TP53 and LRP1B Co-Wild Predicts Improved Survival for Patients with LUSC Receiving Anti-PD-L1 Immunotherapy
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Tumor Samples
2.2. NGS Sequencing and Bioinformatics Analysis
2.3. IHC for PD-L1
2.4. LUSC Data Collect
2.5. TMB
2.6. Oncogenic Signaling Pathway Analysis
2.7. CNV Analysis
2.8. Differentially Expressed Genes Analysis and Pathway Enrichment
2.9. GSEA
2.10. Immune Microenvironment Assessment
2.11. Statistical Analysis
3. Results
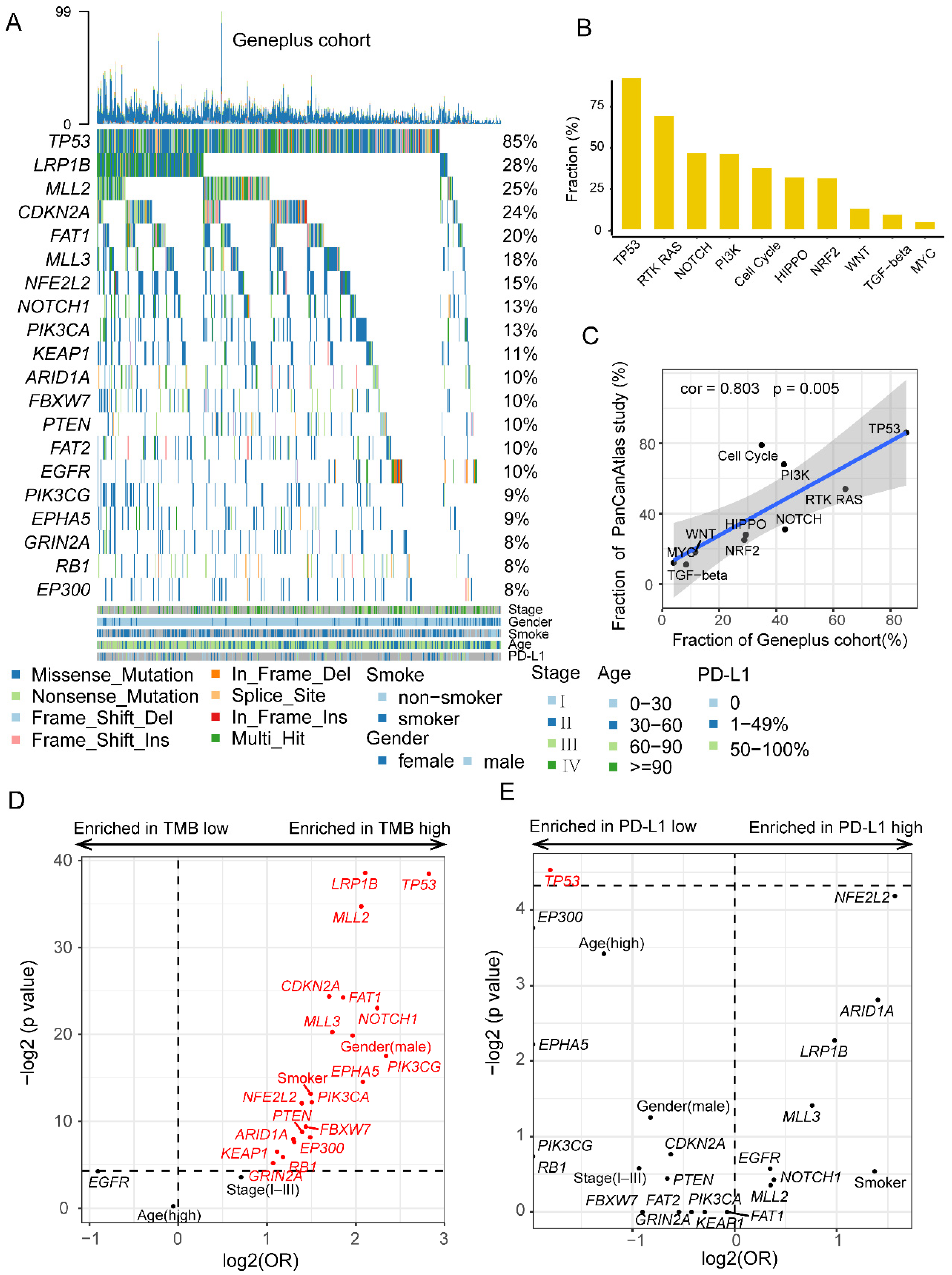
3.1. Comprehensive Genomic Profiles of Patients with LUSC
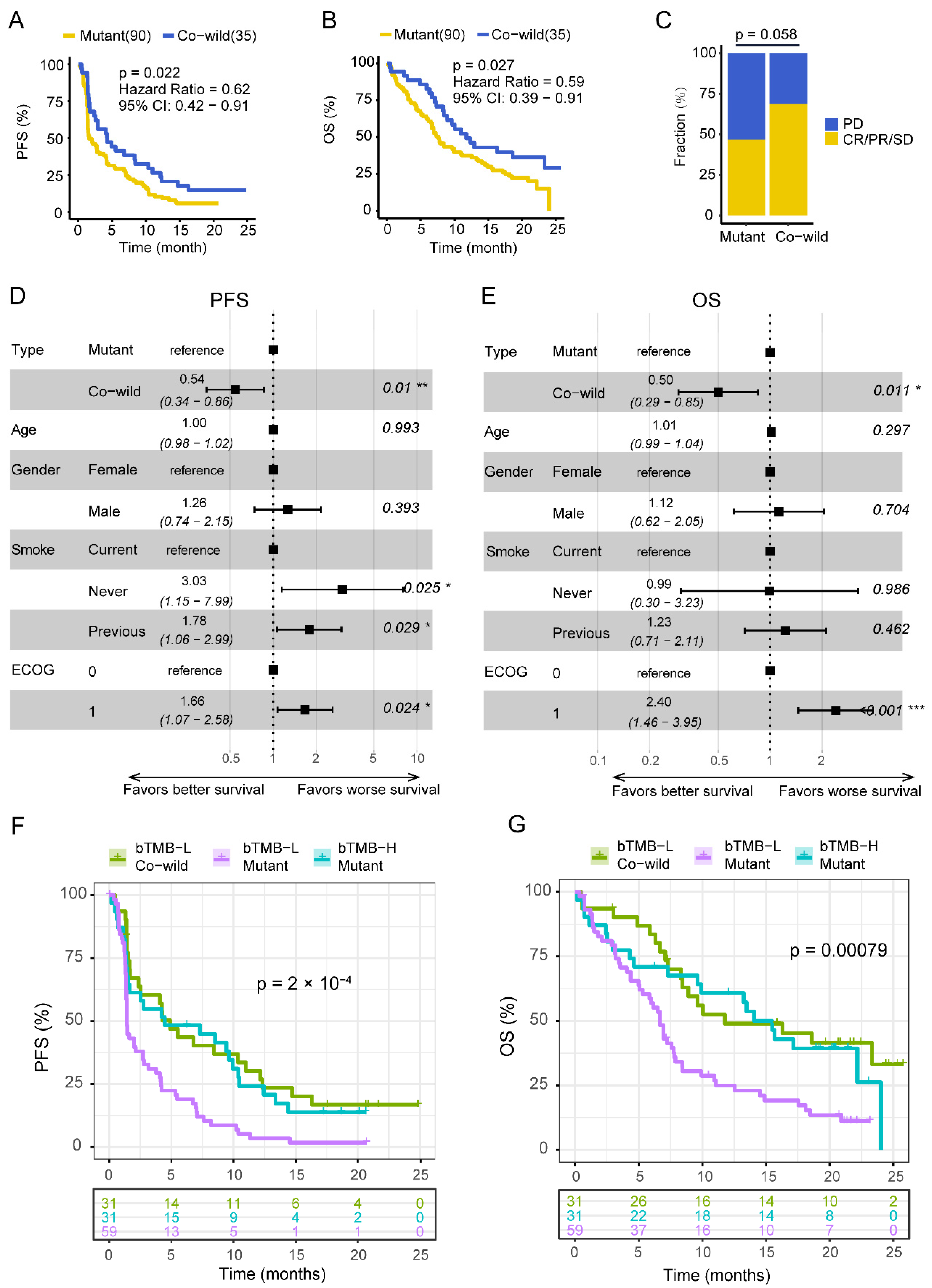
3.2. TP53/LRP1B Co-Wild LUSC Had an Improved Outcome in Receiving Anti-PD-L1 Immunotherapy
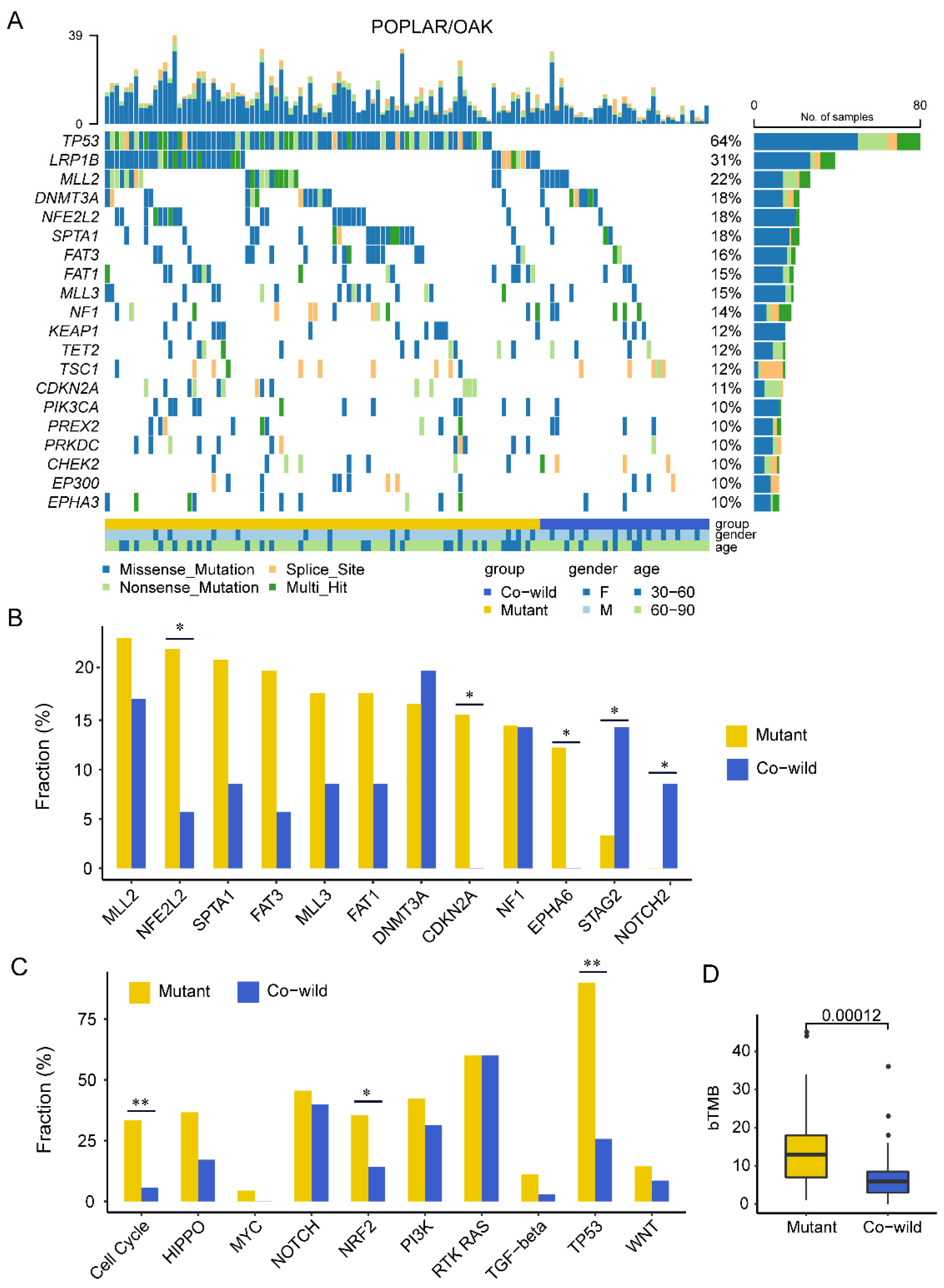
3.3. TP53/LRP1B Co-Wild LUSC Had Unequal Mutational Characteristics Compared with Mutant Type
3.4. Copy-Number Variation Profile Revealed a Higher Level of Chromosome Stability of TP53/LRP1B Co-Wild LUSC Compared with Mutant LUSC
3.5. TP53/LRP1B Co-Wild LUSC Had Expressional Signatures of Leukocyte Activation and Differentiation
3.6. TP53/LRP1B Co-Wild LUSC Was Associated with an Activated Immuno-Phenotype
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Langer, C.J.; Obasaju, C.; Bunn, P.; Bonomi, P.; Gandara, D.; Hirsch, F.R.; Kim, E.S.; Natale, R.B.; Novello, S.; Paz-Ares, L.; et al. Incremental Innovation and Progress in Advanced Squamous Cell Lung Cancer: Current Status and Future Impact of Treatment. J. Thorac. Oncol. 2016, 11, 2066–2081. [Google Scholar] [CrossRef]
- Gandara, D.R.; Hammerman, P.S.; Sos, M.L.; Lara, P.N.; Hirsch, F.R. Squamous Cell Lung Cancer: From Tumor Genomics to Cancer Therapeutics. Clin. Cancer Res. 2015, 21, 2236–2243. [Google Scholar] [CrossRef]
- Khuder, S.A.; Mutgi, A.B. Effect of Smoking Cessation on Major Histologic Types of Lung Cancer. Chest 2001, 120, 1577–1583. [Google Scholar] [CrossRef][Green Version]
- Herbst, R.S.; Morgensztern, D.; Boshoff, C. The biology and management of non-small cell lung cancer. Nature 2018, 553, 446–454. [Google Scholar] [CrossRef]
- Reck, M.; Rabe, K.F. Precision Diagnosis and Treatment for Advanced Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 849–861. [Google Scholar] [CrossRef]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crinò, L.; Eberhardt, W.E.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef]
- Herbst, R.S.; Baas, P.; Kim, D.-W.; Felip, E.; Pérez-Gracia, J.L.; Han, J.-Y.; Molina, J.; Kim, J.-H.; Arvis, C.D.; Ahn, M.-J.; et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): A randomised controlled trial. Lancet 2015, 387, 1540–1550. [Google Scholar] [CrossRef]
- Paz-Ares, L.; Luft, A.; Vicente, D.; Tafreshi, A.; Gümüş, M.; Mazières, J.; Hermes, B.; Çay Şenler, F.; Csőszi, T.; Fülöp, A.; et al. Pembrolizumab plus Chemotherapy for Squamous Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2040–2051. [Google Scholar] [CrossRef]
- Reck, M.; Rodríguez-Abreu, D.; Robinson, A.G.; Hui, R.; Csőszi, T.; Fülöp, A.; Gottfried, M.; Peled, N.; Tafreshi, A.; Cuffe, S.; et al. Pembrolizumab versus Chemotherapy for PD-L1–Positive Non–Small-Cell Lung Cancer. N. Engl. J. Med. 2016, 375, 1823–1833. [Google Scholar] [CrossRef]
- Rittmeyer, A.; Barlesi, F.; Waterkamp, D.; Park, K.; Ciardiello, F.; von Pawel, J.; Gadgeel, S.M.; Hida, T.; Kowalski, D.M.; Dols, M.C.; et al. Atezolizumab versus docetaxel in patients with previously treated non-small-cell lung cancer (OAK): A phase 3, open-label, multicentre randomised controlled trial. Lancet 2017, 389, 255–265. [Google Scholar] [CrossRef]
- Lee, J.S.; Ruppin, E. Multiomics Prediction of Response Rates to Therapies to Inhibit Programmed Cell Death 1 and Programmed Cell Death 1 Ligand 1. JAMA Oncol. 2019, 5, 1614–1618. [Google Scholar] [CrossRef]
- Wang, J.; Lu, S.; Yu, X.; Hu, Y.; Sun, Y.; Wang, Z.; Zhao, J.; Yu, Y.; Hu, C.; Yang, K.; et al. RATIONALE-307: Tislelizumab plus chemotherapy versus chemotherapy alone as first-line treatment for advanced squamous NSCLC in patients aged ≥ 65. J. Clin. Oncol. 2021, 39, 9102. [Google Scholar] [CrossRef]
- Mok, T.S.K.; Wu, Y.-L.; Kudaba, I.; Kowalski, D.M.; Cho, B.C.; Turna, H.Z.; Castro, G., Jr.; Srimuninnimit, V.; Laktionov, K.K.; Bondarenko, I.; et al. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): A randomised, open-label, controlled, phase 3 trial. Lancet 2019, 393, 1819–1830. [Google Scholar] [CrossRef]
- Hellmann, M.D.; Ciuleanu, T.-E.; Pluzanski, A.; Lee, J.S.; Otterson, G.A.; Audigier-Valette, C.; Minenza, E.; Linardou, H.; Burgers, S.; Salman, P.; et al. Nivolumab plus Ipilimumab in Lung Cancer with a High Tumor Mutational Burden. N. Engl. J. Med. 2018, 378, 2093–2104. [Google Scholar] [CrossRef]
- Sacher, A.G.; Gandhi, L. Biomarkers for the Clinical Use of PD-1/PD-L1 Inhibitors in Non–Small-Cell Lung Cancer. JAMA Oncol. 2016, 2, 1217–1222. [Google Scholar] [CrossRef]
- McGrail, D.; Pilié, P.; Rashid, N.; Voorwerk, L.; Slagter, M.; Kok, M.; Jonasch, E.; Khasraw, M.; Heimberger, A.; Lim, B.; et al. High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann. Oncol. 2021, 32, 661–672. [Google Scholar] [CrossRef]
- Nong, J.; Gong, Y.; Guan, Y.; Yi, X.; Yi, Y.; Chang, L.; Yang, L.; Lv, J.; Guo, Z.; Jia, H.; et al. Circulating tumor DNA analysis depicts subclonal architecture and genomic evolution of small cell lung cancer. Nat. Commun. 2018, 9, 3114. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows—Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.B.; Sougnez, C.; Gabriel, S.B.; Meyerson, M.L.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Yang, X.; Chu, Y.; Zhang, R.; Han, Y.; Zhang, L.; Fu, Y.; Li, D.; Peng, R.; Li, D.; Ding, J.; et al. Technical Validation of a Next-Generation Sequencing Assay for Detecting Clinically Relevant Levels of Breast Cancer–Related Single-Nucleotide Variants and Copy Number Variants Using Simulated Cell-Free DNA. J. Mol. Diagn. 2017, 19, 525–536. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Fehrenbacher, L.; Spira, A.; Ballinger, M.; Kowanetz, M.; Vansteenkiste, J.; Mazieres, J.; Park, K.; Smith, D.; Artal-Cortes, A.; Lewanski, C.; et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): A multicentre, open-label, phase 2 randomised controlled trial. Lancet 2016, 387, 1837–1846. [Google Scholar] [CrossRef]
- Gandara, D.R.; Paul, S.M.; Kowanetz, M.; Schleifman, E.; Zou, W.; Li, Y.; Rittmeyer, A.; Fehrenbacher, L.; Otto, G.; Malboeuf, C.; et al. Blood-based tumor mutational burden as a predictor of clinical benefit in non-small-cell lung cancer patients treated with atezolizumab. Nat. Med. 2018, 24, 1441–1448. [Google Scholar] [CrossRef]
- Sanchez-Vega, F.; Mina, M.; Armenia, J.; Chatila, W.K.; Luna, A.; La, K.C.; Dimitriadoy, S.; Liu, D.L.; Kantheti, H.S.; Saghafinia, S.; et al. Oncogenic Signaling Pathways in The Cancer Genome Atlas. Cell 2018, 173, 321–337.e310. [Google Scholar] [CrossRef]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef]
- Quinton, R.J.; DiDomizio, A.; Vittoria, M.A.; Kotýnková, K.; Ticas, C.J.; Patel, S.; Koga, Y.; Vakhshoorzadeh, J.; Hermance, N.; Kuroda, T.S.; et al. Whole-genome doubling confers unique genetic vulnerabilities on tumour cells. Nature 2021, 590, 492–497. [Google Scholar] [CrossRef]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef]
- Hu, J.; Yu, A.; Othmane, B.; Qiu, D.; Li, H.; Li, C.; Liu, P.; Ren, W.; Chen, M.; Gong, G.; et al. Siglec15 shapes a non-inflamed tumor microenvironment and predicts the molecular subtype in bladder cancer. Theranostics 2021, 11, 3089–3108. [Google Scholar] [CrossRef]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef]
- Cristescu, R.; Mogg, R.; Ayers, M.; Albright, A.; Murphy, E.; Yearley, J.; Sher, X.; Liu, X.Q.; Lu, H.; Nebozhyn, M.; et al. Pan-tumor genomic biomarkers for PD-1 checkpoint blockade–based immunotherapy. Science 2018, 362, eaar3593. [Google Scholar] [CrossRef]
- Arrieta, V.A.; Cacho-Díaz, B.; Zhao, J.; Rabadan, R.; Chen, L.; Sonabend, A.M. The possibility of cancer immune editing in gliomas. A critical review. OncoImmunology 2018, 7, e1445458. [Google Scholar] [CrossRef]
- Nagarsheth, N.; Wicha, M.S.; Zou, W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat. Rev. Immunol. 2017, 17, 559–572. [Google Scholar] [CrossRef]
- Jiang, T.; Shi, J.; Dong, Z.; Hou, L.; Zhao, C.; Li, X.; Mao, B.; Zhu, W.; Guo, X.; Zhang, H.; et al. Genomic landscape and its correlations with tumor mutational burden, PD-L1 expression, and immune cells infiltration in Chinese lung squamous cell carcinoma. J. Hematol. Oncol. 2019, 12, 75. [Google Scholar] [CrossRef]
- Assoun, S.; Theou-Anton, N.; Nguenang, M.; Cazes, A.; Danel, C.; Abbar, B.; Pluvy, J.; Gounant, V.; Khalil, A.; Namour, C.; et al. Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer 2019, 132, 65–71. [Google Scholar] [CrossRef]
- Lan, S.; Li, H.; Liu, Y.; Ma, L.; Liu, X.; Liu, Y.; Yan, S.; Cheng, Y. Somatic mutation of LRP1B is associated with tumor mutational burden in patients with lung cancer. Lung Cancer 2019, 132, 154–156. [Google Scholar] [CrossRef]
- Chen, H.; Chong, W.; Wu, Q.; Yao, Y.; Mao, M.; Wang, X. Association of LRP1B Mutation with Tumor Mutation Burden and Outcomes in Melanoma and Non-small Cell Lung Cancer Patients Treated With Immune Check-Point Blockades. Front. Immunol. 2019, 10, 1113. [Google Scholar] [CrossRef]
- Xu, Y.; Li, H.; Huang, Z.; Chen, K.; Yu, X.; Sheng, J.; Zhang, H.-H.; Fan, Y. Predictive values of genomic variation, tumor mutational burden, and PD-L1 expression in advanced lung squamous cell carcinoma treated with immunotherapy. Transl. Lung Cancer Res. 2020, 9, 2367. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Dong, Z.-Y.; Zhong, W.-Z.; Zhang, X.-C.; Su, J.; Xie, Z.; Liu, S.-Y.; Tu, H.-Y.; Chen, H.-J.; Sun, Y.-L.; Zhou, Q.; et al. Potential Predictive Value of TP53 and KRAS Mutation Status for Response to PD-1 Blockade Immunotherapy in Lung Adenocarcinoma. Clin. Cancer Res. 2017, 23, 3012–3024. [Google Scholar] [CrossRef]
- Lin, X.; Wang, L.; Xie, X.; Qin, Y.; Xie, Z.; Ouyang, M.; Zhou, C. Prognostic Biomarker TP53 Mutations for Immune Checkpoint Blockade Therapy and Its Association with Tumor Microenvironment of Lung Adenocarcinoma. Front. Mol. Biosci. 2020, 7, 602328. [Google Scholar] [CrossRef]
- Takeda, H.; Rust, A.G.; Ward, J.M.; Yew, C.C.K.; Jenkins, N.A.; Copeland, N.G. Sleeping Beauty transposon mutagenesis identifies genes that cooperate with mutant Smad4 in gastric cancer development. Proc. Natl. Acad. Sci. USA 2016, 113, E2057–E2065. [Google Scholar] [CrossRef] [PubMed]
- Ding, D.; Lou, X.; Hua, D.; Yu, W.; Li, L.; Wang, J.; Gao, F.; Zhao, N.; Ren, G.; Li, L.; et al. Recurrent Targeted Genes of Hepatitis B Virus in the Liver Cancer Genomes Identified by a Next-Generation Sequencing–Based Approach. PLoS Genet. 2012, 8, e1003065. [Google Scholar] [CrossRef] [PubMed]
- Asano, Y.; Takeuchi, T.; Okubo, H.; Saigo, C.; Kito, Y.; Iwata, Y.; Futamura, M.; Yoshida, K. Nuclear localization of LDL receptor-related protein 1B in mammary gland carcinogenesis. J. Mol. Med. 2019, 97, 257–268. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-X.; Li, Y.; Obermoeller-McCormick, L.M.; Schwartz, A.L.; Bu, G. The Putative Tumor Suppressor LRP1B, a Novel Member of the Low Density Lipoprotein (LDL) Receptor Family, Exhibits Both Overlapping and Distinct Properties with the LDL Receptor-related Protein. J. Biol. Chem. 2001, 276, 28889–28896. [Google Scholar] [CrossRef]
- Cotterchio, M.; Lowcock, E.; Bider-Canfield, Z.; Lemire, M.; Greenwood, C.; Gallinger, S.; Hudson, T. Association between Variants in Atopy-Related Immunologic Candidate Genes and Pancreatic Cancer Risk. PLoS ONE 2015, 10, e0125273. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Hou, W.; Liang, J.; Zhu, L.; Luo, C. LRP1B mutation: A novel independent prognostic factor and a predictive tumor mutation burden in hepatocellular carcinoma. J. Cancer 2021, 12, 4039–4048. [Google Scholar] [CrossRef]
- Wang, L.; Yan, K.; He, X.; Zhu, H.; Song, J.; Chen, S.; Cai, S.; Zhao, Y.; Wang, L. LRP1B or TP53 mutations are associated with higher tumor mutational burden and worse survival in hepatocellular carcinoma. J. Cancer 2021, 12, 217–223. [Google Scholar] [CrossRef]
- Brown, L.C.; Tucker, M.D.; Sedhom, R.; Schwartz, E.B.; Zhu, J.; Kao, C.; Labriola, M.K.; Gupta, R.T.; Marin, D.; Wu, Y.; et al. LRP1B mutations are associated with favorable outcomes to immune checkpoint inhibitors across multiple cancer types. J. Immunother. Cancer 2021, 9, e001792. [Google Scholar] [CrossRef]
- Sansregret, L.; Vanhaesebroeck, B.; Swanton, C. Determinants and clinical implications of chromosomal instability in cancer. Nat. Rev. Clin. Oncol. 2018, 15, 139–150. [Google Scholar] [CrossRef]
- Davoli, T.; Uno, H.; Wooten, E.C.; Elledge, S.J. Tumor aneuploidy correlates with markers of immune evasion and with reduced response to immunotherapy. Science 2017, 355, eaaf8399. [Google Scholar] [CrossRef]
- Dewhurst, S.M.; McGranahan, N.; Burrell, R.A.; Rowan, A.J.; Grönroos, E.; Endesfelder, D.; Joshi, T.; Mouradov, D.; Gibbs, P.; Ward, R.L.; et al. Tolerance of Whole-Genome Doubling Propagates Chromosomal Instability and Accelerates Cancer Genome Evolution. Cancer Discov. 2014, 4, 175–185. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.; Nguyen, T.H.M.; Moka, S.B.; Ellis, J.J.; Grady, J.P.; Oey, H.; Cristino, A.S.; Khanna, K.K.; Kroese, D.P.; Krause, L.; et al. Chromosome arm aneuploidies shape tumour evolution and drug response. Nat. Commun. 2020, 11, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ben-David, U.; Amon, A. Context is everything: Aneuploidy in cancer. Nat. Rev. Genet. 2019, 21, 44–62. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Wang, D.; Yang, X.; Wang, A.; Lin, Y.; Zheng, M.; Zhang, H.; Sang, X.; Wang, H.; Hu, K.; et al. Identification of NOTCH4 mutation as a response biomarker for immune checkpoint inhibitor therapy. BMC Med. 2021, 19, 154. [Google Scholar] [CrossRef] [PubMed]
- Long, J.; Wang, D.; Wang, A.; Chen, P.; Lin, Y.; Bian, J.; Yang, X.; Zheng, M.; Zhang, H.; Zheng, Y.; et al. A mutation-based gene set predicts survival benefit after immunotherapy across multiple cancers and reveals the immune response landscape. Genome Med. 2022, 14, 1–21. [Google Scholar] [CrossRef]
- Correale, P.; Rotundo, M.S.; Del Vecchio, M.T.; Remondo, C.; Migali, C.; Ginanneschi, C.; Tsang, K.Y.; Licchetta, A.; Mannucci, S.; Loiacono, L.; et al. Regulatory (FoxP3+) T-cell Tumor Infiltration Is a Favorable Prognostic Factor in Advanced Colon Cancer Patients Undergoing Chemo or Chemoimmunotherapy. J. Immunother. 2010, 33, 435–441. [Google Scholar] [CrossRef]
- Koh, J.; Hur, J.Y.; Lee, K.Y.; Kim, M.S.; Heo, J.Y.; Ku, B.M.; Sun, J.-M.; Lee, S.-H.; Ahn, J.S.; Park, K.; et al. Regulatory (FoxP3+) T cells and TGF-β predict the response to anti-PD-1 immunotherapy in patients with non-small cell lung cancer. Sci. Rep. 2020, 10, 18994. [Google Scholar] [CrossRef]
- West, N.; Kost, S.E.; Martin, S.; Milne, K.; DeLeeuw, R.J.; Nelson, B.; Watson, P.H. Tumour-infiltrating FOXP3+ lymphocytes are associated with cytotoxic immune responses and good clinical outcome in oestrogen receptor-negative breast cancer. Br. J. Cancer 2012, 108, 155–162. [Google Scholar] [CrossRef]
- Wu, S.-P.; Liao, R.-Q.; Tu, H.-Y.; Wang, W.-J.; Dong, Z.-Y.; Huang, S.-M.; Guo, W.-B.; Gou, L.-Y.; Sun, H.-W.; Zhang, Q.; et al. Stromal PD-L1–Positive Regulatory T cells and PD-1–Positive CD8-Positive T cells Define the Response of Different Subsets of Non–Small Cell Lung Cancer to PD-1/PD-L1 Blockade Immunotherapy. J. Thorac. Oncol. 2017, 13, 521–532. [Google Scholar] [CrossRef]



| Characteristics | All (n = 125) | Co-Wild (n = 35, 28%) | Mutant (n = 90, 72%) | P (Fisher Test) | |
|---|---|---|---|---|---|
| Age (years) | |||||
| 30–60 | 33 (26%) | 6 (17%) | 27 (30%) | 0.18 | |
| 60–90 | 92 (74%) | 29 (83%) | 63 (70%) | ||
| Sex | |||||
| Male | 103 (82%) | 25 (71%) | 78 (87%) | 0.07 | |
| Female | 22 (18%) | 10 (29%) | 12 (13%) | ||
| Smoking | |||||
| Current | 23 (18%) | 5 (14%) | 18 (20%) | 0.22 | |
| Previous | 95 (76%) | 26 (74%) | 69 (77%) | ||
| Never | 7 (6%) | 4 (11%) | 3 (3%) | ||
| ECOG-PS | |||||
| 0 | 37 (30%) | 11 (31%) | 26 (29%) | 0.83 | |
| 1 | 88 (70%) | 24 (69%) | 64 (71%) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yu, J.; Fan, Z.; Zhou, Z.; Zhang, P.; Bai, J.; Li, X.; Tang, M.; Fan, N.; Wu, X.; Nie, X.; et al. TP53 and LRP1B Co-Wild Predicts Improved Survival for Patients with LUSC Receiving Anti-PD-L1 Immunotherapy. Cancers 2022, 14, 3382. https://doi.org/10.3390/cancers14143382
Yu J, Fan Z, Zhou Z, Zhang P, Bai J, Li X, Tang M, Fan N, Wu X, Nie X, et al. TP53 and LRP1B Co-Wild Predicts Improved Survival for Patients with LUSC Receiving Anti-PD-L1 Immunotherapy. Cancers. 2022; 14(14):3382. https://doi.org/10.3390/cancers14143382
Chicago/Turabian StyleYu, Jiangyong, Zaiwen Fan, Zhipeng Zhou, Ping Zhang, Jing Bai, Xu Li, Min Tang, Nannan Fan, Xiaonan Wu, Xin Nie, and et al. 2022. "TP53 and LRP1B Co-Wild Predicts Improved Survival for Patients with LUSC Receiving Anti-PD-L1 Immunotherapy" Cancers 14, no. 14: 3382. https://doi.org/10.3390/cancers14143382
APA StyleYu, J., Fan, Z., Zhou, Z., Zhang, P., Bai, J., Li, X., Tang, M., Fan, N., Wu, X., Nie, X., Chen, X., Ma, D., Chen, X., Cui, L., Xia, X., Yang, L., Yi, X., & Li, L. (2022). TP53 and LRP1B Co-Wild Predicts Improved Survival for Patients with LUSC Receiving Anti-PD-L1 Immunotherapy. Cancers, 14(14), 3382. https://doi.org/10.3390/cancers14143382






