Machine Learning Models and Multiparametric Magnetic Resonance Imaging for the Prediction of Pathologic Response to Neoadjuvant Chemotherapy in Breast Cancer
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Patient Population
2.2. Definition of Pathological Complete Response
2.3. MRI Acquisition
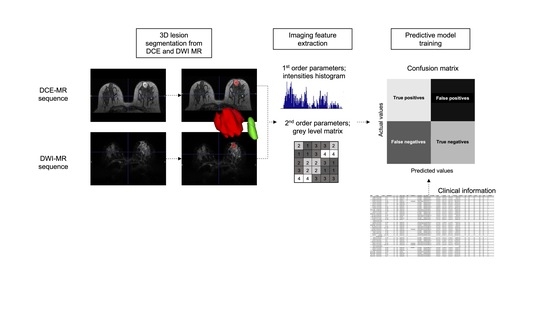
2.4. Lesion Segmentation
2.5. Imaging Feature Analysis
- DCE:
- o
- Initial Area under the Curve at 60 seconds (iAUC60) [mM]: area under the concentration curve increment 60 s after the start of contrast administration;
- o
- Initial slope: represents the initial ascending slope of the concentration curve once the contrast begins to enter the analysed region;
- o
- Peak [mM]: maximum value of the concentration curve;
- o
- Time-to-peak (TTP) [s]: time to reach the maximum value of the curve.
- DWI:
- o
- ADC [mm2/s]: Diffusion was quantified using a Gaussian mono exponential diffusion model to obtain;
- o
- Diffusion Coefficient (D) [mm2/s]: Pure diffusion coefficient derived from bi-exponential model of intra-voxel incoherent motions (IVIM);
- o
- Perfusion Coefficient (D*) [mm2/s]: Fast diffusion coefficient derived from bi-exponential model of intra-voxel incoherent motions (IVIM);
- o
- Vascular fraction (f) [%]: Percentage of the voxel diffusion signal corresponding to fast diffusion.
2.6. Predictive Model
- K-Nearest Neighbour (K-NN): stores all training data and classifies a new data point according to the class of the majority of its k nearest neighbours in the given dataset. To obtain the nearest neighbours for each data, K-NN uses a measure to compute the distance between pairs of data items [36];
- Decision Tree (DT): organises the knowledge extracted from data in a recursive hierarchical structure composed of nodes and branches. Each internal node represents an attribute and is associated to a test relevant for data classification. Leaf nodes of the tree correspond to classes. Branches represent each of the possible results of the applied tests. A new example can be classified following the nodes and branches accordingly until a leaf node is reached [36];
- Random Forest (RF): a method consisting of combinations of tree predictors. Each tree votes for its preferred class and the most voted class gives the final prediction [36];
- Adaptive Boosting (AdaBoost): an ensemble learning method in which a number of weak learners are combined together to form a strong learner. This method focuses on training upon misclassified observations. It alters the distribution of the training dataset to increase weights on sample observations that are difficult to classify [37];
- Gradient Boosting (GBoost): an ensemble learning method in which a number of weak learners are combined together to form a strong learner. This approach trains learners based upon minimising the loss function of a learner (i.e., training on the residuals of the model) [38];
- Gaussian Naïve Bayes (GNB): probabilistic classifier based on the Bayes theorem for conditional probabilities. It builds a function, to be optimised, using a narrow (naïve) assumption that all attributes in a dataset are independent. It follows Gaussian normal distribution and supports continuous data [36];
- Linear Discriminant Analysis (LDA): a common technique used for dimensionality reduction and classification. LDA provides class separability by drawing a decision region between the different classes. LDA tries to maximize the ratio of the between-class variance and the within-class variance. LDA assumes the feature covariance matrices of both classes are the same [39];
- Quadratic Discriminant Analysis (QDA): a generative model that uses a quadratic decision surface to separate measurements of two or more classes of objects or events. It is a variant of the LDA [39];
- Multi-Layer Perceptron (MLP): a neural network algorithm that learns the relationships between linear and non-linear data. It consists of three different layers in which neurons are trained with the back propagation learning algorithm [40];
- Logistic Regression (LR): statistical models in which a logistic curve is fitted to the dataset, modelling the probability of occurrence of a class. The first step in LR consists of building a logit variable, containing the natural log of the odds of the class occurring or not. A maximum likelihood estimation algorithm is then applied to estimate the probabilities [36].
2.7. Statistical Analysis
3. Results
3.1. Clinical Characteristics
3.2. Feature Analysis
3.3. Predictive Model
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Hyder, T.; Bhattacharya, S.; Gade, K.; Nasrazadani, A.; Brufsky, A.M. Approaching neoadjuvant therapy in the management of early-stage breast cancer. Breast Cancer 2021, 13, 199–211. [Google Scholar] [CrossRef]
- Bear, H.D.; Anderson, S.; Brown, A.; Smith, R.; Mamounas, E.P.; Fisher, B.; Margolese, R.; Theoret, H.; Soran, A.; Wickerham, D.L.; et al. The effect on tumor response of adding sequential preoperative docetaxel to preoperative doxorubicin and cyclophosphamide: Preliminary results from National Surgical Adjuvant Breast and Bowel Project Protocol B-27. J. Clin. Oncol. 2003, 21, 4165–4174. [Google Scholar] [CrossRef] [PubMed]
- Pernaut, C.; Lopez, F.; Ciruelos, E. Standard neoadjuvant treatment in early/locally advanced breast cancer. Breast Care 2018, 13, 244–249. [Google Scholar] [CrossRef]
- Gollamudi, J.; Parvani, J.G.; Schiemann, W.P.; Vinayak, S. Neoadjuvant therapy for early-stage breast cancer: The clinical utility of pertuzumab. Cancer Manag. Res. 2016, 8, 21–31. [Google Scholar]
- Willems, A.; Gauger, K.; Henrichs, C.; Harbeck, N. Antibody therapy for breast cancer. Anticancer Res. 2005, 25, 1483–1489. [Google Scholar] [PubMed]
- Colomer, R.; Saura, C.; Sánchez-Rovira, P.; Pascual, T.; Rubio, I.T.; Burgués, O.; Marcos, L.; Rodríguez, C.A.; Martín, M.; Lluch, A. Neoadjuvant management of early breast cancer: A clinical and investigational position statement. Oncologist 2019, 24, 603–611. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnston, S.R. Dual HER2 targeting for early breast cancer. Lancet Oncol. 2013, 14, 1145–1146. [Google Scholar] [CrossRef]
- Cortazar, P.; Zhang, L.; Untch, M.; Mehta, K.; Costantino, J.P.; Wolmark, N.; Bonnefoi, H.; Cameron, D.; Gianni, L.; Valagussa, P.; et al. Pathological complete response and long-term clinical benefit in breast cancer: The CTNeoBC pooled analysis. Lancet 2014, 384, 164–172. [Google Scholar] [CrossRef] [Green Version]
- Kuerer, H.M.; Newman, L.A.; Smith, T.L.; Ames, F.C.; Hunt, K.K.; Dhingra, K.; Theriault, R.L.; Singh, G.; Binkley, S.M.; Sneige, N.; et al. Clinical course of breast cancer patients with complete pathologic primary tumor and axillary lymph node response to doxorubicin-based neoadjuvant chemotherapy. J. Clin. Oncol. 1999, 17, 460–469. [Google Scholar] [CrossRef]
- Rastogi, P.; Anderson, S.J.; Bear, H.D.; Geyer, C.E.; Kahlenberg, M.S.; Robidoux, A.; Margolese, R.G.; Hoehn, J.L.; Vogel, V.G.; Dakhil, S.R.; et al. Preoperative chemotherapy: Updates of National Surgical Adjuvant Breast and Bowel Project Protocols B-18 and B-27. J. Clin. Oncol. 2008, 26, 778–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholl, S.M.; Pierga, J.Y.; Asselain, B.; Beuzeboc, P.; Dorval, T.; Garcia-Giralt, E.; Jouve, M.; Palangié, T.; Remvikos, Y.; Durand, J.C.; et al. Breast tumour response to primary chemotherapy predicts local and distant control as well as survival. Eur. J. Cancer 1995, 31, 1969–1975. [Google Scholar] [CrossRef]
- Shuai, Y.; Ma, L. Prognostic value of pathologic complete response and the alteration of breast cancer immunohistochemical biomarkers after neoadjuvant chemotherapy. Pathol. Res. Pract. 2019, 215, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Mao, X. Evaluation of the efficacy of neoadjuvant chemotherapy for breast cancer. Drug Des. Devel. Ther. 2020, 14, 2423–2433. [Google Scholar] [CrossRef] [PubMed]
- Precht, L.M.; Lowe, K.A.; Atwood, M.; Beatty, J.D. Neoadjuvant chemotherapy of breast cancer: Tumor markers as predictors of pathologic response, recurrence, and survival. Breast J. 2010, 16, 362–368. [Google Scholar] [CrossRef]
- Sachelarie, I.; Grossbard, M.L.; Chadha, M.; Feldman, S.; Ghesani, M.; Blum, R.H. Primary systemic therapy of breast cancer. Oncologist 2006, 11, 574–589. [Google Scholar] [CrossRef] [Green Version]
- Le-Petross, H.T.; Lim, B. Role of MR imaging in neoadjuvant therapy monitoring. Magn. Reson. Imaging Clin. N. Am. 2018, 26, 207–220. [Google Scholar] [CrossRef]
- Bouzón, A.; Acea, B.; Soler, R.; Iglesias, Á.; Santiago, P.; Mosquera, J.; Calvo, L.; Seoane-Pillado, T.; García, A. Diagnostic accuracy of MRI to evaluate tumour response and residual tumour size after neoadjuvant chemotherapy in breast cancer patients. Radiol. Oncol. 2016, 50, 73–79. [Google Scholar] [CrossRef]
- Cavallo Marincola, B.; Telesca, M.; Zaccagna, F.; Riemer, F.; Anzidei, M.; Catalano, C.; Pediconi, F. Can unenhanced MRI of the breast replace contrast-enhanced MRI in assessing response to neoadjuvant chemotherapy? Acta Radiol. 2019, 60, 35–44. [Google Scholar] [CrossRef]
- Li, X.; Arlinghaus, L.R.; Ayers, G.D.; Chakravarthy, A.B.; Abramson, R.G.; Abramson, V.G.; Atuegwu, N.; Farley, J.; Mayer, I.A.; Kelley, M.C.; et al. DCE-MRI analysis methods for predicting the response of breast cancer to neoadjuvant chemotherapy: Pilot study findings. Magn. Reson. Med. 2014, 71, 1592–1602. [Google Scholar] [CrossRef] [Green Version]
- Manton, D.J.; Chaturvedi, A.; Hubbard, A.; Lind, M.J.; Lowry, M.; Maraveyas, A.; Pickles, M.D.; Tozer, D.J.; Turnbull, L.W. Neoadjuvant chemotherapy in breast cancer: Early response prediction with quantitative MR imaging and spectroscopy. Br. J. Cancer 2006, 94, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Sharma, U.; Danishad, K.K.; Seenu, V.; Jagannathan, N.R. Longitudinal study of the assessment by MRI and diffusion-weighted imaging of tumor response in patients with locally advanced breast cancer undergoing neoadjuvant chemotherapy. NMR Biomed. 2009, 22, 104–113. [Google Scholar] [CrossRef] [PubMed]
- Yankeelov, T.E.; Lepage, M.; Chakravarthy, A.; Broome, E.E.; Niermann, K.J.; Kelley, M.C.; Meszoely, I.; Mayer, I.A.; Herman, C.R.; McManus, K.; et al. Integration of quantitative DCE-MRI and ADC mapping to monitor treatment response in human breast cancer: Initial results. Magn. Reson. Imaging 2007, 25, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.-L.; Pan, S.-M.; Ren, J.; Yang, Z.-X.; Jiang, G.-Q. Role of magnetic resonance imaging in detection of pathologic complete remission in breast cancer patients treated with neoadjuvant chemotherapy: A meta-analysis. Clin. Breast Cancer 2017, 17, 245–255. [Google Scholar] [CrossRef] [Green Version]
- Lindenberg, M.A.; Miquel-Cases, A.; Retèl, V.P.; Sonke, G.S.; Wesseling, J.; Stokkel, M.P.M.; van Harten, W.H. Imaging performance in guiding response to neoadjuvant therapy according to breast cancer subtypes: A systematic literature review. Crit. Rev. Oncol./Hematol. 2017, 112, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Chamming’s, F.; Ueno, Y.; Ferré, R.; Kao, E.; Jannot, A.S.; Chong, J.; Omeroglu, A.; Mesurolle, B.; Reinhold, C.; Gallix, B. Features from computerized texture analysis of breast cancers at pretreatment MR imaging are associated with response to neoadjuvant chemotherapy. Radiology 2018, 286, 412–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, M.; Wu, G.; Cheng, H.; Zhang, J.; Shao, G.; Li, L. Radiomic analysis of DCE-MRI for prediction of response to neoadjuvant chemotherapy in breast cancer patients. Eur. J. Radiol. 2017, 94, 140–147. [Google Scholar] [CrossRef]
- Lambin, P.; Rios-Velazquez, E.; Leijenaar, R.; Carvalho, S.; van Stiphout, R.G.; Granton, P.; Zegers, C.M.; Gillies, R.; Boellard, R.; Dekker, A.; et al. Radiomics: Extracting more information from medical images using advanced feature analysis. Eur. J. Cancer 2012, 48, 441–446. [Google Scholar] [CrossRef] [Green Version]
- Aerts, H.J.; Velazquez, E.R.; Leijenaar, R.T.; Parmar, C.; Grossmann, P.; Carvalho, S.; Bussink, J.; Monshouwer, R.; Haibe-Kains, B.; Rietveld, D.; et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat. Commun. 2014, 5, 4006. [Google Scholar] [CrossRef]
- Davnall, F.; Yip, C.S.; Ljungqvist, G.; Selmi, M.; Ng, F.; Sanghera, B.; Ganeshan, B.; Miles, K.A.; Cook, G.J.; Goh, V. Assessment of tumor heterogeneity: An emerging imaging tool for clinical practice? Insights Imaging 2012, 3, 573–589. [Google Scholar] [CrossRef] [Green Version]
- Granzier, R.W.Y.; van Nijnatten, T.J.A.; Woodruff, H.C.; Smidt, M.L.; Lobbes, M.B.I. Exploring breast cancer response prediction to neoadjuvant systemic therapy using MRI-based radiomics: A systematic review. Eur. J. Radiol. 2019, 121, 108736. [Google Scholar] [CrossRef] [Green Version]
- von Minckwitz, G.; Untch, M.; Blohmer, J.-U.; Costa, S.D.; Eidtmann, H.; Fasching, P.A.; Gerber, B.; Eiermann, W.; Hilfrich, J.; Huober, J.; et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J. Clin. Oncol. 2012, 30, 1796–1804. [Google Scholar] [CrossRef] [Green Version]
- Mazouni, C.; Peintinger, F.; Wan-Kau, S.; Andre, F.; Gonzalez-Angulo, A.M.; Symmans, W.F.; Meric-Bernstam, F.; Valero, V.; Hortobagyi, G.N.; Pusztai, L. Residual ductal carcinoma in situ in patients with complete eradication of invasive breast cancer after neoadjuvant chemotherapy does not adversely affect patient outcome. J. Clin. Oncol. 2007, 25, 2650–2655. [Google Scholar] [CrossRef]
- Choi, M.; Park, Y.H.; Ahn, J.S.; Im, Y.-H.; Nam, S.J.; Cho, S.Y.; Cho, E.Y. Evaluation of Pathologic Complete Response in Breast Cancer Patients Treated with Neoadjuvant Chemotherapy: Experience in a Single Institution over a 10-Year Period. J. Pathol. Transl. Med. 2017, 51, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Zwanenburg, A.; Vallières, M.; Abdalah, M.A.; Aerts, H.; Andrearczyk, V.; Apte, A.; Ashrafinia, S.; Bakas, S.; Beukinga, R.J.; Boellaard, R.; et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020, 295, 328–338. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lorena, A.C.; Jacintho, L.F.O.; Siqueira, M.F.; Giovanni, R.D.; Lohmann, L.G.; de Carvalho, A.C.P.L.F.; Yamamoto, M. Comparing machine learning classifiers in potential distribution modelling. Expert Syst. Appl. 2011, 38, 5268–5275. [Google Scholar] [CrossRef]
- Freund, Y.; Schapire, R.E. A Decision-Theoretic Generalization of On-Line Learning and an Application to Boosting. J. Comput. Syst. Sci. 1997, 55, 119–139. [Google Scholar] [CrossRef] [Green Version]
- Natekin, A.; Knoll, A. Gradient boosting machines, a tutorial. Front. Neurorobotics 2013, 7, 21. [Google Scholar] [CrossRef] [Green Version]
- Rezaei, N.; Jabbari, P. Chapter 8—Linear and quadratic discriminant analysis in R. In Immunoinformatics of Cancers; Rezaei, N., Jabbari, P., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 127–142. [Google Scholar]
- Abirami, S.; Chitra, P. Chapter Fourteen—Energy-efficient edge based real-time healthcare support system. In Advances in Computers; Raj, P., Evangeline, P., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 339–368. [Google Scholar]
- Romero, A.; García-Sáenz, J.A.; Fuentes-Ferrer, M.; López Garcia-Asenjo, J.A.; Furió, V.; Román, J.M.; Moreno, A.; de la Hoya, M.; Díaz-Rubio, E.; Martín, M.; et al. Correlation between response to neoadjuvant chemotherapy and survival in locally advanced breast cancer patients. Ann. Oncol. 2013, 24, 655–661. [Google Scholar] [CrossRef]
- Zhou, J.; Lu, J.; Gao, C.; Zeng, J.; Zhou, C.; Lai, X.; Cai, W.; Xu, M. Predicting the response to neoadjuvant chemotherapy for breast cancer: Wavelet transforming radiomics in MRI. BMC Cancer 2020, 20, 100. [Google Scholar] [CrossRef]
- Braman, N.M.; Etesami, M.; Prasanna, P.; Dubchuk, C.; Gilmore, H.; Tiwari, P.; Plecha, D.; Madabhushi, A. Intratumoral and peritumoral radiomics for the pretreatment prediction of pathological complete response to neoadjuvant chemotherapy based on breast DCE-MRI. Breast Cancer Res. 2017, 19, 57. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.; Chen, Y.; Li, X.; Arlinghaus, L.; Chakravarthy, A.B.; Abramson, V.; Bhave, S.R.; Levy, M.A.; Xu, H.; Yankeelov, T.E. Machine learning for predicting the response of breast cancer to neoadjuvant chemotherapy. J. Am. Med. Inform. Assoc. 2013, 20, 688–695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tahmassebi, A.; Wengert, G.J.; Helbich, T.H.; Bago-Horvath, Z.; Alaei, S.; Bartsch, R.; Dubsky, P.; Baltzer, P.; Clauser, P.; Kapetas, P.; et al. Impact of machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy and survival outcomes in breast cancer patients. Investig. Radiol. 2019, 54, 110–117. [Google Scholar] [CrossRef]
- Bian, T.; Wu, Z.; Lin, Q.; Wang, H.; Ge, Y.; Duan, S.; Fu, G.; Cui, C.; Su, X. Radiomic signatures derived from multiparametric MRI for the pretreatment prediction of response to neoadjuvant chemotherapy in breast cancer. Br. J. Radiol. 2020, 93, 20200287. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Shu, Z.; Li, Y.; Chen, B.; Tang, L.; Mo, W.; Shao, G.; Shao, F. Machine learning-Bbased radiomics nomogram using magnetic resonance images for prediction of neoadjuvant chemotherapy efficacy in breast cancer patients. Front. Oncol. 2020, 10, 1410. [Google Scholar]
- Liu, Z.; Li, Z.; Qu, J.; Zhang, R.; Zhou, X.; Li, L.; Sun, K.; Tang, Z.; Jiang, H.; Li, H.; et al. Radiomics of multiparametric MRI for pretreatment prediction of pathologic complete response to neoadjuvant chemotherapy in breast cancer: A multicenter study. Clin. Cancer Res. 2019, 25, 3538–3547. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.; Yu, X.; Gao, T. Machine learning with magnetic resonance imaging for prediction of response to neoadjuvant chemotherapy in breast cancer: A systematic review and meta-analysis. Eur. J. Radiol. 2022, 150, 110247. [Google Scholar] [CrossRef]
- Lindgren, A.; Anttila, M.; Arponen, O.; Rautiainen, S.; Könönen, M.; Vanninen, R.; Sallinen, H. Prognostic value of preoperative dynamic contrast-enhanced magnetic resonance imaging in epithelial ovarian cancer. Eur. J. Radiol. 2019, 115, 66–73. [Google Scholar] [CrossRef]
- Yi, B.; Kang, D.K.; Yoon, D.; Jung, Y.S.; Kim, K.S.; Yim, H.; Kim, T.H. Is there any correlation between model-based perfusion parameters and model-free parameters of time-signal intensity curve on dynamic contrast enhanced MRI in breast cancer patients? Eur. Radiol. 2014, 24, 1089–1096. [Google Scholar] [CrossRef]
- Kuo, W.H.; Chen, C.N.; Hsieh, F.J.; Shyu, M.K.; Chang, L.Y.; Lee, P.H.; Liu, L.Y.; Cheng, C.H.; Wang, J.; Chang, K.J. Vascularity change and tumor response to neoadjuvant chemotherapy for advanced breast cancer. Ultrasound Med. Biol. 2008, 34, 857–866. [Google Scholar] [CrossRef]
- Pan, J.H.; Zhu, S.; Huang, J.; Liang, J.; Zhang, D.; Zhao, X.; Ding, H.; Qin, L.; Shi, C.; Luo, L.; et al. Monitoring the process of endostar-induced tumor vascular normalization by non-contrast intravoxel incoherent motion diffusion-weighted MRI. Front. Oncol. 2018, 8, 524. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Wang, K.; Chan, Q.; Liu, Z.; Zhang, J.; He, H.; Zhang, S.; Liang, C. Intravoxel incoherent motion MR imaging for breast lesions: Comparison and correlation with pharmacokinetic evaluation from dynamic contrast-enhanced MR imaging. Eur. Radiol. 2016, 26, 3888–3898. [Google Scholar] [CrossRef] [PubMed]
- Le Bihan, D.; Breton, E.; Lallemand, D.; Aubin, M.L.; Vignaud, J.; Laval-Jeantet, M. Separation of diffusion and perfusion in intravoxel incoherent motion MR imaging. Radiology 1988, 168, 497–505. [Google Scholar] [CrossRef]
- Andreou, A.; Koh, D.M.; Collins, D.J.; Blackledge, M.; Wallace, T.; Leach, M.O.; Orton, M.R. Measurement reproducibility of perfusion fraction and pseudodiffusion coefficient derived by intravoxel incoherent motion diffusion-weighted MR imaging in normal liver and metastases. Eur. Radiol. 2013, 23, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Kuai, Z.X.; Liu, W.Y.; Zhu, Y.M. Effect of multiple perfusion components on pseudo-diffusion coefficient in intravoxel incoherent motion imaging. Phys. Med. Biol. 2017, 62, 8197–8209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sigmund, E.E.; Cho, G.Y.; Kim, S.; Finn, M.; Moccaldi, M.; Jensen, J.H.; Sodickson, D.K.; Goldberg, J.D.; Formenti, S.; Moy, L. Intravoxel incoherent motion imaging of tumor microenvironment in locally advanced breast cancer. Magn. Reson. Med. 2011, 65, 1437–1447. [Google Scholar] [CrossRef] [Green Version]
- Thibault, G.; Tudorica, A.; Afzal, A.; Chui, S.Y.C.; Naik, A.; Troxell, M.L.; Kemmer, K.A.; Oh, K.Y.; Roy, N.; Jafarian, N.; et al. DCE-MRI texture features for early prediction of breast cancer therapy response. Tomography (Ann. Arbor Mich.) 2017, 3, 23–32. [Google Scholar] [CrossRef]
- Lubner, M.G.; Smith, A.D.; Sandrasegaran, K.; Sahani, D.V.; Pickhardt, P.J. CT Texture Analysis: Definitions, Applications, Biologic Correlates, and Challenges. Radiographics 2017, 37, 1483–1503. [Google Scholar] [CrossRef]
- Teruel, J.R.; Heldahl, M.G.; Goa, P.E.; Pickles, M.; Lundgren, S.; Bathen, T.F.; Gibbs, P. Dynamic contrast-enhanced MRI texture analysis for pretreatment prediction of clinical and pathological response to neoadjuvant chemotherapy in patients with locally advanced breast cancer. NMR Biomed. 2014, 27, 887–896. [Google Scholar] [CrossRef]
- Ahmed, A.; Gibbs, P.; Pickles, M.; Turnbull, L. Texture analysis in assessment and prediction of chemotherapy response in breast cancer. J. Magn. Reson. Imaging 2013, 38, 89–101. [Google Scholar] [CrossRef]
- Mohammed, R.A.A.; Ellis, I.O.; Mahmmod, A.M.; Hawkes, E.C.; Green, A.R.; Rakha, E.A.; Martin, S.G. Lymphatic and blood vessels in basal and triple-negative breast cancers: Characteristics and prognostic significance. Mod. Pathol. 2011, 24, 774–785. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Characteristic | Patients(N = 58) n (%) |
|---|---|
| Premenopausal | 23 (40) |
| Clinical TNM stage | |
| IIA | 19 (32) |
| IIB | 10 (18) |
| IIIA | 9 (16) |
| IIIB | 13 (22) |
| IIIC | 7 (12) |
| Histology | |
| Ductal | 52 (90) |
| Others | 6 (10) |
| Associated DISC | 20 (34) |
| Nottingham Histologic Score (Grade II) | 37 (64) |
| Lymphovascular invasion | 18 (30) |
| Molecular subtype | |
| Luminal A | 15 (26) |
| Luminal B | 9 (15) |
| HER2+ | 23 (40) |
| Triple-negative | 11 (19) |
| Neoadjuvant chemotherapy | |
| ddAC + paclitaxel weekly | 35 (60) |
| ddAC + paclitaxel + CBDCA AUC2 | 8 (14) |
| NAC with trastuzumab + pertuzumab | 19 (32) |
| Clinical trial with trastuzumab + pertuzumab | 4 (7) |
| Conservative surgery | 26 (45) |
| Negative margins | 57 (99) |
| Residual tumour | |
| No | 17 (29) |
| Yes | 41 (71) |
| I | 31 (54) |
| II | 5 (8) |
| III | 5 (8) |
| Systemic adjuvant treatment | |
| Adjuvant chemotherapy | 10 (18) |
| Adjuvant hormone therapy | 34 (59) |
| Adjuvant trastuzumab | 23 (40) |
| Adjuvant radiation therapy | 52 (90) |
| Type of Response | n (%) |
|---|---|
| Clinical response | |
| SD | 15 (26) |
| PR | 24 (42) |
| CR | 19 (32) |
| Radiological response | |
| SD | 7 (12) |
| PR | 25 (44) |
| CR | 25 (44) |
| pCR (ypT0/is ypN0) | |
| Yes | 12 (21) |
| No | 46 (79) |
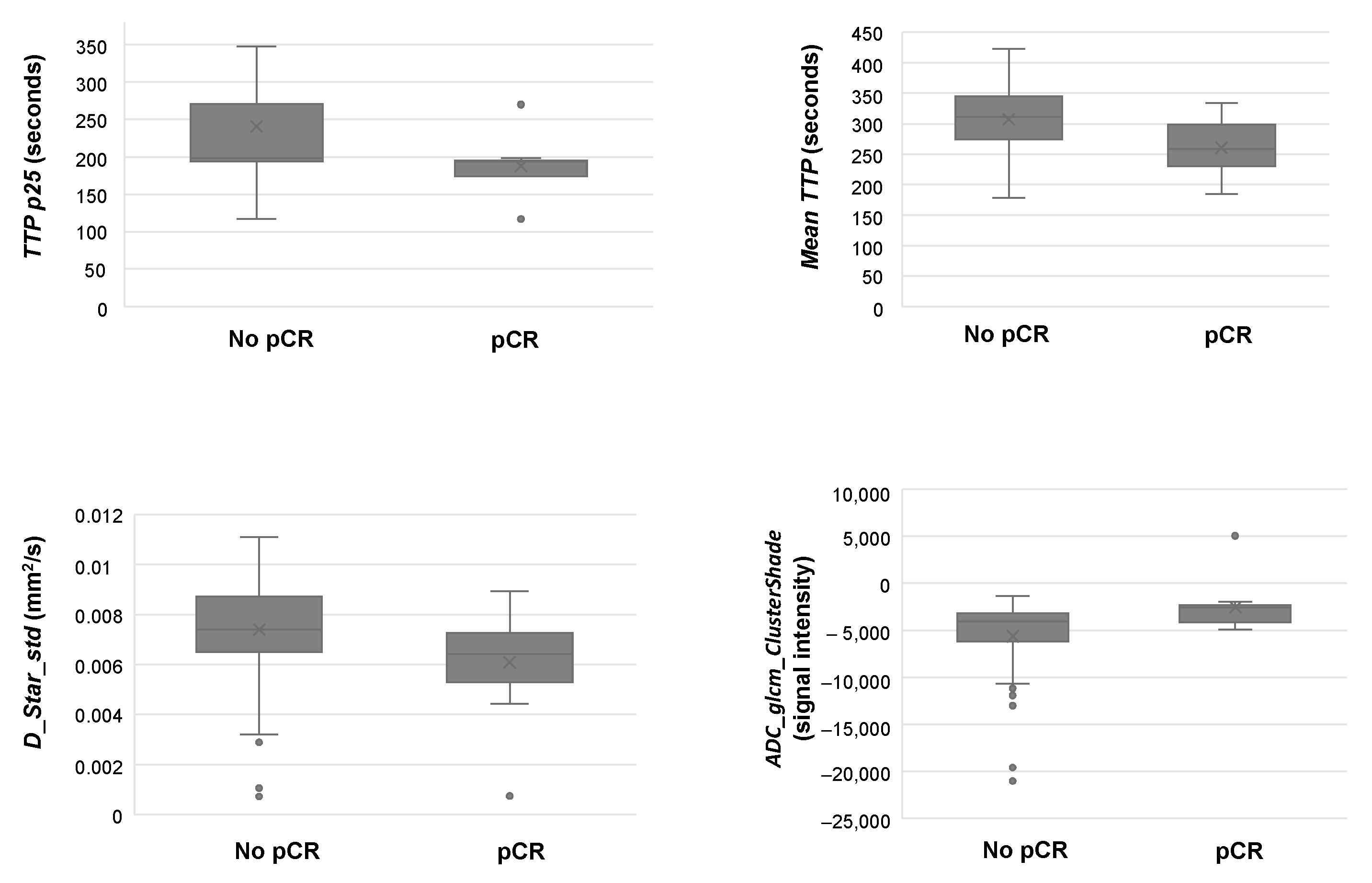
| Imaging Feature | Mean (SD) | p-Value |
|---|---|---|
| TTP p25 (s) | ||
| No pCR | 237.67 (79.20) | 0.004 |
| pCR | 187.47 (51.27) | |
| meanTTP (s) | ||
| No pCR | 302.40 (56.67) | 0.026 |
| pCR | 260.52 (46.61) | |
| D_star_std (mm2/s) | ||
| No pCR | 0.007 (0.002) | 0.012 |
| pCR | 0.006 (0.002) | |
| ADC_glcm_ClusterShade (signal intensity; absolute value) | ||
| No pCR | −5140.14 (3644.81) | 0.035 |
| pCR | −2542.84 (2607.65) |
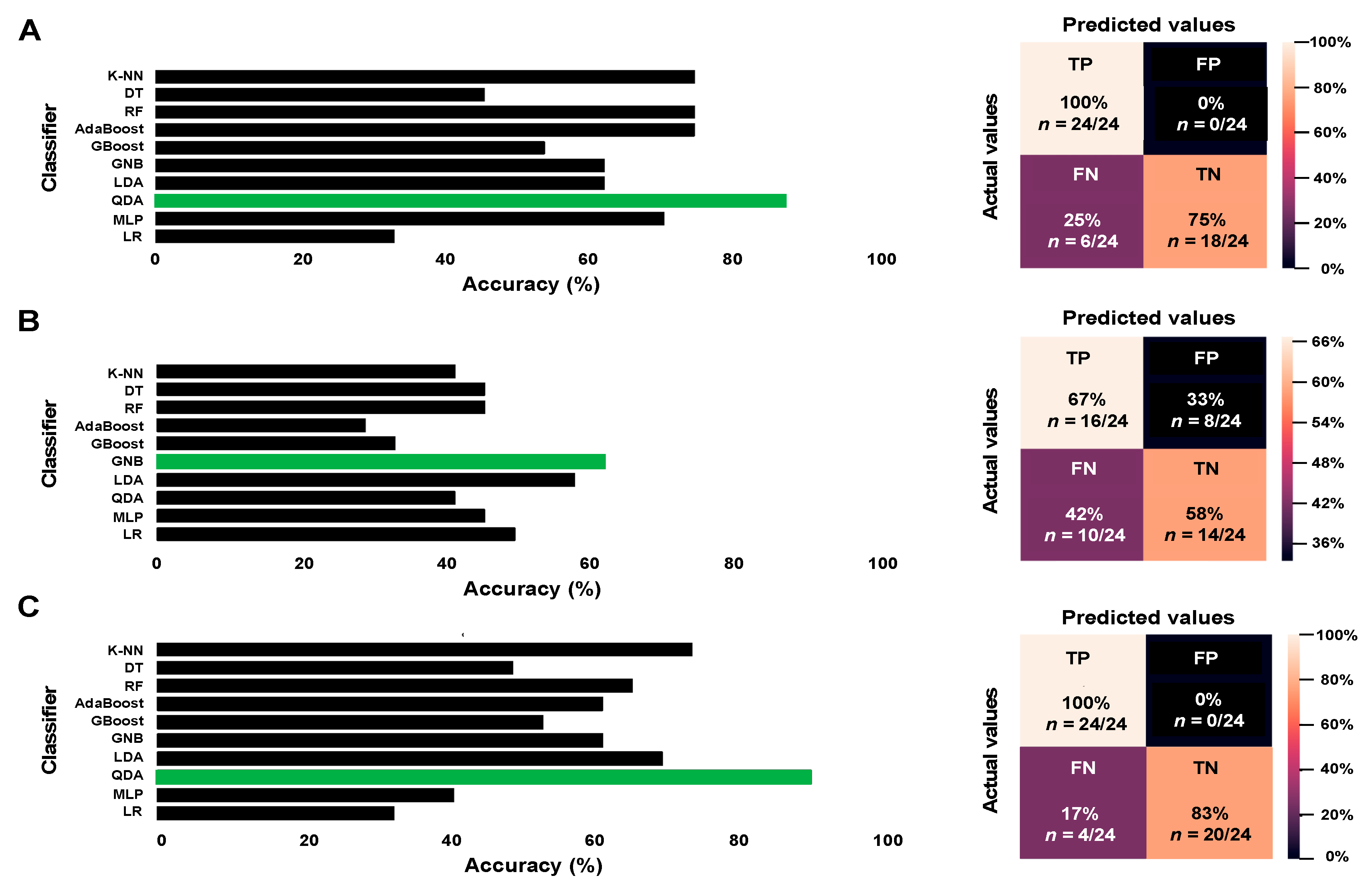
| Predictive Models | Imaging Data QDA Classifier | Clinical Data GNB Classifier | Imaging + Clinical DataQDA Classifier | |
|---|---|---|---|---|
| Performance | ||||
| Sensitivity | 100% | 63% | 100% | |
| Specificity | 80% | 61.5% | 85.5% | |
| Error rate | 12.5% | 37.5% | 8.5% | |
| Accuracy | 87.5% | 62.5% | 91.5% | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herrero Vicent, C.; Tudela, X.; Moreno Ruiz, P.; Pedralva, V.; Jiménez Pastor, A.; Ahicart, D.; Rubio Novella, S.; Meneu, I.; Montes Albuixech, Á.; Santamaria, M.Á.; et al. Machine Learning Models and Multiparametric Magnetic Resonance Imaging for the Prediction of Pathologic Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancers 2022, 14, 3508. https://doi.org/10.3390/cancers14143508
Herrero Vicent C, Tudela X, Moreno Ruiz P, Pedralva V, Jiménez Pastor A, Ahicart D, Rubio Novella S, Meneu I, Montes Albuixech Á, Santamaria MÁ, et al. Machine Learning Models and Multiparametric Magnetic Resonance Imaging for the Prediction of Pathologic Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancers. 2022; 14(14):3508. https://doi.org/10.3390/cancers14143508
Chicago/Turabian StyleHerrero Vicent, Carmen, Xavier Tudela, Paula Moreno Ruiz, Víctor Pedralva, Ana Jiménez Pastor, Daniel Ahicart, Silvia Rubio Novella, Isabel Meneu, Ángela Montes Albuixech, Miguel Ángel Santamaria, and et al. 2022. "Machine Learning Models and Multiparametric Magnetic Resonance Imaging for the Prediction of Pathologic Response to Neoadjuvant Chemotherapy in Breast Cancer" Cancers 14, no. 14: 3508. https://doi.org/10.3390/cancers14143508
APA StyleHerrero Vicent, C., Tudela, X., Moreno Ruiz, P., Pedralva, V., Jiménez Pastor, A., Ahicart, D., Rubio Novella, S., Meneu, I., Montes Albuixech, Á., Santamaria, M. Á., Fonfria, M., Fuster-Matanzo, A., Olmos Antón, S., & Martínez de Dueñas, E. (2022). Machine Learning Models and Multiparametric Magnetic Resonance Imaging for the Prediction of Pathologic Response to Neoadjuvant Chemotherapy in Breast Cancer. Cancers, 14(14), 3508. https://doi.org/10.3390/cancers14143508