C8orf76 Modulates Ferroptosis in Liver Cancer via Transcriptionally Up-Regulating SLC7A11
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Human Samples
2.2. Immunohistochemistry
2.3. Lentiviral Vector Construction and Transfection
2.4. Cell Lines and Regents
2.5. Cell Viability
2.6. Cell Growth Assay
2.7. Colony Formation Measurements
2.8. Cell Cycle
2.9. PI Staining
2.10. Tumor Xenografts
2.11. Measurement of Lipid ROS and GSH
2.12. Glutamate Release Assays
2.13. Fluorescence Microscope Assay
2.14. Western Blot Analysis and Antibodies
2.15. Transmission Electron Microscopy
2.16. RNA Extraction and Quantitative Real-Time PCR
2.17. Immunofluorescent Staining
2.18. Chromatin Immunoprecipitation (ChIP)
2.19. Luciferase Assay
2.20. Statistical Analysis
3. Results
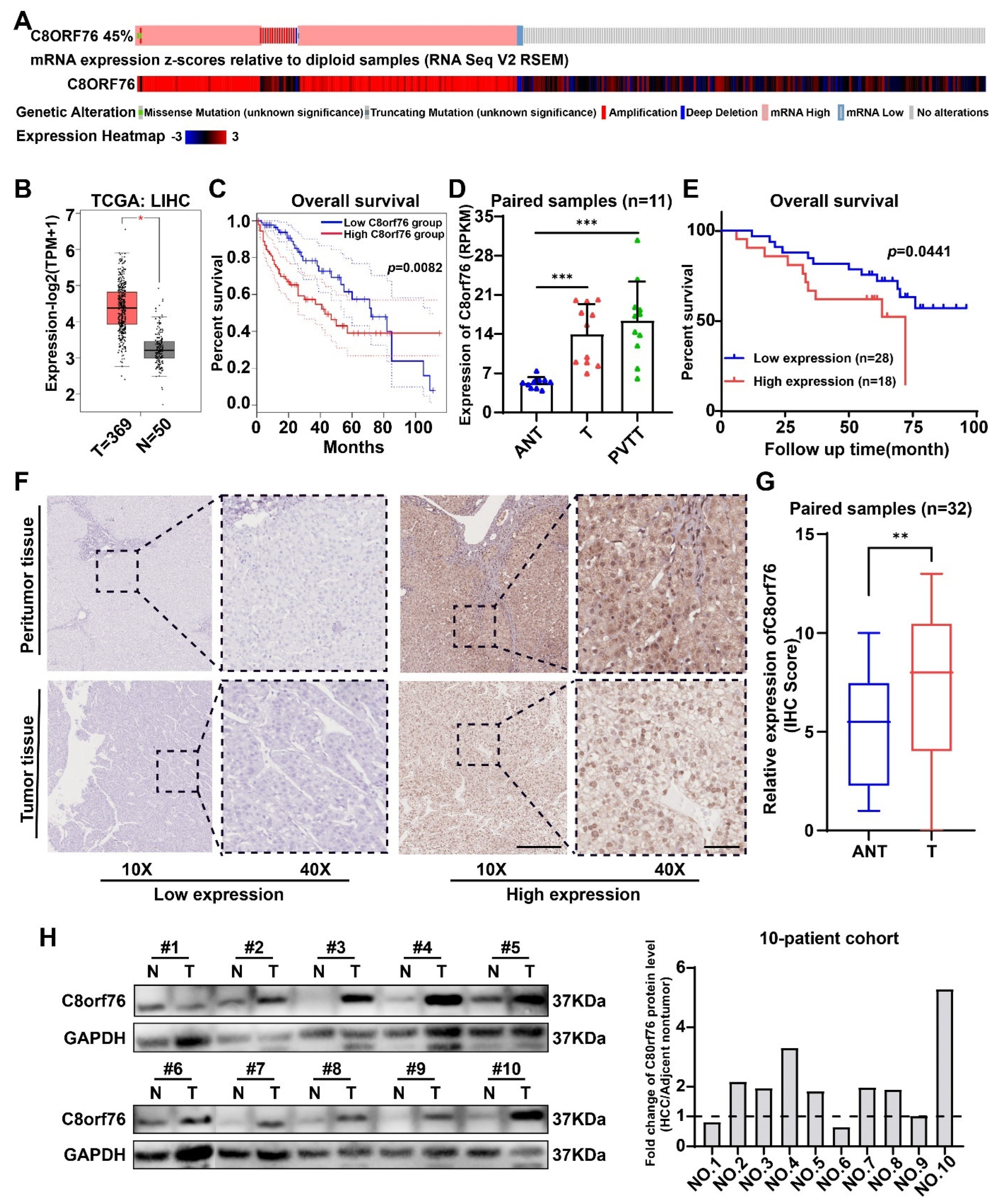
3.1. C8orf76 Expression Was Enhanced in HCC Patients and Related to HCC Progression and Poor Clinical Outcomes
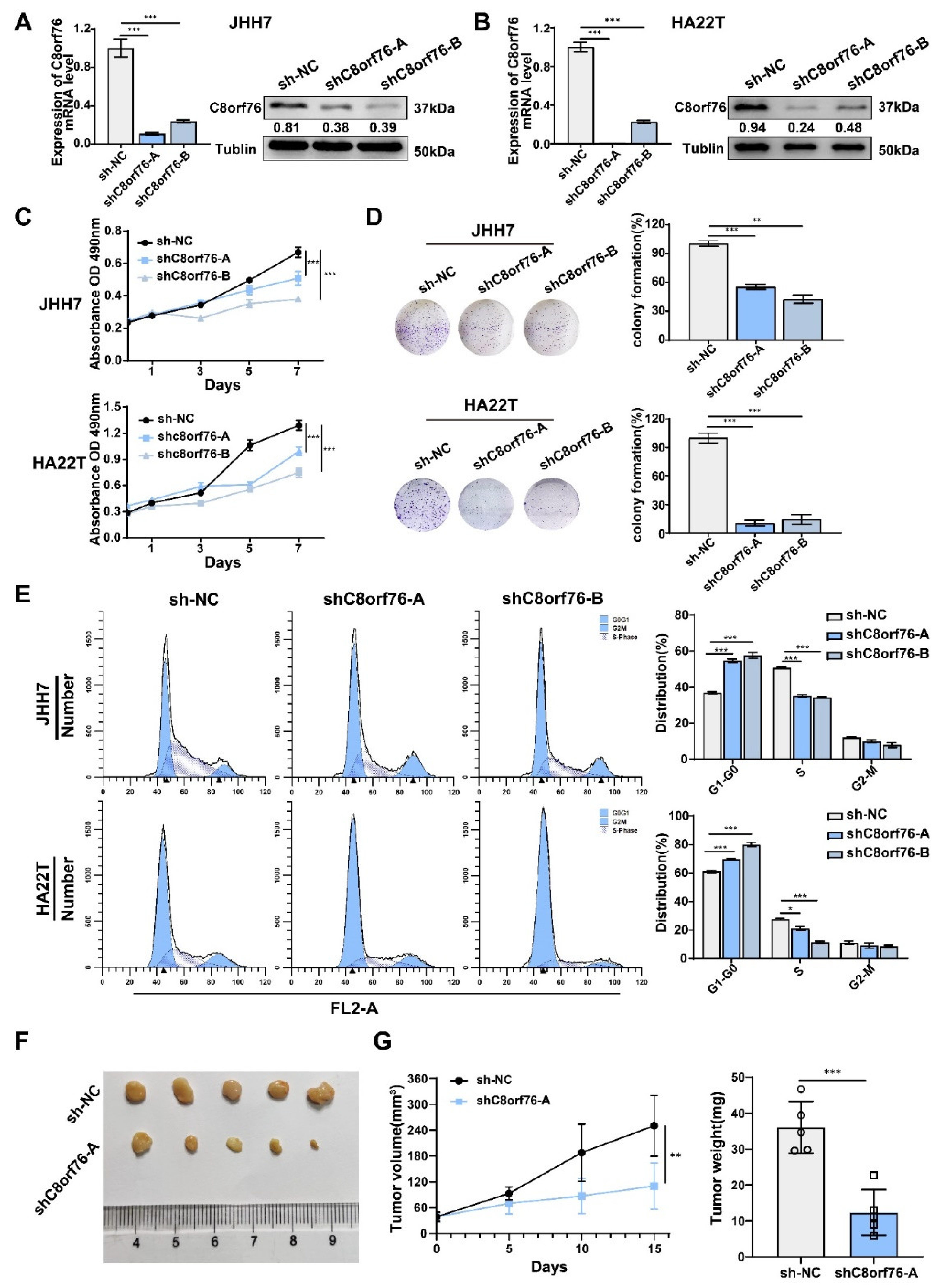
3.2. C8orf76 Knockdown Inhibited HCC Cell Growth and Cell-Cycle Progression
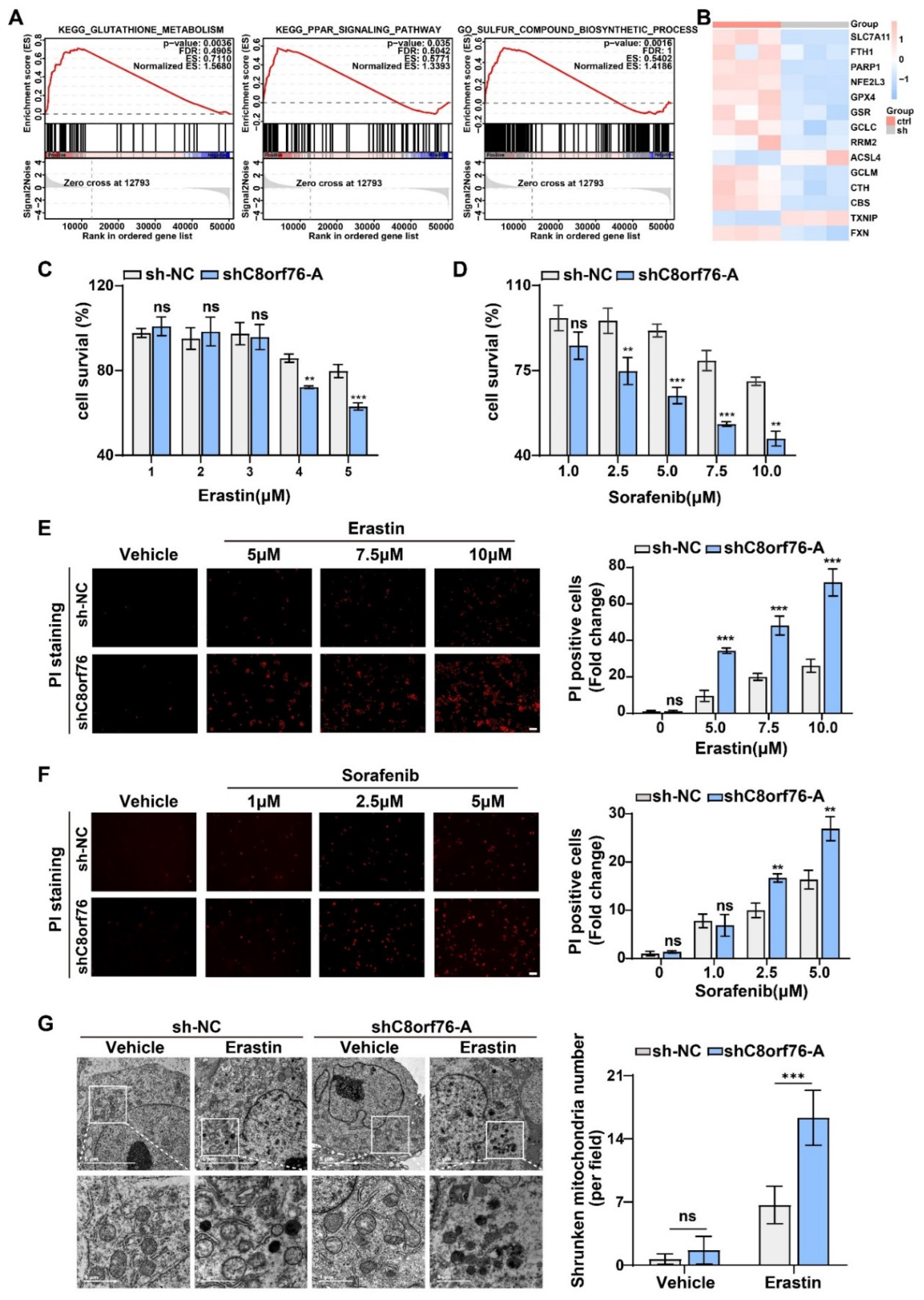
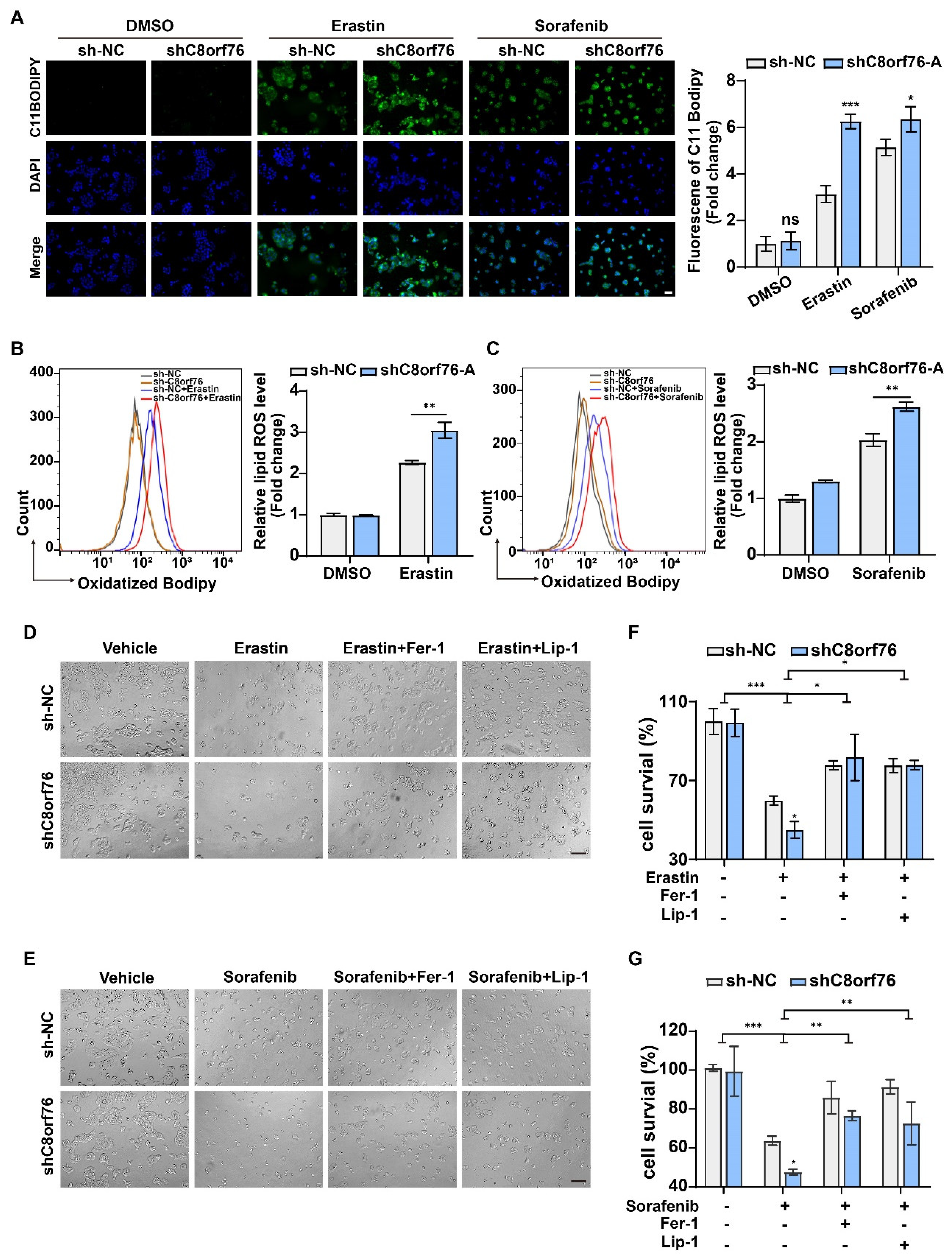
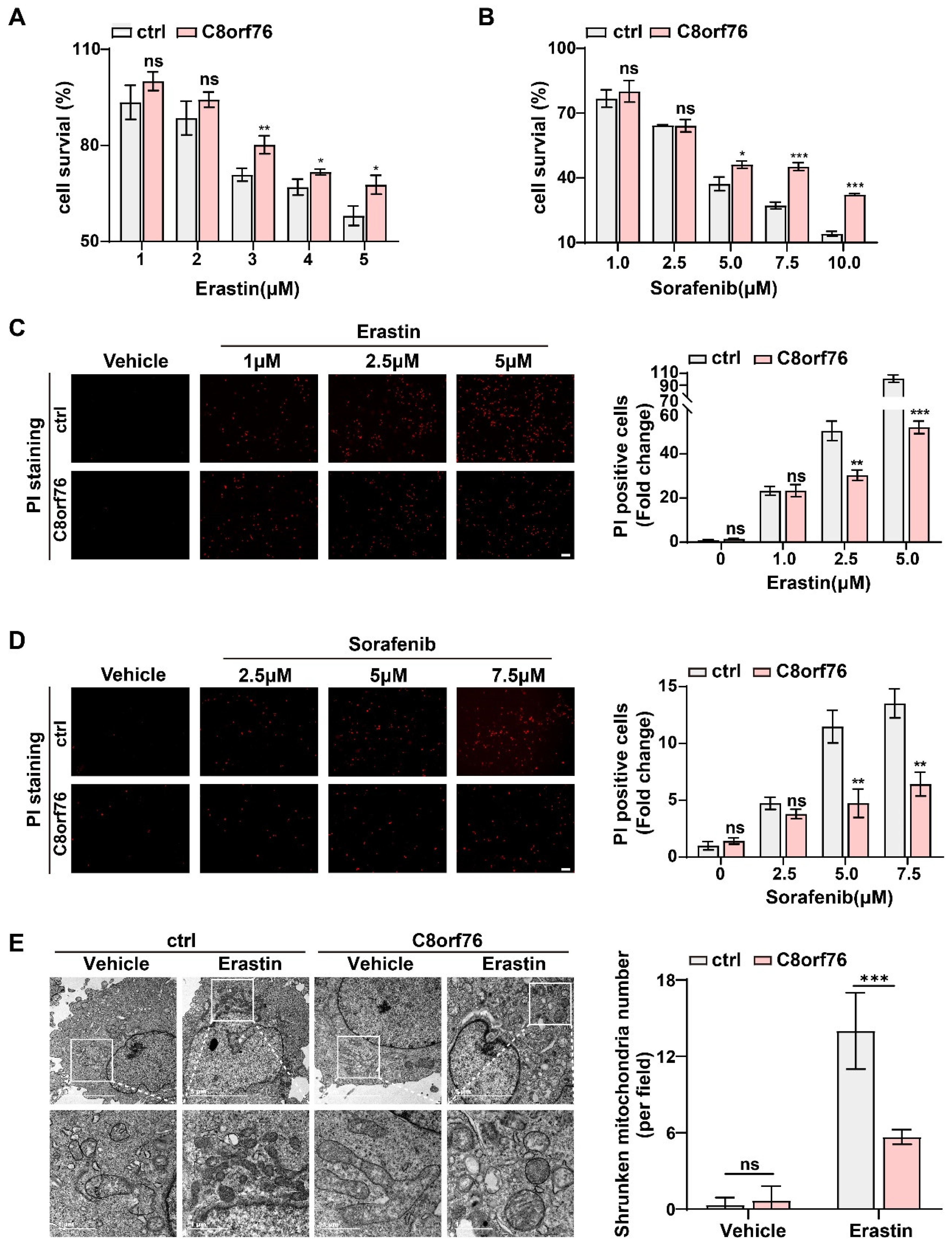
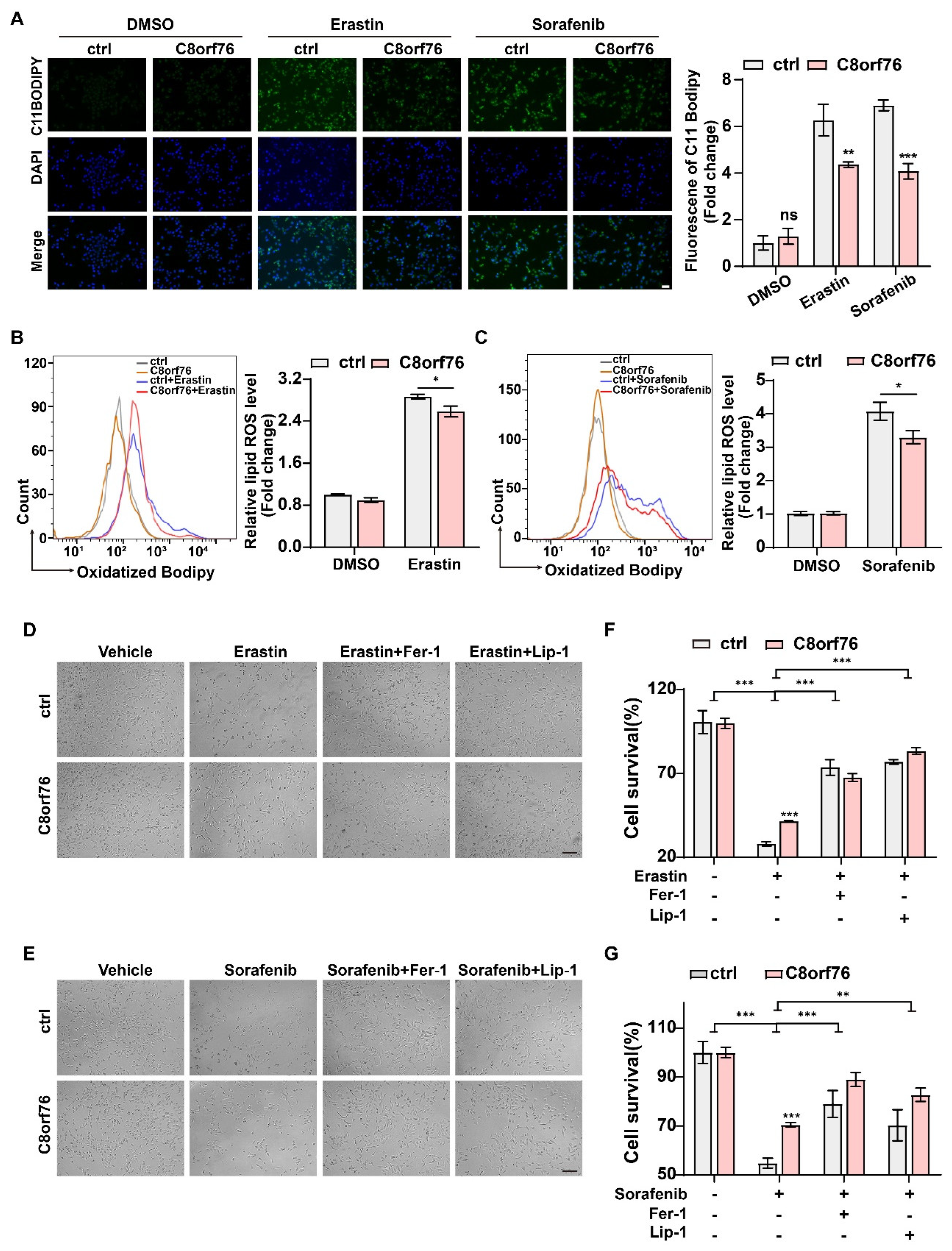
3.3. C8orf76 Knockdown Rendered HCC Cells Susceptible for Ferroptosis
3.4. C8orf76 Overexpression Mediated Tumor Ferroptosis Resistance
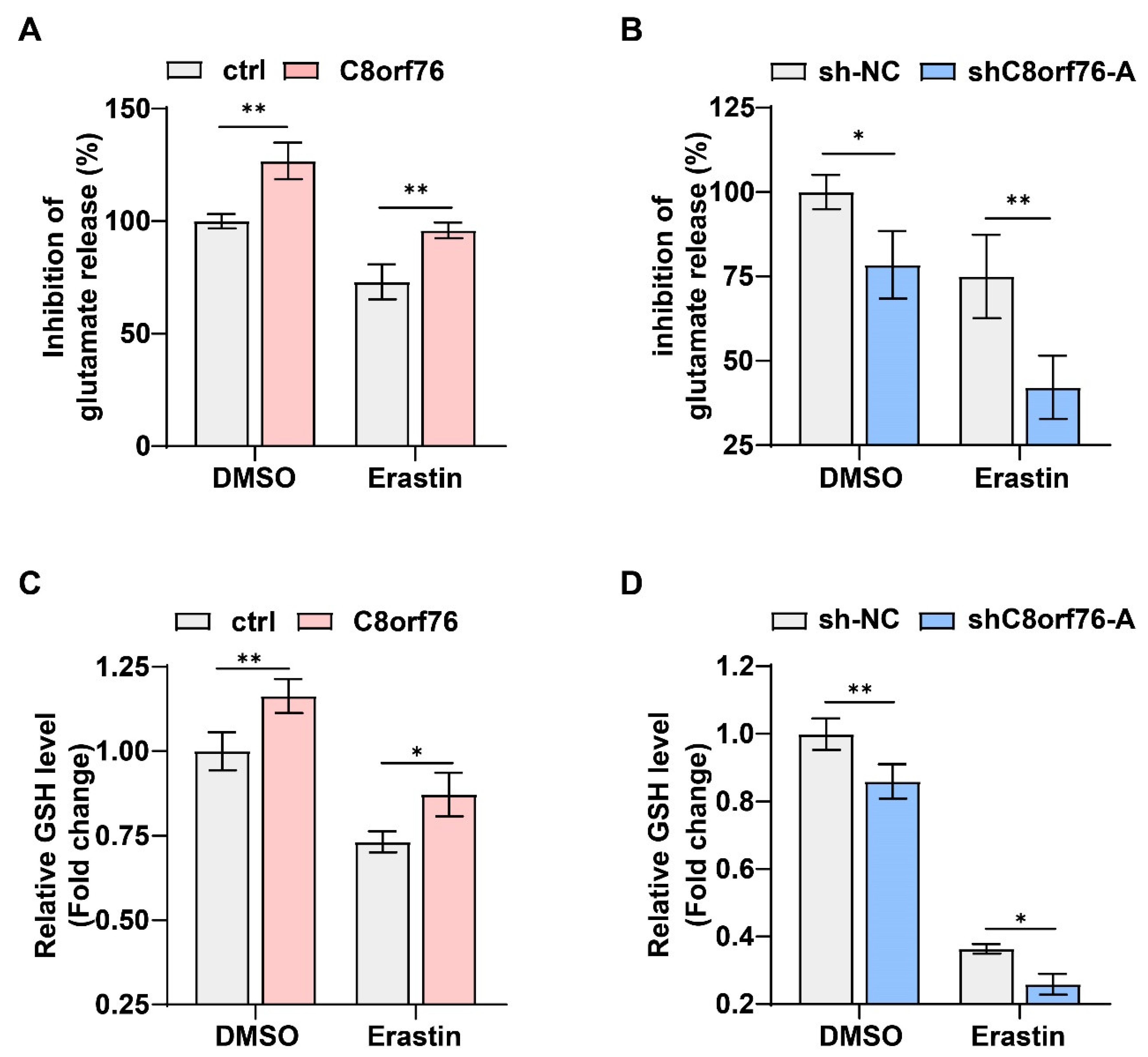
3.5. C8orf76 Regulated xCT Activity and Glutamate Secretion
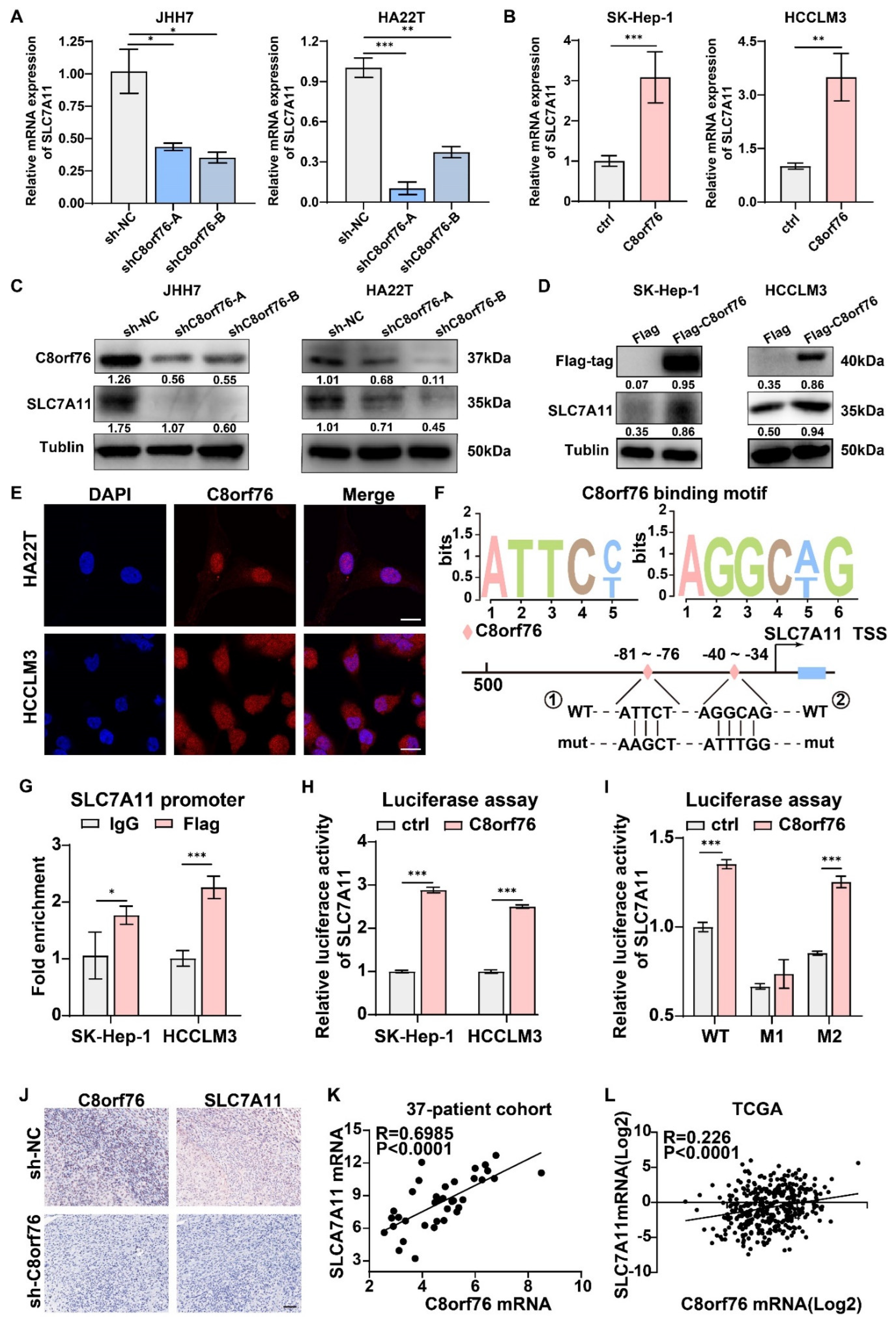
3.6. C8orf76 Bound to the Promoter of SLC7A11 and Promoted Its Transcription
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 7–30. [Google Scholar] [CrossRef]
- Gomaa, A.I.; Khan, S.A.; Toledano, M.B.; Waked, I.; Taylor-Robinson, S.D. Hepatocellular carcinoma: Epidemiology, risk factors and pathogenesis. World J. Gastroenterol. 2008, 14, 4300–4308. [Google Scholar] [CrossRef]
- Wong, C.M.; Tsang, F.H.; Ng, I.O. Non-coding RNAs in hepatocellular carcinoma: Molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 137–151. [Google Scholar] [CrossRef] [PubMed]
- Gollin, S.M. Chromosomal instability. Curr. Opin. Oncol. 2004, 16, 25–31. [Google Scholar] [CrossRef] [PubMed]
- Patil, M.A.; Gütgemann, I.; Zhang, J.; Ho, C.; Cheung, S.T.; Ginzinger, D.; Li, R.; Dykema, K.J.; So, S.; Fan, S.T.; et al. Array-based comparative genomic hybridization reveals recurrent chromosomal aberrations and Jab1 as a potential target for 8q gain in hepatocellular carcinoma. Carcinogenesis 2005, 26, 2050–2057. [Google Scholar] [CrossRef] [PubMed]
- Walker, L.C.; McDonald, M.; Wells, J.E.; Harris, G.C.; Robinson, B.A.; Morris, C.M. Dual-color fluorescence in situ hybridization reveals an association of chromosome 8q22 but not 8p21 imbalance with high grade invasive breast carcinoma. PLoS ONE 2013, 8, e70790. [Google Scholar] [CrossRef][Green Version]
- Okamoto, H.; Yasui, K.; Zhao, C.; Arii, S.; Inazawa, J. PTK2 and EIF3S3 genes may be amplification targets at 8q23-q24 and are associated with large hepatocellular carcinomas. Hepatology 2003, 38, 1242–1249. [Google Scholar] [CrossRef]
- Zhao, K.; Zhao, Y.; Zhu, J.Y.; Dong, H.; Cong, W.M.; Yu, Y.; Wang, H.; Zhu, Z.Z.; Xu, Q. A Panel of Genes Identified as Targets for 8q24.13-24.3 Gain Contributing to Unfavorable Overall Survival in Patients with Hepatocellular Carcinoma. Curr. Med. Sci. 2018, 38, 590–596. [Google Scholar] [CrossRef]
- Wang, X.; Liang, Q.; Zhang, L.; Gou, H.; Li, Z.; Chen, H.; Dong, Y.; Ji, J.; Yu, J. C8orf76 Promotes Gastric Tumorigenicity and Metastasis by Directly Inducing lncRNA DUSP5P1 and Associates with Patient Outcomes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019, 25, 3128–3140. [Google Scholar] [CrossRef]
- Stockwell, B.R.; Friedmann Angeli, J.P.; Bayir, H.; Bush, A.I.; Conrad, M.; Dixon, S.J.; Fulda, S.; Gascón, S.; Hatzios, S.K.; Kagan, V.E.; et al. Ferroptosis: A Regulated Cell Death Nexus Linking Metabolism, Redox Biology, and Disease. Cell 2017, 171, 273–285. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef] [PubMed]
- Dixon, S.J.; Lemberg, K.M.; Lamprecht, M.R.; Skouta, R.; Zaitsev, E.M.; Gleason, C.E.; Patel, D.N.; Bauer, A.J.; Cantley, A.M.; Yang, W.S.; et al. Ferroptosis: An iron-dependent form of nonapoptotic cell death. Cell 2012, 149, 1060–1072. [Google Scholar] [CrossRef] [PubMed]
- Koppula, P.; Zhang, Y.; Zhuang, L.; Gan, B. Amino acid transporter SLC7A11/xCT at the crossroads of regulating redox homeostasis and nutrient dependency of cancer. Cancer Commun. 2018, 38, 12. [Google Scholar] [CrossRef]
- Liu, J.; Xia, X.; Huang, P. xCT: A Critical Molecule That Links Cancer Metabolism to Redox Signaling. Mol. Ther. J. Am. Soc. Gene Ther. 2020, 28, 2358–2366. [Google Scholar] [CrossRef]
- Feng, H.; Stockwell, B.R. Unsolved mysteries: How does lipid peroxidation cause ferroptosis? PLoS Biol. 2018, 16, e2006203. [Google Scholar] [CrossRef]
- Imai, H.; Matsuoka, M.; Kumagai, T.; Sakamoto, T.; Koumura, T. Lipid Peroxidation-Dependent Cell Death Regulated by GPx4 and Ferroptosis. Curr. Top. Microbiol. Immunol. 2017, 403, 143–170. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Wang, C.; Liu, G.; Bi, C.; Wang, X.; Zhou, Q.; Jin, H. SLC7A11/xCT in cancer: Biological functions and therapeutic implications. Am. J. Cancer Res. 2020, 10, 3106–3126. [Google Scholar]
- Li, T.; Kon, N.; Jiang, L.; Tan, M.; Ludwig, T.; Zhao, Y.; Baer, R.; Gu, W. Tumor suppression in the absence of p53-mediated cell-cycle arrest, apoptosis, and senescence. Cell 2012, 149, 1269–1283. [Google Scholar] [CrossRef]
- Jiang, L.; Kon, N.; Li, T.; Wang, S.J.; Su, T.; Hibshoosh, H.; Baer, R.; Gu, W. Ferroptosis as a p53-mediated activity during tumour suppression. Nature 2015, 520, 57–62. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Du, T.; Yang, H.; Lei, L.; Guo, M.; Ding, H.F.; Zhang, J.; Wang, H.; Chen, X.; et al. ATF3 promotes erastin-induced ferroptosis by suppressing system Xc(). Cell Death Differ. 2020, 27, 662–675. [Google Scholar] [CrossRef]
- Sato, H.; Nomura, S.; Maebara, K.; Sato, K.; Tamba, M.; Bannai, S. Transcriptional control of cystine/glutamate transporter gene by amino acid deprivation. Biochem. Biophys. Res. Commun. 2004, 325, 109–116. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, H.; Sato, H.; Kuriyama-Matsumura, K.; Sato, K.; Maebara, K.; Wang, H.; Tamba, M.; Itoh, K.; Yamamoto, M.; Bannai, S. Electrophile response element-mediated induction of the cystine/glutamate exchange transporter gene expression. J. Biol. Chem. 2002, 277, 44765–44771. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Guo, H.; Dai, B.; He, L.; Zhou, D.; Lin, H. The association between methylation patterns of DNAH17 and clinicopathological factors in hepatocellular carcinoma. Cancer Med. 2019, 8, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Li, Y.; Yi, X.; Chen, G.; Jin, S.; Dai, Y.; Cui, B.; Dai, B.; Lin, H.; Zhou, D. Epigenome-wide DNA methylation profiling of portal vein tumor thrombosis (PVTT) tissues in hepatocellular carcinoma patients. Neoplasia 2020, 22, 630–643. [Google Scholar] [CrossRef]
- Niu, B.; Liao, K.; Zhou, Y.; Wen, T.; Quan, G.; Pan, X.; Wu, C. Application of glutathione depletion in cancer therapy: Enhanced ROS-based therapy, ferroptosis, and chemotherapy. Biomaterials 2021, 277, 121110. [Google Scholar] [CrossRef]
- Venkatesh, D.; O’Brien, N.A.; Zandkarimi, F.; Tong, D.R.; Stokes, M.E.; Dunn, D.E.; Kengmana, E.S.; Aron, A.T.; Klein, A.M.; Csuka, J.M.; et al. MDM2 and MDMX promote ferroptosis by PPARα-mediated lipid remodeling. Genes Dev. 2020, 34, 526–543. [Google Scholar] [CrossRef]
- Seco-Cervera, M.; González-Cabo, P.; Pallardó, F.V.; Romá-Mateo, C.; García-Giménez, J.L. Thioredoxin and Glutaredoxin Systems as Potential Targets for the Development of New Treatments in Friedreich’s Ataxia. Antioxidants 2020, 9, 1257. [Google Scholar] [CrossRef]
- Wang, H.; Lin, D.; Yu, Q.; Li, Z.; Lenahan, C.; Dong, Y.; Wei, Q.; Shao, A. A Promising Future of Ferroptosis in Tumor Therapy. Front. Cell Dev. Biol. 2021, 9, 629150. [Google Scholar] [CrossRef]
- Cao, J.Y.; Dixon, S.J. Mechanisms of ferroptosis. Cell. Mol. Life Sci. CMLS 2016, 73, 2195–2209. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Y.; Jiang, R.; Xue, R.; Yin, X.; Wu, M.; Meng, Q. Ferroptosis in liver disease: New insights into disease mechanisms. Cell Death Discov. 2021, 7, 276. [Google Scholar] [CrossRef]
- Chen, X.; Kang, R.; Kroemer, G.; Tang, D. Broadening horizons: The role of ferroptosis in cancer. Nat. Rev. Clin. Oncol. 2021, 18, 280–296. [Google Scholar] [CrossRef] [PubMed]
- Hassannia, B.; Vandenabeele, P.; Vanden Berghe, T. Targeting Ferroptosis to Iron Out Cancer. Cancer Cell 2019, 35, 830–849. [Google Scholar] [CrossRef] [PubMed]
- Louandre, C.; Ezzoukhry, Z.; Godin, C.; Barbare, J.C.; Mazière, J.C.; Chauffert, B.; Galmiche, A. Iron-dependent cell death of hepatocellular carcinoma cells exposed to sorafenib. Int. J. Cancer 2013, 133, 1732–1742. [Google Scholar] [CrossRef] [PubMed]
- Koppula, P.; Zhuang, L.; Gan, B. Cystine transporter SLC7A11/xCT in cancer: Ferroptosis, nutrient dependency, and cancer therapy. Protein Cell 2021, 12, 599–620. [Google Scholar] [CrossRef]
- Buettner, R.; Mora, L.B.; Jove, R. Activated STAT signaling in human tumors provides novel molecular targets for therapeutic intervention. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2002, 8, 945–954. [Google Scholar]
- Linher-Melville, K.; Haftchenary, S.; Gunning, P.; Singh, G. Signal transducer and activator of transcription 3 and 5 regulate system Xc- and redox balance in human breast cancer cells. Mol. Cell. Biochem. 2015, 405, 205–221. [Google Scholar] [CrossRef]
- Ogiwara, H.; Takahashi, K.; Sasaki, M.; Kuroda, T.; Yoshida, H.; Watanabe, R.; Maruyama, A.; Makinoshima, H.; Chiwaki, F.; Sasaki, H.; et al. Targeting the Vulnerability of Glutathione Metabolism in ARID1A-Deficient Cancers. Cancer Cell 2019, 35, 177–190.e178. [Google Scholar] [CrossRef]
| Variable | C8orf76 | |||
|---|---|---|---|---|
| Sum | Low Expression | High Expression | p-Value a | |
| Age | 0.607 | |||
| <50 | 15 | 8 | 7 | |
| ≥50 | 31 | 19 | 12 | |
| Gender | 0.355 | |||
| Male | 39 | 24 | 15 | |
| Female | 7 | 3 | 4 | |
| HBV | 0.685 | |||
| Negative | 21 | 13 | 8 | |
| Positive | 25 | 14 | 11 | |
| AFP (ng/mL) | 0.036 | |||
| <400 | 32 | 22 | 10 | |
| ≥400 | 14 | 5 | 9 | |
| Cirrhosis | 0.655 | |||
| No | 26 | 16 | 10 | |
| Yes | 20 | 11 | 9 | |
| Tumor size (cm) | 0.362 | |||
| T < 3 | 13 | 9 | 4 | |
| T ≥ 3 | 33 | 18 | 15 | |
| Vascular invasion | 0.025 | |||
| No | 40 | 26 | 14 | |
| Yes | 6 | 1 | 5 | |
| LNM | 0.810 | |||
| No | 38 | 22 | 16 | |
| Yes | 8 | 5 | 3 | |
| TNM stage | 0.655 | |||
| I–II | 20 | 11 | 9 | |
| III–IV | 26 | 16 | 10 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, D.; Pan, J.; Zhang, Y.; Li, Y.; Jin, S.; Zhong, C.; Chen, P.; Ma, J.; Hu, W.; Fan, X.; et al. C8orf76 Modulates Ferroptosis in Liver Cancer via Transcriptionally Up-Regulating SLC7A11. Cancers 2022, 14, 3410. https://doi.org/10.3390/cancers14143410
Li D, Pan J, Zhang Y, Li Y, Jin S, Zhong C, Chen P, Ma J, Hu W, Fan X, et al. C8orf76 Modulates Ferroptosis in Liver Cancer via Transcriptionally Up-Regulating SLC7A11. Cancers. 2022; 14(14):3410. https://doi.org/10.3390/cancers14143410
Chicago/Turabian StyleLi, Duguang, Junhai Pan, Yiyin Zhang, Yirun Li, Shengxi Jin, Cheng Zhong, Peng Chen, Jingjing Ma, Wendi Hu, Xiaoxiao Fan, and et al. 2022. "C8orf76 Modulates Ferroptosis in Liver Cancer via Transcriptionally Up-Regulating SLC7A11" Cancers 14, no. 14: 3410. https://doi.org/10.3390/cancers14143410
APA StyleLi, D., Pan, J., Zhang, Y., Li, Y., Jin, S., Zhong, C., Chen, P., Ma, J., Hu, W., Fan, X., & Lin, H. (2022). C8orf76 Modulates Ferroptosis in Liver Cancer via Transcriptionally Up-Regulating SLC7A11. Cancers, 14(14), 3410. https://doi.org/10.3390/cancers14143410





