Immunophenotypic and Molecular Features of Acute Myeloid Leukemia with Plasmacytoid Dendritic Cell Differentiation Are Distinct from Blastic Plasmacytoid Dendritic Cell Neoplasm
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort
2.2. Immunophenotypic Analysis by Flow Cytometry
2.3. Immunophenotypic Analysis by Immunohistochemistry
2.4. Next Generation Sequencing Analysis
2.5. Statistical Analysis
3. Results
3.1. Clinicopathologic Characteristics
3.2. Histopathologic Features of pDCs in pDC-AML
3.3. Immunophenotype of pDCs in pDC-AML
3.4. Immunophenotypic Differences of pDCs in pDC-AML and BPDCN
3.5. Immunophenotype of Myeloblasts in pDC-AML by Flow Cytometry
3.6. Immunophenotype of pDCs and Myeloblasts in pDC-AML by Immunohistochemistry
3.7. Molecular Profile of pDC-AML
3.8. Genetic Differences between pDC-AML and BPDCN
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Reizis, B. Plasmacytoid Dendritic Cells: Development, Regulation, and Function. Immunity 2019, 50, 37–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodrigues, P.; Alberti-Servera, L.; Eremin, A.; Grajales-Reyes, G.E.; Ivanek, R.; Tussiwand, R. Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat. Immunol. 2018, 19, 711–722. [Google Scholar] [CrossRef]
- Sathe, P.; Vremec, D.; Wu, L.; Corcoran, L.; Shortman, K. Convergent differentiation: Myeloid and lymphoid pathways to murine plasmacytoid dendritic cells. Blood 2013, 121, 11–19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shigematsu, H.; Reizis, B.; Iwasaki, H.; Mizuno, S.-I.; Hu, D.; Traver, D.; Leder, P.; Sakaguchi, N.; Akashi, K. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity 2004, 21, 43–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, J.; Pucella, J.N.; Jang, G.; Alcántara-Hernández, M.; Upadhaya, S.; Adams, N.M.; Khodadadi-Jamayran, A.; Lau, C.M.; Stoeckius, M.; Hao, S.; et al. Clonal lineage tracing reveals shared origin of conventional and plasmacytoid dendritic cells. Immunity 2022, 55, 405–422.e11. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Chan, A.; Waarts, M.R.; Mishra, T.; Liu, Y.; Cai, S.F.; Yao, J.; Gao, Q.; Bowman, R.L.; Koche, R.P.; et al. Plasmacytoid dendritic cell expansion defines a distinct subset of RUNX1-mutated acute myeloid leukemia. Blood 2021, 137, 1377–1391. [Google Scholar] [CrossRef]
- Hamadeh, F.; Awadallah, A.; Meyerson, H.J.; Beck, R.C. Flow Cytometry Identifies a Spectrum of Maturation in Myeloid Neoplasms Having Plasmacytoid Dendritic Cell Differentiation. Cytom. Part B Clin. Cytom. 2019, 98, 43–51. [Google Scholar] [CrossRef]
- Zalmaï, L.; Viailly, P.-J.; Biichle, S.; Cheok, M.; Soret, L.; Angelot-Delettre, F.; Petrella, T.; Collonge-Rame, M.-A.; Seilles, E.; Geffroy, S.; et al. Plasmacytoid dendritic cells proliferation associated with acute myeloid leukemia: Phenotype profile and mutation landscape. Haematologica 2021, 106, 3056–3066. [Google Scholar] [CrossRef]
- Martín-Martín, L.; López, A.; Vidriales, B.; Caballero, M.D.; Rodrigues, A.S.; Ferreira, S.I.; Lima, M.; Almeida, S.; Valverde, B.; Martínez, P.; et al. Classification and clinical behavior of blastic plasmacytoid dendritic cell neoplasms according to their maturation-associated immunophenotypic profile. Oncotarget 2015, 6, 19204–19216. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Wang, Y.; Chang, Y.; Yuan, X.; Hao, L.; Shi, H.; Lai, Y.; Huang, X.; Liu, Y. Myeloid Neoplasms with Elevated Plasmacytoid Dendritic Cell Differentiation Reflect the Maturation Process of Dendritic Cells. Cytom. Part A 2019, 97, 61–69. [Google Scholar] [CrossRef]
- Wang, W.; Khoury, J.D.; Miranda, R.N.; Jorgensen, J.L.; Xu, J.; Loghavi, S.; Li, S.; Pemmaraju, N.; Nguyen, T.; Medeiros, L.J.; et al. Immunophenotypic characterization of reactive and neoplastic plasmacytoid dendritic cells permits establishment of a 10-color flow cytometric panel for initial workup and residual disease evaluation of blastic plasmacytoid dendritic cell neoplasm. Haematologica 2021, 106, 1047–1055. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, C.C.; Pemmaraju, N.; You, M.J.; Li, S.; Xu, J.; Wang, W.; Tang, Z.; Alswailmi, O.; Bhalla, K.N.; Qazilbash, M.H.; et al. Integrated Clinical Genotype-Phenotype Characteristics of Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancers 2021, 13, 5888. [Google Scholar] [CrossRef] [PubMed]
- Sukswai, N.; Aung, P.; Yin, C.C.; Li, S.; Wang, W.; Wang, S.A.; Ortega, V.; Lyapichev, K.; Nagarajan, P.; Alfattal, R.; et al. Dual Expression of TCF4 and CD123 Is Highly Sensitive and Specific For Blastic Plasmacytoid Dendritic Cell Neoplasm. Am. J. Surg. Pathol. 2019, 43, 1429–1437. [Google Scholar] [CrossRef] [PubMed]
- Patel, K.P.; Ruiz-Cordero, R.; Chen, W.; Routbort, M.J.; Floyd, K.; Rodriguez, S.; Galbincea, J.; Barkoh, B.A.; Hatfield, D.; Khogeer, H.; et al. Ultra-Rapid Reporting of GENomic Targets (URGENTseq): Clinical Next-Generation Sequencing Results within 48 Hours of Sample Collection. J. Mol. Diagn. 2019, 21, 89–98. [Google Scholar] [CrossRef]
- Martín-Martín, L.; Almeida, J.; Hernández-Campo, P.M.; Sánchez, M.L.; Lécrevisse, Q.; Orfao, A. Immunophenotypical, morphologic, and functional characterization of maturation-associated plasmacytoid dendritic cell subsets in normal adult human bone marrow. Transfusion 2009, 49, 1692–1708. [Google Scholar] [CrossRef] [PubMed]
- Lucas, N.; Duchmann, M.; Rameau, P.; Noël, F.; Michea, P.; Saada, V.; Kosmider, O.; Pierron, G.; Fernandez-Zapico, M.E.; Howard, M.T.; et al. Biology and prognostic impact of clonal plasmacytoid dendritic cells in chronic myelomonocytic leukemia. Leukemia 2019, 33, 2466–2480. [Google Scholar] [CrossRef]
- William, V.; Fabio, F.; Stefano, R.; Federica, V.; Elisa, R.; Silvana, F.; Daniele, R.; Piergiovanni, G.; Giovannino, M.; Glauco, F. Nodal and extranodal tumor-forming accumulation of plasmacytoid monocytes/interferon-producing cells associated with myeloid disorders. Am. J. Surg. Pathol. 2004, 28, 585–595. [Google Scholar]
- Dress, R.J.; Dutertre, C.-A.; Giladi, A.; Schlitzer, A.; Low, I.; Shadan, N.B.; Tay, A.; Lum, J.; Kairi, M.F.B.M.; Hwang, Y.Y.; et al. Plasmacytoid dendritic cells develop from Ly6D(+) lymphoid progenitors distinct from the myeloid lineage. Nat. Immunol. 2019, 20, 852–864. [Google Scholar] [CrossRef]
- Chaperot, L.; Bendriss, N.; Manches, O.; Gressin, R.; Maynadie, M.; Trimoreau, F.; Orfeuvre, H.; Corront, B.; Feuillard, J.; Sotto, J.-J.; et al. Identification of a leukemic counterpart of the plasmacytoid dendritic cells. Blood 2001, 97, 3210–3217. [Google Scholar] [CrossRef] [Green Version]
- Sapienza, M.R.; Fuligni, F.; Agostinelli, C.; Tripodo, C.; Righi, S.; Laginestra, M.A.; Pileri, A., Jr.; Mancini, M.; Rossi, M.; Ricci, F.; et al. Molecular profiling of blastic plasmacytoid dendritic cell neoplasm reveals a unique pattern and suggests selective sensitivity to NF-kB pathway inhibition. Leukemia 2014, 28, 1606–1616. [Google Scholar] [CrossRef] [Green Version]
- Pagano, L.; Valentini, C.G.; Grammatico, S.; Pulsoni, A. Blastic plasmacytoid dendritic cell neoplasm: Diagnostic criteria and therapeutical approaches. Br. J. Haematol. 2016, 174, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Alayed, K.; Patel, K.P.; Konoplev, S.; Singh, R.R.; Routbort, M.J.; Reddy, N.; Pemmaraju, N.; Zhang, L.; Al Shaikh, A.; Aladily, T.N.; et al. TET2 mutations, myelodysplastic features, and a distinct immunoprofile characterize blastic plasmacytoid dendritic cell neoplasm in the bone marrow. Am. J. Hematol. 2013, 88, 1055–1061. [Google Scholar] [CrossRef] [PubMed]
- Khoury, J.D.; Medeiros, L.J.; Manning, J.T.; Sulak, L.E.; Bueso-Ramos, C.; Jones, D. CD56(+) TdT(+) blastic natural killer cell tumor of the skin: A primitive systemic malignancy related to myelomonocytic leukemia. Cancer 2002, 94, 2401–2408. [Google Scholar] [CrossRef]
- Khanlari, M.; Yin, C.C.; Takahashi, K.; Lachowiez, C.; Tang, G.; Loghavi, S.; Bah, I.; Wang, W.; Konoplev, S.; Medeiros, L.J.; et al. Bone marrow clonal hematopoiesis is highly prevalent in blastic plasmacytoid dendritic cell neoplasm and frequently sharing a clonal origin in elderly patients. Leukemia 2022, 36, 1343–1350. [Google Scholar] [CrossRef]
- Batta, K.; Bossenbroek, H.M.; Pemmaraju, N.; Wilks, D.P.; Chasty, R.; Dennis, M.; Milne, P.; Collin, M.; Beird, H.C.; Taylor, J.; et al. Divergent clonal evolution of blastic plasmacytoid dendritic cell neoplasm and chronic myelomonocytic leukemia from a shared TET2-mutated origin. Leukemia 2021, 35, 3299–3303. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, L.; Di Battista, V.; Venanzi, A.; Schiavoni, G.; Martelli, M.P.; Ascani, S.; Mecucci, C.; Tiacci, E.; Falini, B. Blastic plasmacytoid dendritic cell neoplasm and chronic myelomonocytic leukemia: A shared clonal origin. Leukemia 2017, 31, 1238–1240. [Google Scholar] [CrossRef]
- Patnaik, M.M.; Lasho, T.; Howard, M.; Finke, C.; Ketterling, R.L.; Al-Kali, A.; Pardanani, A.; Droin, N.; Gangat, N.; Tefferi, A.; et al. Biallelic inactivation of the retinoblastoma gene results in transformation of chronic myelomonocytic leukemia to a blastic plasmacytoid dendritic cell neoplasm: Shared clonal origins of two aggressive neoplasms. Blood Cancer J. 2018, 8, 82. [Google Scholar] [CrossRef]

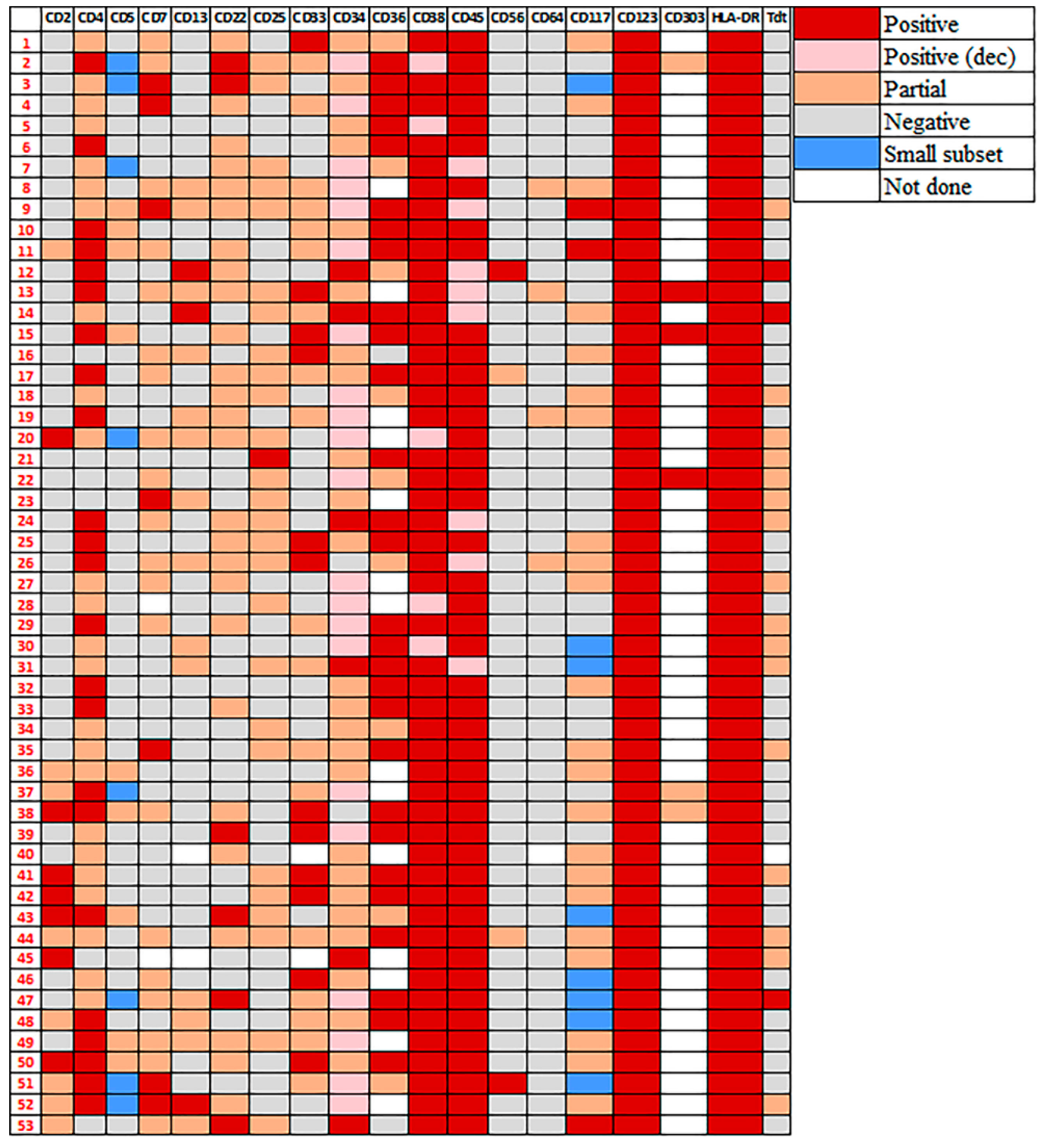
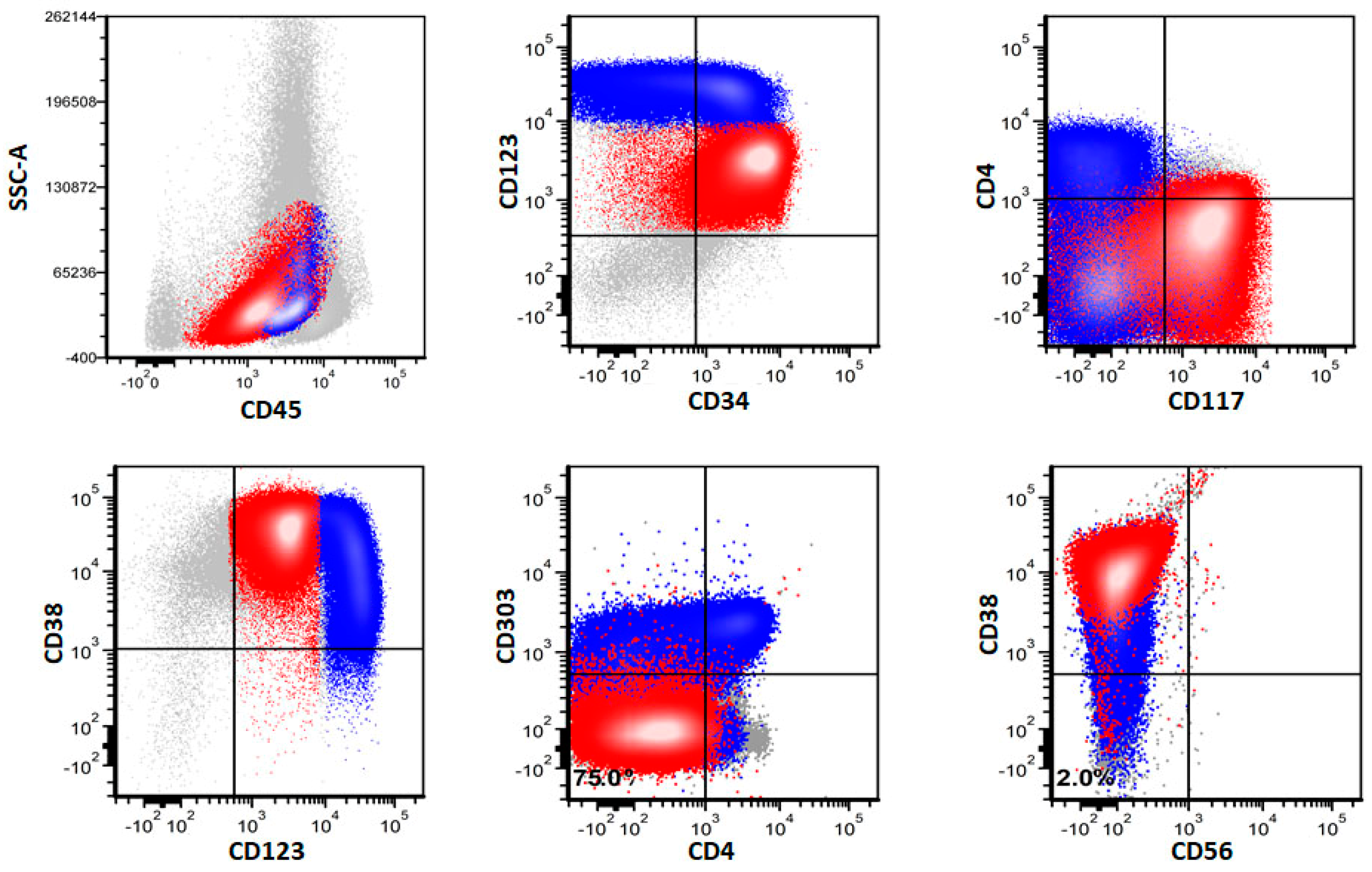

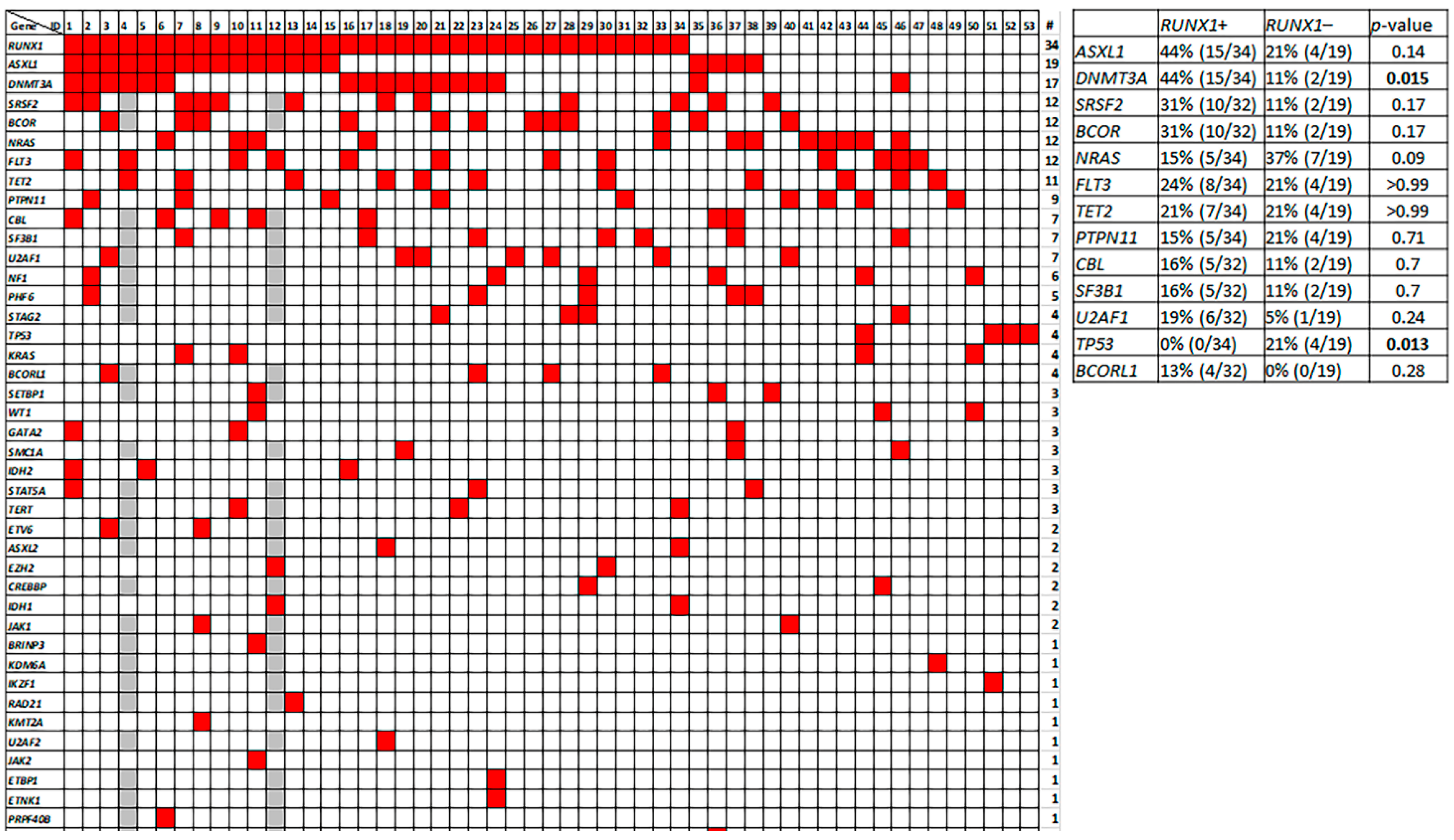
| Markers | Positivity (%) | p Value | |
|---|---|---|---|
| pDC-AML | BPDCN | ||
| CD2 | 28% (15/53) | 19% (5/27) | 0.42 |
| cCD3 | 1.9% (1/53) | 0% (0/15) | >0.99 |
| CD4 | 89% (47/53) | 100% (38/38) | 0.039 |
| CD5 * | 17% (9/53) | 3% (1/30) | 0.086 |
| CD7 | 53% (27/51) | 64% (21/33) | 0.37 |
| CD13 | 33% (17/51) | 0% (0/30) | 0.0001 |
| CD14 | 0% (0/53) | 3% (1/34) | 0.39 |
| CD15 | 0% (0/53) | 0% (0/28) | >0.99 |
| CD19 | 0% (0/53) | 0% (0/30) | >0.99 |
| CD22 | 60% (32/53) | 0% (0/26) | <0.0001 |
| CD25 | 51% (27/53) | 0% (0/22) | <0.0001 |
| CD33 | 59% (30/51) | 48% (16/33) | 0.38 |
| CD34 | 96% (51/53) | 0% (0/27) | <0.0001 |
| CD36 | 95% (37/39) | 57% (17/30) | 0.0002 |
| CD38 | 100% (53/53) | 88% (30/34) | 0.021 |
| CD45 | 100% (53/53) | 100% (39/39) | >0.99 |
| CD56 | 8% (4/53) | 97% (36/37) | <0.0001 |
| CD64 | 8% (4/52) | 0% (0/36) | 0.14 |
| CD117 * | 47% (25/53) | 9% (3/34) | 0.0001 |
| CD123 | 100% (53/53) | 100% (36/36) | >0.99 |
| CD303 | 100% (6/6) | 44% (7/16) | 0.046 |
| HLA-DR | 100% (53/53) | 100% (36/36) | >0.99 |
| MPO | 0% (0/52) | 0% (0/16) | >0.99 |
| TdT | 37% (19/52) | 25% (4/16) | 0.55 |
| TCL1 * | 12% (2/17) | 98% (46/47) | <0.0001 |
| pDC-AML | BPDCN | p Value | |
|---|---|---|---|
| RUNX1 | 64% (34/53) | 2% (1/50) | <0.0001 |
| ASXL1 | 36% (19/53) | 46% (23/50) | 0.32 |
| DNMT3A | 32% (17/53) | 10% (5/50) | 0.0079 |
| SRSF2 | 24% (12/51) | 9% (3/32) | 0.14 |
| BCOR | 24% (12/51) | 0% (0/32) | 0.0026 |
| NRAS | 23% (12/53) | 10% (5/50) | 0.11 |
| FLT3 | 23% (12/53) | 0% (0/50) | 0.0003 |
| TET2 | 21% (11/53) | 56% (28/50) | 0.0003 |
| ZRSR2 | 2% (1/51) | 16% (5/32) | 0.030 |
| IKZF1 | 2% (1/51) | 9% (3/32) | 0.29 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, W.; Xu, J.; Khoury, J.D.; Pemmaraju, N.; Fang, H.; Miranda, R.N.; Yin, C.C.; Hussein, S.E.; Jia, F.; Tang, Z.; et al. Immunophenotypic and Molecular Features of Acute Myeloid Leukemia with Plasmacytoid Dendritic Cell Differentiation Are Distinct from Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancers 2022, 14, 3375. https://doi.org/10.3390/cancers14143375
Wang W, Xu J, Khoury JD, Pemmaraju N, Fang H, Miranda RN, Yin CC, Hussein SE, Jia F, Tang Z, et al. Immunophenotypic and Molecular Features of Acute Myeloid Leukemia with Plasmacytoid Dendritic Cell Differentiation Are Distinct from Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancers. 2022; 14(14):3375. https://doi.org/10.3390/cancers14143375
Chicago/Turabian StyleWang, Wei, Jie Xu, Joseph D. Khoury, Naveen Pemmaraju, Hong Fang, Roberto N. Miranda, C. Cameron Yin, Siba El Hussein, Fuli Jia, Zhenya Tang, and et al. 2022. "Immunophenotypic and Molecular Features of Acute Myeloid Leukemia with Plasmacytoid Dendritic Cell Differentiation Are Distinct from Blastic Plasmacytoid Dendritic Cell Neoplasm" Cancers 14, no. 14: 3375. https://doi.org/10.3390/cancers14143375
APA StyleWang, W., Xu, J., Khoury, J. D., Pemmaraju, N., Fang, H., Miranda, R. N., Yin, C. C., Hussein, S. E., Jia, F., Tang, Z., Hu, S., Konopleva, M., Medeiros, L. J., & Wang, S. A. (2022). Immunophenotypic and Molecular Features of Acute Myeloid Leukemia with Plasmacytoid Dendritic Cell Differentiation Are Distinct from Blastic Plasmacytoid Dendritic Cell Neoplasm. Cancers, 14(14), 3375. https://doi.org/10.3390/cancers14143375









