differ. 2011, 18, 1692–1701. [Google Scholar] [CrossRef] [PubMed]
- da Silva, G.H.G.; dos Santos, M.G.P.; Nagasse, H.Y.; Coltri, P.P. Human Antigen R (HuR) Facilitates miR-19 Synthesis and Affects Cellular Kinetics in Papillary Thyroid Cancer. Cell. Physiol. Biochem. 2022, 56, 105–119. [Google Scholar] [CrossRef]
- Zhou, B.; Xu, J.; Chen, Y.; Gao, S.; Feng, X.; Lu, X. miR-200b/c-RAP1B axis represses tumorigenesis and malignant progression of papillary thyroid carcinoma through inhibiting the NF-κB/Twist1 pathway. Exp. Cell Res. 2020, 387, 111785. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Z.; Zhu, X.; Chen, L.; Ma, Y.; Wang, J.; Yang, X.; Liu, Z. Exosomal miR-1246 in serum as a potential biomarker for early diagnosis of gastric cancer. Int. J. Clin. Oncol. 2020, 25, 89–99. [Google Scholar] [CrossRef]
- Peng, W.; Li, J.; Chen, R.; Gu, Q.; Yang, P.; Qian, W.; Ji, D.; Wang, Q.; Zhang, Z.; Tang, J.; et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 393. [Google Scholar] [CrossRef]
- Guo, S.; Chen, J.; Chen, F.; Zeng, Q.; Liu, W.-L.; Zhang, G. Exosomes derived from Fusobacterium nucleatum-infected colorectal cancer cells facilitate tumour metastasis by selectively carrying miR-1246/92b-3p/27a-3p and CXCL16. Gut 2020, 70, 1507–1519. [Google Scholar] [CrossRef]
- Si, G.; Li, S.; Zheng, Q.; Zhu, S.; Zhou, C. miR-1246 shuttling from fibroblasts promotes colorectal cancer cell migration. Neoplasma 2021, 68, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Del Vecchio, G.; De Vito, F.; Saunders, S.J.; Risi, A.; Mannironi, C.; Bozzoni, I.; Presutti, C. RNA-binding protein HuR and the members of the miR-200 family play an unconventional role in the regulation of c-Jun mRNA. RNA 2016, 22, 1510–1521. [Google Scholar] [CrossRef] [PubMed]
- Prislei, S.; Martinelli, E.; Mariani, M.; Raspaglio, G.; Sieber, S.; Ferrandina, G.; Shahabi, S.; Scambia, G.; Ferlini, C. MiR-200c and HuR in ovarian cancer. BMC Cancer 2013, 13, 72. [Google Scholar] [CrossRef] [PubMed]
- Tian, Z.; Luo, Y.; Zhu, J.; Hua, X.; Xu, J.; Huang, C.; Jin, H.; Huang, H.; Huang, C. Transcriptionally elevation of miR-494 by new ChlA-F compound via a HuR/JunB axis inhibits human bladder cancer cell invasion. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 822–833. [Google Scholar] [CrossRef]
- Rossi, F.; Beltran, M.; Damizia, M.; Grelloni, C.; Colantoni, A.; Setti, A.; Di Timoteo, G.; Dattilo, D.; Centrón-Broco, A.; Nicoletti, C.; et al. Circular RNA ZNF609/CKAP5 mRNA interaction regulates microtubule dynamics and tumorigenicity. Mol. Cell 2022, 82, 75–89.e9. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Qu, H.; Han, M.; Ding, Y.; Xie, M.; Hu, J.; Chen, Y.; Dong, H. MSC-induced lncRNA AGAP2-AS1 promotes stemness and trastuzumab resistance through regulating CPT1 expression and fatty acid oxidation in breast cancer. Oncogene 2021, 40, 833–847. [Google Scholar] [CrossRef]
- Shi, J.; Guo, C.; Ma, J. CCAT2 enhances autophagy-related invasion and metastasis via regulating miR-4496 and ELAVL1 in hepatocellular carcinoma. J. Cell. Mol. Med. 2021, 25, 8985–8996. [Google Scholar] [CrossRef]
- Li, X.; Wang, C.; Chen, G.; Zou, W.; Deng, Y.; Zhou, F. EIF4A3-induced circCCNB1 (hsa_circ_0001495) promotes glioma progression by elevating CCND1 through interacting miR-516b-5p and HuR. Metab. Brain Dis. 2022, 37, 819–833. [Google Scholar] [CrossRef]
- D’Angelo, D.; Arra, C.; Fusco, A. RPSAP52 lncRNA Inhibits p21Waf1/CIP Expression by Interacting With the RNA Binding Protein HuR. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2020, 28, 191–201. [Google Scholar] [CrossRef]
- Ehses, J.; Fernández-Moya, S.M.; Schröger, L.; Kiebler, M.A. Synergistic regulation of Rgs4 mRNA by HuR and miR-26/RISC in neurons. RNA Biol. 2021, 18, 988–998. [Google Scholar] [CrossRef]
- Taylor, G.A.; Lai, W.S.; Oakey, R.J.; Seldin, M.F.; Shows, T.B.; Eddy, R.L.; Blackshear, P.J. The human TTP protein: Sequence, alignment with related proteins, and chromosomal localization of the mouse and human genes. Nucleic Acids Res. 1991, 19, 3454. [Google Scholar] [CrossRef] [PubMed]
- Varnum, B.C.; Ma, Q.F.; Chi, T.H.; Fletcher, B.; Herschman, H.R. The TIS11 primary response gene is a member of a gene family that encodes proteins with a highly conserved sequence containing an unusual Cys-His repeat. Mol. Cell. Biol. 1991, 11, 1754–1758. [Google Scholar] [CrossRef] [PubMed]
- Saini, Y.; Chen, J.; Patial, S. The Tristetraprolin Family of RNA-Binding Proteins in Cancer: Progress and Future Prospects. Cancers 2020, 12, 1539. [Google Scholar] [CrossRef]
- Ogawa, K.; Chen, F.; Kim, Y.-J.; Chen, Y. Transcriptional Regulation of Tristetraprolin by Transforming Growth Factor-β in Human T Cells. J. Biol. Chem. 2003, 278, 30373–30381. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.F.; Sanduja, S.; Deane, N.G.; Blackshear, P.; Dixon, D.A. Transforming Growth Factor β Regulates P-Body Formation through Induction of the mRNA Decay Factor Tristetraprolin. Mol. Cell. Biol. 2014, 34, 180–195. [Google Scholar] [CrossRef]
- Chen, Y.-L.; Jiang, Y.-W.; Su, Y.-L.; Lee, S.-C.; Chang, M.-S.; Chang, C.-J. Transcriptional regulation of tristetraprolin by NF-κB signaling in LPS-stimulated macrophages. Mol. Biol. Rep. 2013, 40, 2867–2877. [Google Scholar] [CrossRef]
- Lai, W.S.; Stumpo, D.J.; Blackshear, P.J. Rapid insulin-stimulated accumulation of an mRNA encoding a proline-rich protein. J. Biol. Chem. 1990, 265, 16556–16563. [Google Scholar] [CrossRef]
- Cao, H.; Urban, J.F.; Anderson, R.A. Insulin Increases Tristetraprolin and Decreases VEGF Gene Expression in Mouse 3T3-L1 Adipocytes. Obesity 2008, 16, 1208–1218. [Google Scholar] [CrossRef]
- Carballo, E.; Blackshear, P. Roles of tumor necrosis factor-α receptor subtypes in the pathogenesis of the tristetraprolin-deficiency syndrome. Blood 2001, 98, 2389–2395. [Google Scholar] [CrossRef]
- Sanduja, S.; Blanco, F.F.; Young, L.E.; Kaza, V.; Dixon, D.A. The role of tristetraprolin in cancer and inflammation. Front. Biosci. 2012, 17, 174–188. [Google Scholar] [CrossRef]
- A Taylor, G.; Carballo, E.; Lee, D.M.; Lai, W.S.; Thompson, M.J.; Patel, D.D.; I Schenkman, D.; Gilkeson, G.S.; E Broxmeyer, H.; Haynes, B.F.; et al. A Pathogenetic Role for TNFα in the Syndrome of Cachexia, Arthritis, and Autoimmunity Resulting from Tristetraprolin (TTP) Deficiency. Immunity 1996, 4, 445–454. [Google Scholar] [CrossRef]
- Dolicka, D.; Sobolewski, C.; Gjorgjieva, M.; de Sousa, M.C.; Berthou, F.; De Vito, C.; Colin, D.J.; Bejuy, O.; Fournier, M.; Maeder, C.; et al. Tristetraprolin Promotes Hepatic Inflammation and Tumor Initiation but Restrains Cancer Progression to Malignancy. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 597–621. [Google Scholar] [CrossRef] [PubMed]
- Jing, Q.; Huang, S.; Guth, S.; Zarubin, T.; Motoyama, A.; Chen, J.; Di Padova, F.; Lin, S.-C.; Gram, H.; Han, J. Involvement of MicroRNA in AU-Rich Element-Mediated mRNA Instability. Cell 2005, 120, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Kim, C.W.; Vo, M.-T.; Kim, H.K.; Lee, H.H.; Yoon, N.A.; Lee, B.J.; Min, Y.J.; Joo, W.D.; Cha, H.J.; Park, J.W.; et al. Ectopic over-expression of tristetraprolin in human cancer cells promotes biogenesis of let-7 by down-regulation of Lin28. Nucleic Acids Res. 2012, 40, 3856–3869. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Kim, H.J.; Yoon, N.A.; Lee, W.H.; Min, Y.J.; Ko, B.K.; Lee, B.J.; Lee, A.; Cha, H.J.; Cho, W.J.; et al. Tumor suppressor p53 plays a key role in induction of both tristetraprolin and let-7 in human cancer cells. Nucleic Acids Res. 2013, 41, 5614–5625. [Google Scholar] [CrossRef] [PubMed]
- Sobolewski, C.; Sanduja, S.; Blanco, F.F.; Hu, L.; Dixon, D.A. Histone Deacetylase Inhibitors Activate Tristetraprolin Expression through Induction of Early Growth Response Protein 1 (EGR1) in Colorectal Cancer Cells. Biomolecules 2015, 5, 2035–2055. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.S.N.; Ip, C.K.M.; Mak, A.S.C.; Wong, A.S.T. A novel p70 S6 kinase-microRNA biogenesis axis mediates multicellular spheroid formation in ovarian cancer progression. Oncotarget 2016, 7, 38064–38077. [Google Scholar] [CrossRef]
- Bhattacharyya, S.; Kumar, P.; Tsuchiya, M.; Bhattacharyya, A.; Biswas, R. Regulation of miR-155 biogenesis in cystic fibrosis lung epithelial cells: Antagonistic role of two mRNA-destabilizing proteins, KSRP and TTP. Biochem. Biophys. Res. Commun. 2013, 433, 484–488. [Google Scholar] [CrossRef]
- Liu, H.; Han, L.; Liu, Z.; Gao, N. Long noncoding RNA MNX1-AS1 contributes to lung cancer progression through the miR-527/BRF2 pathway. J. Cell. Physiol. 2019, 234, 13843–13850. [Google Scholar] [CrossRef]
- Goutas, D.; Pergaris, A.; Giaginis, C.; Theocharis, S. HuR as Therapeutic Target in Cancer: What the Future Holds. Curr. Med. Chem. 2022, 29, 56–65. [Google Scholar] [CrossRef]
- Li, Q.; Tong, D.; Guo, C.; Wu, F.; Li, F.; Wang, X.; Jiang, Q.; Wei, Y.; Liu, L.; Ni, L.; et al. MicroRNA-145 suppresses gastric cancer progression by targeting Hu-antigen R. Am. J. Physiol. Cell Physiol. 2020, 318, C605–C614. [Google Scholar] [CrossRef] [PubMed]
- Zhou, N.; Zhou, Y.; Tang, Y.; Yu, W. [MiR-519 inhibits gastric cancer cell activity through regulation of HuR expression]. Zhong Nan Da Xue Xue Bao Yi Xue Ban 2016, 41, 19–23. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Yu, T.; Jing, X.; Ma, L.; Fan, Y.; Yang, F.; Ma, P.; Jiang, H.; Wu, X.; Shu, Y.; et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol. Cancer 2020, 19, 112. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, K.; Li, D.; Chen, M.; Li, A.; Wang, J. miR-140-3p is involved in the occurrence and metastasis of gastric cancer by regulating the stability of FAM83B. Cancer Cell Int. 2021, 21, 537. [Google Scholar] [CrossRef] [PubMed]
- Andrieu, N.A.; Motiño, O.; Mayoral, R.; Izquierdo, C.L.; Fernández-Alvarez, A.; Boscá, L.; Casado, M.; Martín-Sanz, P. Cyclooxygenase-2 Is a Target of MicroRNA-16 in Human Hepatoma Cells. PLoS ONE 2012, 7, e50935. [Google Scholar] [CrossRef]
- Xue, F.; Li, Q.R.; Xu, Y.H.; Bin Zhou, H. MicroRNA-139-3p Inhibits The Growth And Metastasis Of Ovarian Cancer By Inhibiting ELAVL1. Onco. Targets Ther. 2019, 12, 8935–8945. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Kim, M.M.; Srikantan, S.; Mercken, E.M.; Brennan, S.E.; Wilson, G.; de Cabo, R.; Gorospe, M. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 2010, 9, 1354–1359. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Srikantan, S.; Kuwano, Y.; Gorospe, M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. USA 2008, 105, 20297–20302. [Google Scholar] [CrossRef]
- Wu, D.; Wang, B.; Shang, J.; Song, J.; Zhang, H. miR-31 Reduces Cell Growth of Papillary Thyroid Carcinoma by RNA-Binding Protein HuR. Clin. Lab. 2015, 61, 1625–1634. [Google Scholar] [CrossRef]
- Schultz, C.W.; Preet, R.; Dhir, T.; Dixon, D.A.; Brody, J.R. Understanding and targeting the disease-related RNA binding protein human antigen R (HuR). Wiley Interdiscip. Rev. RNA 2020, 11, e1581. [Google Scholar] [CrossRef]
- Garinet, S.; Wang, P.; Mansuet-Lupo, A.; Fournel, L.; Wislez, M.; Blons, H. Updated Prognostic Factors in Localized NSCLC. Cancers 2022, 14, 1400. [Google Scholar] [CrossRef] [PubMed]
- Ni, Z.-Z.; He, J.-K.; Tang, X.; Tao, Z.; Zhang, Y.; Xie, B. Identification of ELAVL1 gene and miRNA-139-3p involved in the aggressiveness of NSCLC. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 9453–9464. [Google Scholar] [CrossRef] [PubMed]
- Iacona, J.R.; Monteleone, N.J.; Lemenze, A.D.; Cornett, A.L.; Lutz, C.S. Transcriptomic studies provide insights into the tumor suppressive role of miR-146a-5p in non-small cell lung cancer (NSCLC) cells. RNA Biol. 2019, 16, 1721–1732. [Google Scholar] [CrossRef]
- Xu, H.; Ma, J.; Zheng, J.; Wu, J.; Qu, C.; Sun, F.; Xu, S. MiR-31 Functions as a Tumor Suppressor in Lung Adenocarcinoma Mainly by Targeting HuR. Clin. Lab. 2016, 62, 711–718. [Google Scholar] [CrossRef]
- Liu, Y.; Chen, X.; Cheng, R.; Yang, F.; Yu, M.; Wang, C.; Cui, S.; Hong, Y.; Liang, H.; Liu, M.; et al. The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer. Mol. Cancer 2018, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Zhang, M.; Sun, W.; Dong, C. Upregulation of miR-324-5p Inhibits Proliferation and Invasion of Colorectal Cancer Cells by Targeting ELAVL1. Oncol. Res. Featur. Preclin. Clin. Cancer Ther. 2019, 27, 515–524. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.; Leung, W.; Ng, S.S. Exploiting a novel miR-519c–HuR–ABCG2 regulatory pathway to overcome chemoresistance in colorectal cancer. Exp. Cell Res. 2015, 338, 222–231. [Google Scholar] [CrossRef] [PubMed]
- Al-Haidari, A.; Algaber, A.; Madhi, R.; Syk, I.; Thorlacius, H. MiR-155-5p controls colon cancer cell migration via post-transcriptional regulation of Human Antigen R (HuR). Cancer Lett. 2018, 421, 145–151. [Google Scholar] [CrossRef]
- Wang, D.-D.; Sun, D.-L.; Yang, S.-P.; Song, J.; Wu, M.-Y.; Niu, W.-W.; Song, M.; Zhang, X.-L. Long noncoding RNA TNFRSF10A-AS1 promotes colorectal cancer through upregulation of HuR. World J. Gastroenterol. 2022, 28, 2184–2200. [Google Scholar] [CrossRef]
- Fan, M.; He, P.; Lin, X.; Yang, C.; Li, C.; Xing, L. MicroRNA -324-5p affects the radiotherapy response of cervical cancer via targeting ELAV-like RNA binding protein 1. Kaohsiung J. Med Sci. 2020, 36, 965–972. [Google Scholar] [CrossRef]
- Marasa, B.S.; Srikantan, S.; Martindale, J.L.; Kim, M.M.; Lee, E.K.; Gorospe, M.; Abdelmohsen, K. MicroRNA profiling in human diploid fibroblasts uncovers miR-519 role in replicative senescence. Aging 2010, 2, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Balkhi, M.Y.; Iwenofu, O.H.; Bakkar, N.; Ladner, K.J.; Chandler, D.S.; Houghton, P.J.; London, C.A.; Kraybill, W.; Perrotti, D.; Croce, C.M.; et al. miR-29 Acts as a Decoy in Sarcomas to Protect the Tumor Suppressor A20 mRNA from Degradation by HuR. Sci. Signal. 2013, 6, ra63. [Google Scholar] [CrossRef] [PubMed]
- Pan, B.; Zhao, M.; Xu, L. Long noncoding RNA gastric cancer-associated transcript 3 plays oncogenic roles in glioma through sponging miR-3127-5p. J. Cell. Physiol. 2019, 234, 8825–8833. [Google Scholar] [CrossRef]
- Yang, W.; Yu, H.; Shen, Y.; Liu, Y.; Yang, Z.; Sun, T. MiR-146b-5p overexpression attenuates stemness and radioresistance of glioma stem cells by targeting HuR/lincRNA-p21/β-catenin pathway. Oncotarget 2016, 7, 41505–41526. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, X.; Lei, Y.; Liu, X.; Liu, Z.; Tong, T.; Wang, W. Loss of Repression of HuR Translation by miR-16 May Be Responsible for the Elevation of HuR in Human Breast Carcinoma. J. Cell. Biochem. 2010, 111, 727–734. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wu, Y.; Hartley, R. MicroRNA-125a represses cell growth by targeting HuR in breast cancer. RNA Biol. 2009, 6, 575–583. [Google Scholar] [CrossRef]
- Ren, L.; Li, Y.; Zhao, Q.; Fan, L.; Tan, B.; Zang, A.; Yang, H. miR-519 regulates the proliferation of breast cancer cells via targeting human antigen R. Oncol. Lett. 2020, 19, 1567–1576. [Google Scholar] [CrossRef]
- Al-Ahmadi, W.; Al-Ghamdi, M.; Al-Souhibani, N.; Khabar, K.S. miR -29a inhibition normalizes HuR over-expression and aberrant AU -rich mRNA stability in invasive cancer. J. Pathol. 2013, 230, 28–38. [Google Scholar] [CrossRef]
- Sun, J.; Xu, Y.; Zhou, H.; Zhang, A.; Qi, H. Levels, occurrence and human exposure to novel brominated flame retardants (NBFRs) and Dechlorane Plus (DP) in dust from different indoor environments in Hangzhou, China. Sci. Total Environ. 2018, 631-632, 1212–1220. [Google Scholar] [CrossRef]
- Liu, H.; Song, X.; Hou, J.; Zhao, Z.; Chang, J. Posttranscriptional Regulation of Human Antigen R by miR-133b Enhances Docetaxel Cytotoxicity Through the Inhibition of ATP-Binding Cassette Subfamily G Member 2 in Prostate Cancer Cells. DNA Cell Biol. 2018, 37, 210–219. [Google Scholar] [CrossRef]
- Kojima, K.; Fujita, Y.; Nozawa, Y.; Deguchi, T.; Ito, M. MiR-34a attenuates paclitaxel-resistance of hormone-refractory prostate cancer PC3 cells through direct and indirect mechanisms. Prostate 2010, 70, 1501–1512. [Google Scholar] [CrossRef]
- Hu, W.; Li, H.; Wang, S. LncRNA SNHG7 promotes the proliferation of nasopharyngeal carcinoma by miR-514a-5p/ELAVL1 axis. BMC Cancer 2020, 20, 376. [Google Scholar] [CrossRef] [PubMed]
- House, R.; Majumder, M.; Janakiraman, H.; Ogretmen, B.; Kato, M.; Erkul, E.; Hill, E.; Atkinson, C.; Barth, J.; Day, T.A.; et al. Smoking-induced control of miR-133a-3p alters the expression of EGFR and HuR in HPV-infected oropharyngeal cancer. PLoS ONE 2018, 13, e0205077. [Google Scholar] [CrossRef]
- Shen, Z.; Zhan, G.; Deng, H.; Ren, Y.; Ye, D.; Xiao, B.; Guo, J. MicroRNA-519a demonstrates significant tumour suppressive activity in laryngeal squamous cells by targeting anti-carcinoma HuR gene. J. Laryngol. Otol. 2013, 127, 1194–1202. [Google Scholar] [CrossRef]
- Shen, Z.; Zhan, G.; Deng, H.; Kang, C.; Guo, J. Growth inhibitory effect of microRNA-519b-3p on larynx squamous Hep-2 cells. Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi = CN. J. Otorhinolaryngol. Head Neck Surg. 2014, 49, 151–156. [Google Scholar]
- Huang, K.; Dong, B.; Wang, Y.; Tian, T.; Zhang, B. MicroRNA-519 enhances HL60 human acute myeloid leukemia cell line proliferation by reducing the expression level of RNA-binding protein human antigen R. Mol. Med. Rep. 2015, 12, 7830–7836. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Alemdehy, M.F.; Haanstra, J.R.; de Looper, H.W.J.; van Strien, P.M.H.; Verhagen-Oldenampsen, J.; Caljouw, Y.; Sanders, M.A.; Hoogenboezem, R.; de Ru, A.H.; Janssen, G.M.C.; et al. ICL-induced miR139-3p and miR199a-3p have opposite roles in hematopoietic cell expansion and leukemic transformation. Blood 2015, 125, 3937–3948. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-J.; Chiou, J.-T.; Lee, Y.-C.; Chang, L.-S. Docetaxel-triggered SIDT2/NOX4/JNK/HuR signaling axis is associated with TNF-α-mediated apoptosis of cancer cells. Biochem. Pharmacol. 2022, 195, 114865. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Song, X.; Lan, J.; Wang, X.; Wang, M. Bone marrow stromal cells derived exosomal miR-10a and miR-16 may be involved in progression of patients with multiple myeloma by regulating EPHA8 or IGF1R/CCND1. Medicine 2021, 100, e23447. [Google Scholar] [CrossRef]
- Mariotti, B.; Calabretto, G.; Rossato, M.; Teramo, A.; Castellucci, M.; Barilà, G.; Leoncin, M.; Vicenzetto, C.; Facco, M.; Semenzato, G.; et al. Identification of a miR-146b-Fas ligand axis in the development of neutropenia in T large granular lymphocyte leukemia. Haematologica 2020, 105, 1351–1360. [Google Scholar] [CrossRef]
- Mihailovich, M.; Bremang, M.; Spadotto, V.; Musiani, D.; Vitale, E.; Varano, G.; Zambelli, F.; Mancuso, F.; Cairns, D.A.; Pavesi, G.; et al. miR-17-92 fine-tunes MYC expression and function to ensure optimal B cell lymphoma growth. Nat. Commun. 2015, 6, 8725. [Google Scholar] [CrossRef]
- Sohn, B.H.; Park, I.Y.; Lee, J.J.; Yang, S.; Jang, Y.J.; Park, K.C.; Kim, D.J.; Lee, D.C.; Sohn, H.A.; Kim, T.W.; et al. Functional Switching of TGF-β1 Signaling in Liver Cancer via Epigenetic Modulation of a Single CpG Site in TTP Promoter. Gastroenterology 2010, 138, 1898–1908.e12. [Google Scholar] [CrossRef] [PubMed]
- Al-Souhibani, N.; Al-Ghamdi, M.; Al-Ahmadi, W.; Khabar, K.S. Posttranscriptional control of the chemokine receptor CXCR4 expression in cancer cells. Carcinogenesis 2014, 35, 1983–1992. [Google Scholar] [CrossRef]
- Sun, X.-J.; Liu, B.-Y.; Yan, S.; Jiang, T.-H.; Cheng, H.-Q.; Jiang, H.-S.; Cao, Y.; Mao, A.-W. MicroRNA-29a Promotes Pancreatic Cancer Growth by Inhibiting Tristetraprolin. Cell. Physiol. Biochem. 2015, 37, 707–718. [Google Scholar] [CrossRef]
- Wang, J.-Y.; Zhang, Q.; Wang, D.-D.; Yan, W.; Sha, H.-H.; Zhao, J.-H.; Yang, S.-J.; Zhang, H.-D.; Hou, J.-C.; Xu, H.-Z.; et al. MiR-29a: A potential therapeutic target and promising biomarker in tumors. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Mao, Y.; Lv, M.; Cao, W.; Liu, X.; Cui, J.; Wang, Y.; Wang, Y.; Nie, G.; Liu, X.; Wang, H. Circular RNA 000554 represses epithelial-mesenchymal transition in breast cancer by regulating microRNA-182/ZFP36 axis. FASEB J. 2020, 34, 11405–11420. [Google Scholar] [CrossRef] [PubMed]
- De Luca, L.; Trino, S.; Laurenzana, I.; Tagliaferri, D.; Falco, G.; Grieco, V.; Bianchino, G.; Nozza, F.; Campia, V.; D’Alessio, F.; et al. Knockdown of miR-128a induces Lin28a expression and reverts myeloid differentiation blockage in acute myeloid leukemia. Cell Death Dis. 2017, 8, e2849. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Pu, J.; Zhang, Y.; Yao, T.; Luo, Z.; Li, W.; Xu, G.; Liu, J.; Wei, W.; Deng, Y. Exosome-transmitted long non-coding RNA SENP3-EIF4A1 suppresses the progression of hepatocellular carcinoma. Aging 2020, 12, 11550–11567. [Google Scholar] [CrossRef]
- Perdigão-Henriques, R.; Petrocca, F.; Altschuler, G.; Thomas, M.P.; LE, T.N.M.; Tan, S.M.; Hide, W.; A Lieberman, J. miR-200 promotes the mesenchymal to epithelial transition by suppressing multiple members of the Zeb2 and Snail1 transcriptional repressor complexes. Oncogene 2016, 35, 158–172. [Google Scholar] [CrossRef]
- Feng, X.; Zhou, S.; Cai, W.; Guo, J. The miR-93-3p/ZFP36L1/ZFX axis regulates keratinocyte proliferation and migration during skin wound healing. Mol. Ther. Nucleic Acids 2021, 23, 450–463. [Google Scholar] [CrossRef]
- Guo, X.; Piao, H.; Zhang, Y.; Sun, P.; Yao, B. Overexpression of microRNA-129-5p in glioblastoma inhibits cell proliferation, migration, and colony-forming ability by targeting ZFP36L1. Bosn. J. Basic Med Sci. 2020, 20, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Kou, Z.; Liu, H.; Wang, Y.-C.; Huang, Q.; Wang, Z.-S.; Gu, Z.-L.N.E.; Lang, T.; Nie, Y.-L.; An, L.; Li A, Z.-G.; et al. Expression Level and Target Gene Prediction of miR-181b in Patients with Chronic Lymphocytic Leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2020, 28, 808–814. [Google Scholar] [CrossRef] [PubMed]
- Yonemori, K.; Seki, N.; Kurahara, H.; Osako, Y.; Idichi, T.; Arai, T.; Koshizuka, K.; Kita, Y.; Maemura, K.; Natsugoe, S. ZFP36L2 promotes cancer cell aggressiveness and is regulated by antitumor microRNA-375 in pancreatic ductal adenocarcinoma. Cancer Sci. 2017, 108, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qu, J.; Zhou, L.; Liao, F.; Wang, J. MicroRNA-373 Inhibits Cell Proliferation and Invasion via Targeting BRF2 in Human Non-small Cell Lung Cancer A549 Cell Line. Cancer Res. Treat. 2018, 50, 936–949. [Google Scholar] [CrossRef]
- Li, Y.; Dong, R.; Lu, M.; Cheng, C.; Feng, Z.; Zhao, R.; Liang, J.; Han, J.; Jiang, J.; Xu-Welliver, M.; et al. Let-7b-3p inhibits tumor growth and metastasis by targeting the BRF2-mediated MAPK/ERK pathway in human lung adenocarcinoma. Transl. Lung Cancer Res. 2021, 10, 1841–1856. [Google Scholar] [CrossRef]
- Qu, Y.-L.; Wang, H.-F.; Sun, Z.-Q.; Tang, Y.; Han, X.-N.; Yu, X.-B.; Liu, K. Up-regulated miR-155-5p promotes cell proliferation, invasion and metastasis in colorectal carcinoma. Int. J. Clin. Exp. Pathol. 2015, 8, 6988–6994. [Google Scholar]
- Cheng, G.; Song, Z.; Liu, Y.; Xiao, H.; Ruan, H.; Cao, Q.; Wang, K.; Xiao, W.; Xiong, Z.; Liu, D.; et al. Long noncoding RNA SNHG12 indicates the prognosis of prostate cancer and accelerates tumorigenesis via sponging miR-133b. J. Cell. Physiol. 2020, 235, 1235–1246. [Google Scholar] [CrossRef]
- Huang, S.; Wa, Q.; Pan, J.; Peng, X.; Ren, D.; Li, Q.; Dai, Y.; Yang, Q.; Huang, Y.; Zhang, X.; et al. Transcriptional downregulation of miR-133b by REST promotes prostate cancer metastasis to bone via activating TGF-β signaling. Cell Death Dis. 2018, 9, 779. [Google Scholar] [CrossRef]
- Duan, K.; Ge, Y.-C.; Zhang, X.-P.; Wu, S.-Y.; Feng, J.-S.; Chen, S.-L.; Zhang, L.; Yuan, Z.-H.; Fu, C.-H. miR-34a inhibits cell proliferation in prostate cancer by downregulation of SIRT1 expression. Oncol. Lett. 2015, 10, 3223–3227. [Google Scholar] [CrossRef]
- Russo, A.; Potenza, N. Antiproliferative Activity of microRNA-125a and its Molecular Targets. MicroRNA 2019, 8, 173–179. [Google Scholar] [CrossRef]
- Fontana, A.; Barbano, R.; Dama, E.; Pasculli, B.; Rendina, M.; Morritti, M.G.; Melocchi, V.; Castelvetere, M.; Valori, V.M.; Ravaioli, S.; et al. Combined analysis of miR-200 family and its significance for breast cancer. Sci. Rep. 2021, 11, 2980. [Google Scholar] [CrossRef] [PubMed]
- Niu, M.; Feng, Y.; Zhang, N.; Shao, T.; Zhang, H.; Wang, R.; Yao, Y.; Yao, R.; Wu, Q.; Cao, J.; et al. High expression of miR-25 predicts favorable chemotherapy outcome in patients with acute myeloid leukemia. Cancer Cell Int. 2019, 19, 122. [Google Scholar] [CrossRef] [PubMed]
- Paczkowska, J.; Giefing, M. MicroRNA signature in classical Hodgkin lymphoma. J. Appl. Genet. 2021, 62, 281–288. [Google Scholar] [CrossRef]
- Li, X.; Zeng, L.; Cao, C.; Lu, C.; Lian, W.; Han, J.; Zhang, X.; Zhang, J.; Tang, T.; Li, M. Long noncoding RNA MALAT1 regulates renal tubular epithelial pyroptosis by modulated miR-23c targeting of ELAVL1 in diabetic nephropathy. Exp. Cell Res. 2017, 350, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Shwetha, S.; Sharma, G.; Raheja, H.; Goel, A.; Aggarwal, R.; Das, S. Interaction of miR-125b-5p with Human antigen R mRNA: Mechanism of controlling HCV replication. Virus Res. 2018, 258, 1–8. [Google Scholar] [CrossRef]
- Wang, J.; Hjelmeland, A.B.; Nabors, L.B.; King, P.H. Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells. Cancer Biol. Ther. 2019, 20, 979–988. [Google Scholar] [CrossRef]
- HafezQorani, S.; Lafzi, A.; De Bruin, R.G.; Van Zonneveld, A.J.; Van Der Veer, E.P.; Son, Y.A.; Kazan, H. Modeling the combined effect of RNA-binding proteins and microRNAs in post-transcriptional regulation. Nucleic Acids Res. 2016, 44, e83. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobolewski, C.; Dubuquoy, L.; Legrand, N. MicroRNAs, Tristetraprolin Family Members and HuR: A Complex Interplay Controlling Cancer-Related Processes. Cancers 2022, 14, 3516. https://doi.org/10.3390/cancers14143516
Sobolewski C, Dubuquoy L, Legrand N. MicroRNAs, Tristetraprolin Family Members and HuR: A Complex Interplay Controlling Cancer-Related Processes. Cancers. 2022; 14(14):3516. https://doi.org/10.3390/cancers14143516
Chicago/Turabian StyleSobolewski, Cyril, Laurent Dubuquoy, and Noémie Legrand. 2022. "MicroRNAs, Tristetraprolin Family Members and HuR: A Complex Interplay Controlling Cancer-Related Processes" Cancers 14, no. 14: 3516. https://doi.org/10.3390/cancers14143516
APA StyleSobolewski, C., Dubuquoy, L., & Legrand, N. (2022). MicroRNAs, Tristetraprolin Family Members and HuR: A Complex Interplay Controlling Cancer-Related Processes. Cancers, 14(14), 3516. https://doi.org/10.3390/cancers14143516







