Homo and Heterotypic Cellular Cross-Talk in Epithelial Ovarian Cancer Impart Pro-Tumorigenic Properties through Differential Activation of the Notch3 Pathway
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Reagents and Antibodies
2.2. Cell Lines & Notch3 Reporter Sensor
2.3. Co-Culture Assay
2.4. Immunofluorescence and Western Blotting
2.5. Luciferase Assay and Imaging
2.6. Flow Cytometry
2.7. Cell Invasion Assay
2.8. Cell Cytotoxicity Assay
2.9. VEGFA Enzyme-Linked Immunosorbent Assay (ELISA)
2.10. Notch3 Immunohistochemistry
2.11. RNA Isolation
2.12. Quantitative Real-Time PCR and Notch3 Gene Profiler Array
2.13. Isolation of Mesothelial Cells from Patients
2.14. Statistical Analysis
3. Results
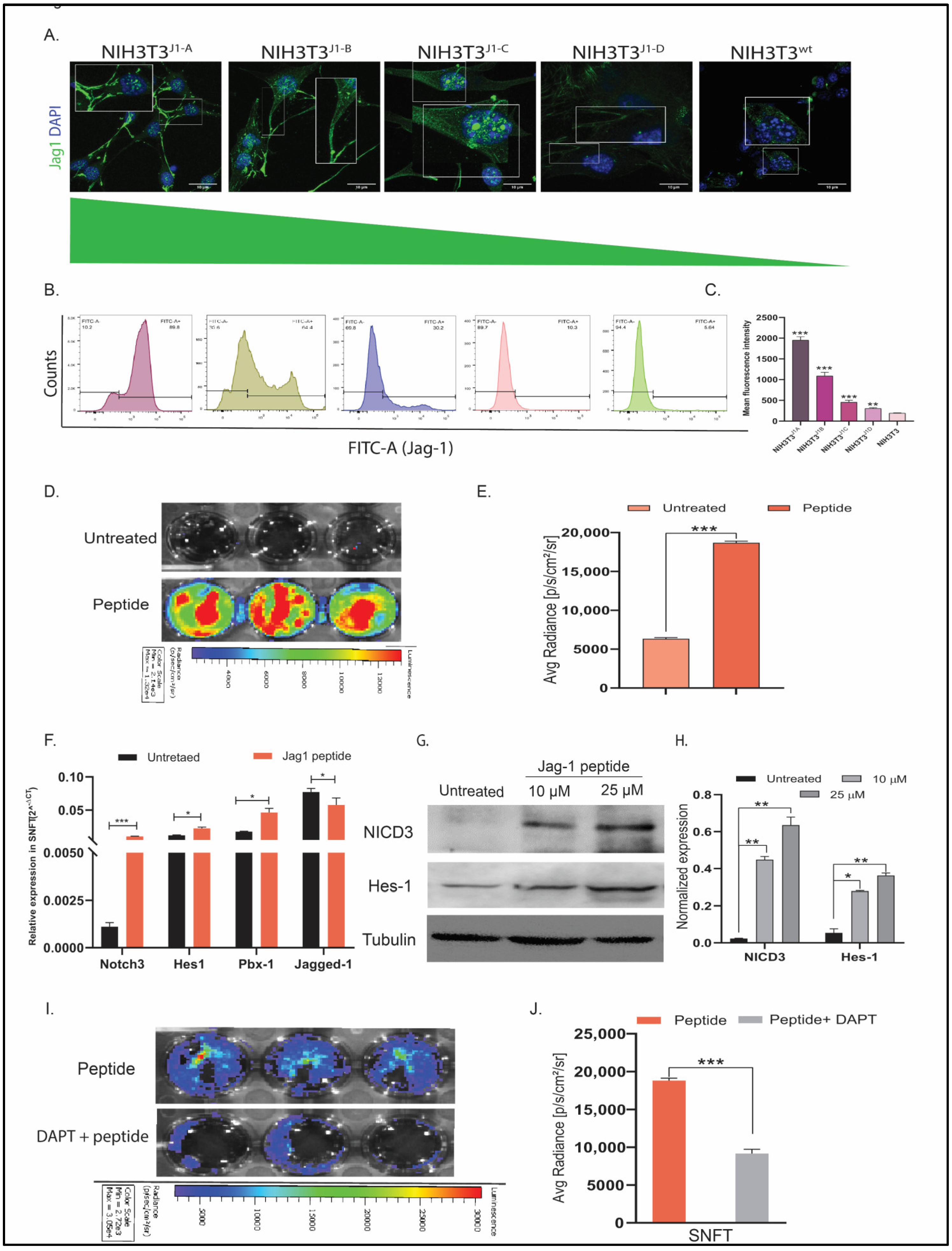
3.1. Establishment of Differential NIH3T3jag1 Clones and Notch3 Reporter Sensor
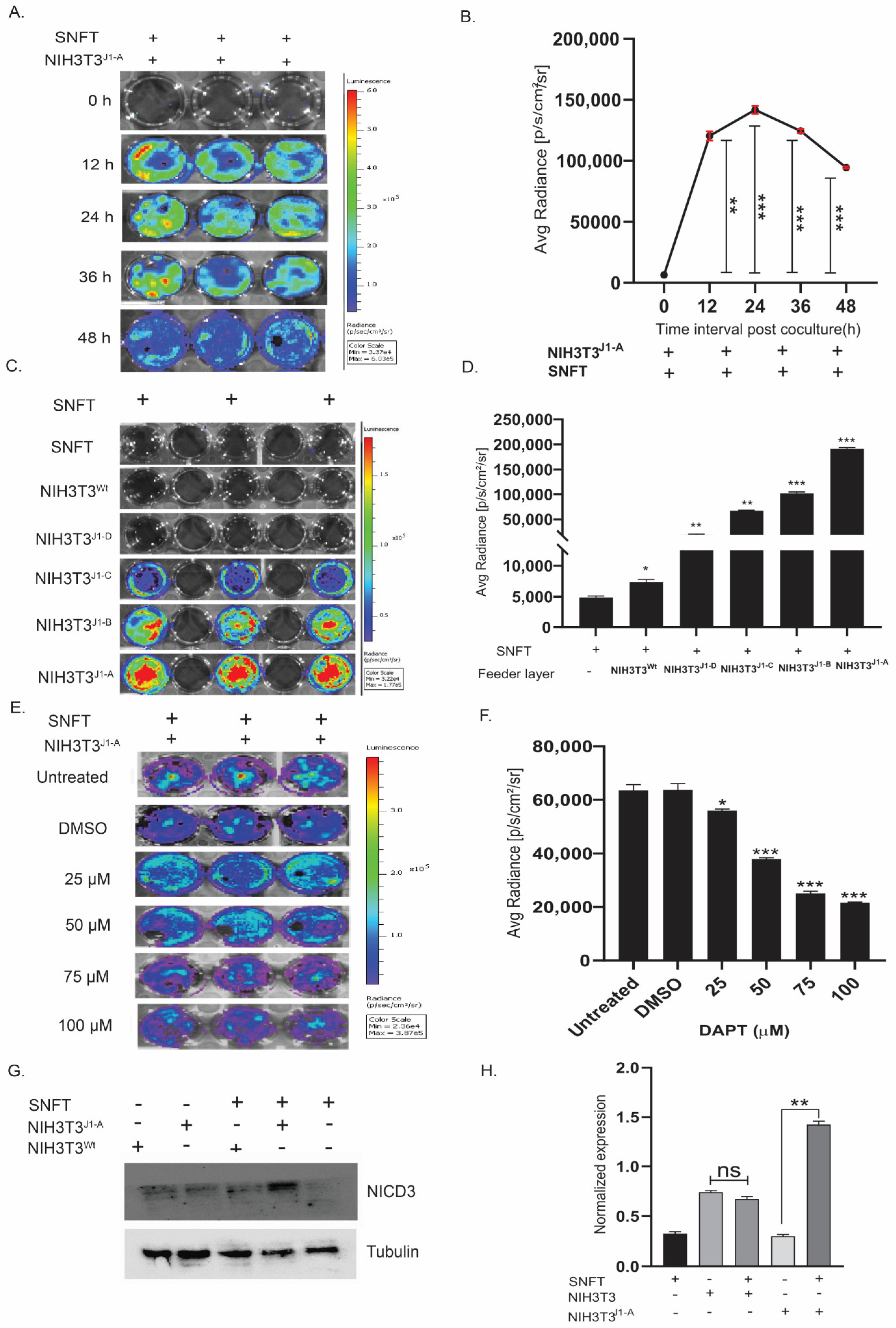
3.2. Establishment of a Unique Co-Culture Model of Differential Notch3 Activation by SNFT and NIH3T3Jag1 Cells
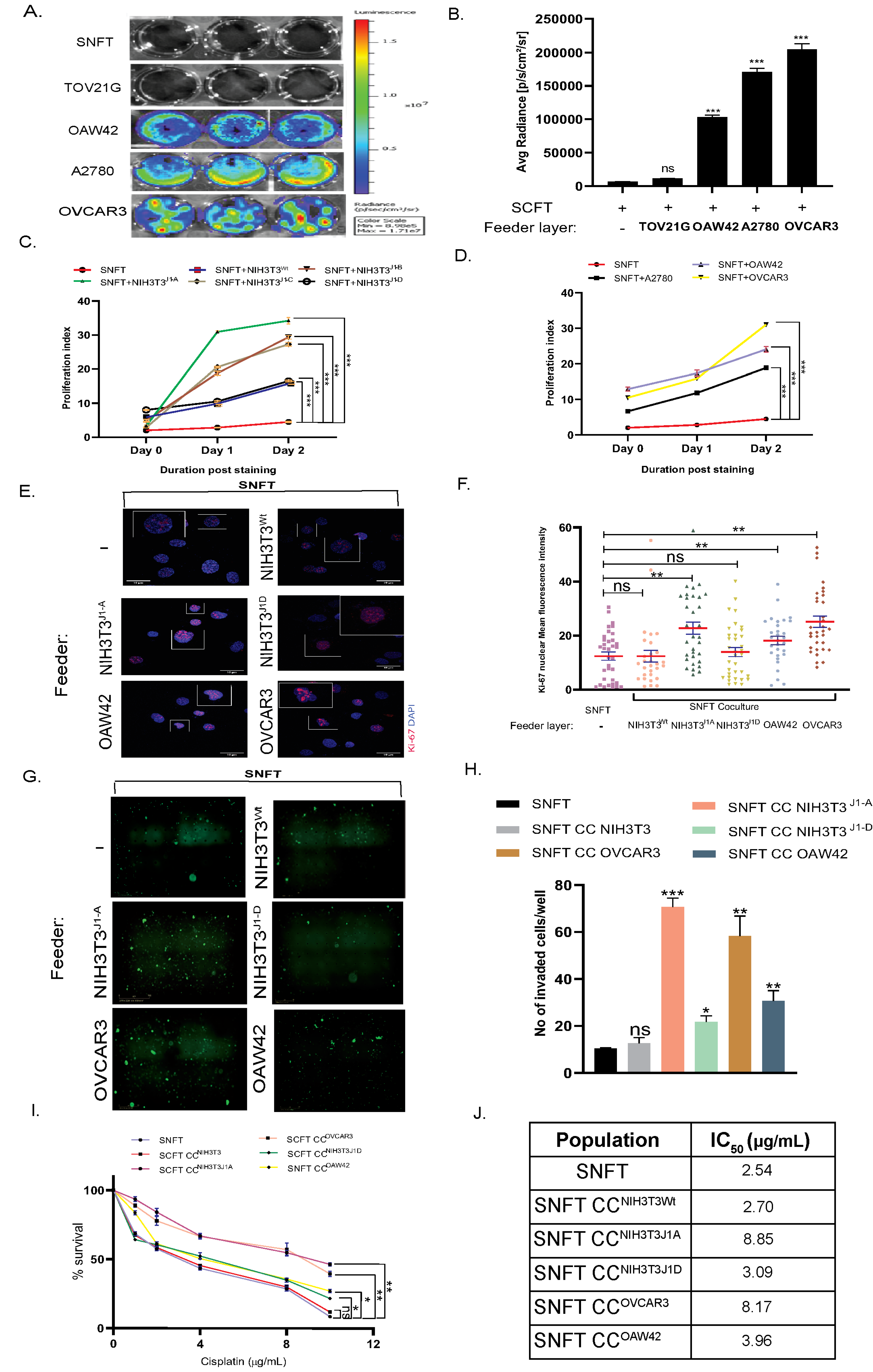
3.3. Differential Induction of Notch3 by Homo/Heterotypic Cellular Interactions Leads to Differential Modulation in Proliferation, Invasiveness, and Cisplatin Sensitivity in SNFT
3.4. Cancer-Associated Fibroblasts (CAF) Induce Notch3 Activation in the Co-Culture Model
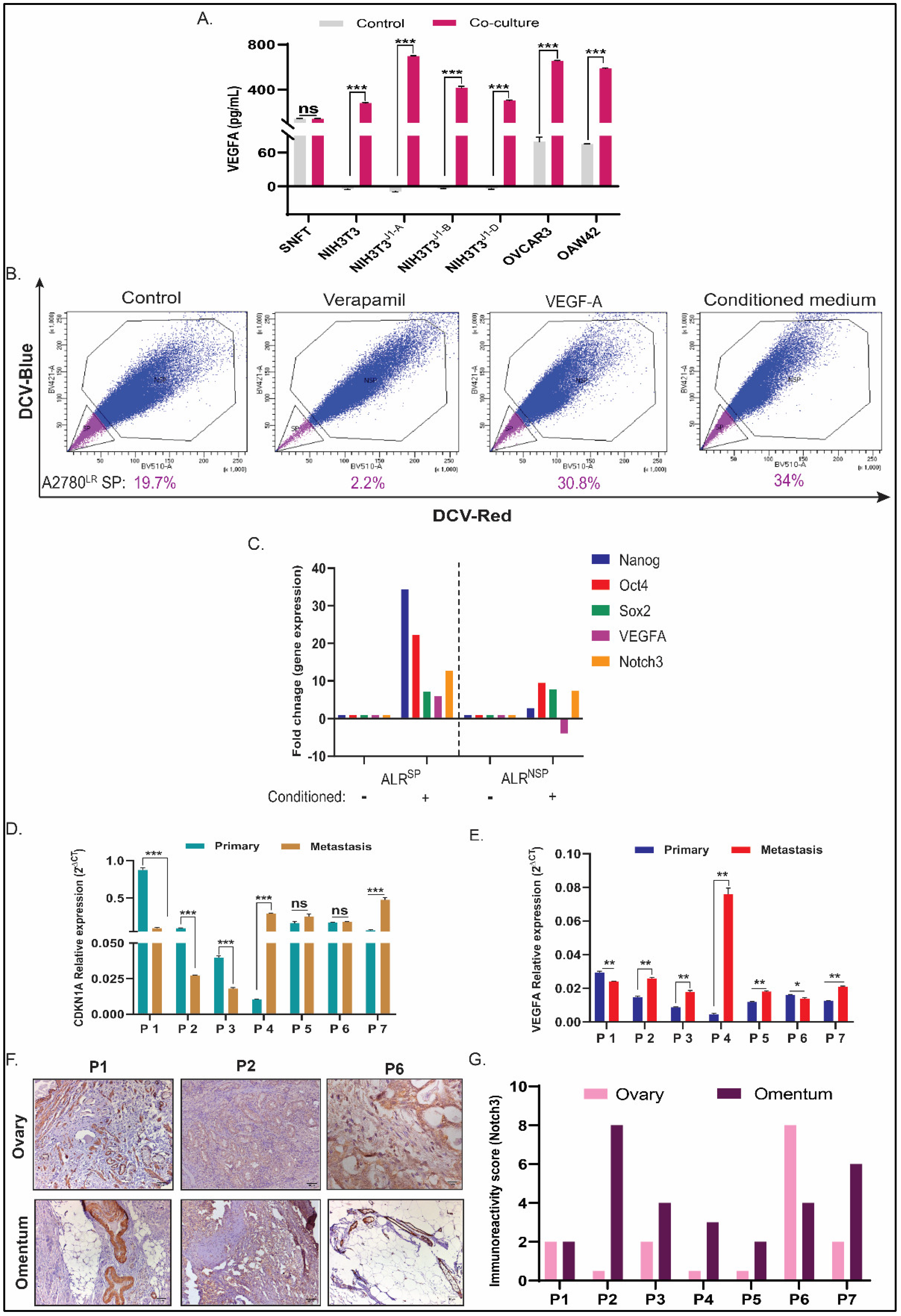
3.5. CDKN1A and VEGFA Are Two Key Differential Genes (DGs) in SNFT Post Homotypic/Heterotypic Activation of the Notch3 Pathway
3.6. VEGFA Regulates CSC-Non CSC Turnover in A2780LR Cells, and Its Expression along with CDK2N1A Correlates with Notch3 in Metastatic HGSOC Tumors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pogge von Strandmann, E.; Reinartz, S.; Wager, U.; Muller, R. Tumor-Host Cell Interactions in Ovarian Cancer: Pathways to Therapy Failure. Trends Cancer 2017, 3, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Armingol, E.; Officer, A.; Harismendy, O.; Lewis, N.E. Deciphering cell-cell interactions and communication from gene expression. Nat. Rev. Genet. 2021, 22, 71–88. [Google Scholar] [CrossRef] [PubMed]
- Kusamura, S.; Baratti, D.; Zaffaroni, N.; Villa, R.; Laterza, B.; Balestra, M.R.; Deraco, M. Pathophysiology and biology of peritoneal carcinomatosis. World J. Gastrointest. Oncol. 2010, 2, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Tallapragada, S.; Schaar, B.; Kamat, K.; Chanana, A.M.; Zhang, Y.; Patel, S.; Parkash, V.; Rinker-Schaeffer, C.; Folkins, A.K.; et al. Omental macrophages secrete chemokine ligands that promote ovarian cancer colonization of the omentum via CCR1. Commun. Biol. 2020, 3, 524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, N.; Garcia, J.R.; Mohamed, A.; Benencia, F.; Rubin, S.C.; Allman, D.; Coukos, G. Generation of a syngeneic mouse model to study the effects of vascular endothelial growth factor in ovarian carcinoma. Am. J. Pathol. 2002, 161, 2295–2309. [Google Scholar] [CrossRef]
- Nieman, K.M.; Kenny, H.A.; Penicka, C.V.; Ladanyi, A.; Buell-Gutbrod, R.; Zillhardt, M.R.; Romero, I.L.; Carey, M.S.; Mills, G.B.; Hotamisligil, G.S. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat. Med. 2011, 17, 1498–1503. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical praofiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Choi, J.H.; Park, J.T.; Davidson, B.; Morin, P.J.; Shih Ie, M.; Wang, T.L. Jagged-1 and Notch3 juxtacrine loop regulates ovarian tumor growth and adhesion. Cancer Res. 2008, 68, 5716–5723. [Google Scholar] [CrossRef]
- Gaikwad, S.M.; Thakur, B.; Sakpal, A.; Singh, R.K.; Ray, P. Differential activation of NF-kappaB signaling is associated with platinum and taxane resistance in MyD88 deficient epithelial ovarian cancer cells. Int. J. Biochem. Cell Biol. 2015, 61, 90–102. [Google Scholar] [CrossRef]
- Hansson, E.M.; Teixeira, A.I.; Gustafsson, M.V.; Dohda, T.; Chapman, G.; Meletis, K.; Muhr, J.; Lendahl, U. Recording Notch signaling in real time. Dev. Neurosci. 2006, 28, 118–127. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Dhadve, A.; Sakpal, A.; De, A.; Ray, P. An active IGF-1R-AKT signaling imparts functional heterogeneity in ovarian CSC population. Sci. Rep. 2016, 6, 36612. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Liu, T.; Ivan, C.; Sun, Y.; Huang, J.; Mangala, L.S.; Miyake, T.; Dalton, H.J.; Pradeep, S.; Rupaimoole, R. Notch3 pathway alterations in ovarian cancer. Cancer Res. 2014, 74, 3282–3293. [Google Scholar] [CrossRef] [PubMed]
- Gao, C.F.; Xie, Q.; Su, Y.L.; Koeman, J.; Khoo, S.K.; Gustafson, M.; Knudsen, B.S.; Hay, R.; Shinomiya, N.; Vande Woude, G.F. Proliferation and invasion: Plasticity in tumor cells. Proc. Natl. Acad. Sci. USA 2005, 102, 10528–10533. [Google Scholar] [CrossRef] [PubMed]
- McAuliffe, S.M.; Morgan, S.L.; Wyant, G.A.; Tran, L.T.; Muto, K.W.; Chen, Y.S.; Chin, K.T.; Partridge, J.C.; Poole, B.B.; Cheng, K.H.; et al. Targeting Notch, a key pathway for ovarian cancer stem cells, sensitizes tumors to platinum therapy. Proc. Natl. Acad. Sci. USA 2012, 109, E2939–E2948. [Google Scholar] [CrossRef]
- Contreras-Cornejo, H.; Saucedo-Correa, G.; Oviedo-Boyso, J.; Valdez-Alarcon, J.J.; Baizabal-Aguirre, V.M.; Cajero-Juarez, M.; Bravo-Patino, A. The CSL proteins, versatile transcription factors and context dependent corepressors of the notch signaling pathway. Cell Div. 2016, 11, 12. [Google Scholar] [CrossRef]
- Zhao, D.; Pan, C.; Sun, J.; Gilbert, C.; Drews-Elger, K.; Azzam, D.J.; Picon-Ruiz, M.; Kim, M.; Ullmer, W.; El-Ashry, D.; et al. VEGF drives cancer-initiating stem cells through VEGFR-2/Stat3 signaling to upregulate Myc and Sox2. Oncogene 2015, 34, 3107–3119. [Google Scholar] [CrossRef]
- Kim, M.; Jang, K.; Miller, P.; Picon-Ruiz, M.; Yeasky, T.M.; El-Ashry, D.; Slingerland, J.M. VEGFA links self-renewal and metastasis by inducing Sox2 to repress miR-452, driving Slug. Oncogene 2017, 36, 5199–5211. [Google Scholar] [CrossRef]
- Burges, A.; Schmalfeldt, B. Ovarian cancer: Diagnosis and treatment. Dtsch. Arztebl. Int. 2011, 108, 635–641. [Google Scholar]
- Geng, X.; Chen, H.; Zhao, L.; Hu, J.; Yang, W.; Li, G.; Cheng, C.; Zhao, Z.; Zhang, T.; Li, L.; et al. Cancer-Associated Fibroblast (CAF) Heterogeneity and Targeting Therapy of CAFs in Pancreatic Cancer. Front. Cell Dev. Biol. 2021, 9, 655152. [Google Scholar] [CrossRef]
- Horowitz, M.; Esakov, E.; Rose, P.; Reizes, O. Signaling within the epithelial ovarian cancer tumor microenvironment: The challenge of tumor heterogeneity. Ann. Transl. Med. 2020, 8, 905. [Google Scholar] [CrossRef] [PubMed]
- Ilagan, M.; Lim, S.; Fulbright, M.; Piwnica-Worms, D.; Kopan, R. Real-time imaging of notch activation with a luciferase complementation-based reporter. Sci. Signal. 2011, 4, rs7. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Sun, W.; Li, X.; Wang, M.; Boyce, B.F.; Hilton, M.J.; Xing, L. Use of Hes1-GFP reporter mice to assess activity of the Hes1 promoter in bone cells under chronic inflammation. Bone 2016, 90, 80–89. [Google Scholar] [CrossRef] [PubMed]
- Basak, O.; Taylor, V. Identification of self-replicating multipotent progenitors in the embryonic nervous system by high Notch activity and Hes5 expression. Eur. J. Neurosci. 2007, 25, 1006–1022. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Huang, J.; Perrimon, N. Development of an optimized synthetic Notch receptor as an in vivo cell-cell contact sensor. Proc. Natl. Acad. Sci. USA 2017, 114, 5467–5472. [Google Scholar] [CrossRef] [PubMed]
- Morsut, L.; Roybal, K.T.; Xiong, X.; Gordley, R.M.; Coyle, S.M.; Thomson, M.; Lim, W.A. Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Cell 2016, 164, 780–791. [Google Scholar] [CrossRef]
- D’Souza, B.; Meloty-Kapella, L.; Weinmaster, G. Canonical and non-canonical Notch ligands. Curr. Top. Dev. Biol. 2010, 92, 73–129. [Google Scholar]
- Park, J.T.; Chen, X.; Trope, C.G.; Davidson, B.; Shih Ie, M.; Wang, T.L. Notch3 overexpression is related to the recurrence of ovarian cancer and confers resistance to carboplatin. Am. J. Pathol. 2010, 177, 1087–1094. [Google Scholar] [CrossRef]
- Zhou, J.X.; Zhou, L.; Li, Q.J.; Feng, W.; Wang, P.M.; Li, E.F.; Gong, W.J.; Kou, M.W.; Gou, W.T.; Yang, Y.L. Association between high levels of Notch3 expression and high invasion and poor overall survival rates in pancreatic ductal adenocarcinoma. Oncol. Rep. 2016, 36, 2893–2901. [Google Scholar] [CrossRef]
- Xiu, M.; Wang, Y.; Li, B.; Wang, X.; Xiao, F.; Chen, S.; Zhang, L.; Zhou, B.; Hua, F. The Role of Notch3 Signaling in Cancer Stemness and Chemoresistance: Molecular Mechanisms and Targeting Strategies. Front. Mol. Biosci. 2021, 8, 694141. [Google Scholar] [CrossRef]
- Pekkonen, P.; Alve, S.; Balistreri, G.; Gramolelli, S.; Tatti-Bugaeva, O.; Paatero, I.; Niiranen, O.; Tuohinto, K.; Perala, N.; Taiwo, A.; et al. Lymphatic endothelium stimulates melanoma metastasis and invasion via MMP14-dependent Notch3 and beta1-integrin activation. eLife 2018, 7, e32490. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.Q.; Liang, Y.K.; Wu, Y.; Chen, M.; Chen, W.L.; Li, R.H.; Zeng, Y.Z.; Huang, W.H.; Wu, J.D.; Zeng, D.; et al. Notch3 inhibits cell proliferation and tumorigenesis and predicts better prognosis in breast cancer through transactivating PTEN. Cell Death Dis. 2021, 12, 502. [Google Scholar] [CrossRef] [PubMed]
- Tai, M.H.; Weng, C.H.; Mon, D.P.; Hu, C.Y.; Wu, M.H. Ultraviolet C irradiation induces different expression of cyclooxygenase 2 in NIH 3T3 cells and A431 cells: The roles of COX-2 are different in various cell lines. Int. J. Mol. Sci. 2012, 13, 4351–4366. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.D.; Pan, L.Y.; Yuan, Y.; Lang, J.H.; Mao, N. Identification of platinum-resistance associated proteins through proteomic analysis of human ovarian cancer cells and their platinum-resistant sublines. J. Proteome Res. 2007, 6, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef]
- Katoh, M. Precision medicine for human cancers with Notch signaling dysregulation (Review). Int. J. Mol. Med. 2020, 45, 279–297. [Google Scholar] [CrossRef]
- Perchet, T.; Petit, M.; Banchi, E.G.; Meunier, S.; Cumano, A.; Golub, R. The Notch Signaling Pathway Is Balancing Type 1 Innate Lymphoid Cell Immune Functions. Front. Immunol. 2018, 9, 1252. [Google Scholar] [CrossRef]
- Gilding, L.N.; Somervaille, T.C.P. The Diverse Consequences of FOXC1 Deregulation in Cancer. Cancers 2019, 11, 184. [Google Scholar] [CrossRef]
- Wang, L.Y.; Li, L.S.; Yang, Z. Correlation of FOXC1 protein with clinicopathological features in serous ovarian tumors. Oncol. Lett. 2016, 11, 933–938. [Google Scholar] [CrossRef]
- Zhou, J.; Cheng, Y.; Tang, L.; Martinka, M.; Kalia, S. Up-regulation of SERPINA3 correlates with high mortality of melanoma patients and increased migration and invasion of cancer cells. Oncotarget 2017, 8, 18712–18725. [Google Scholar] [CrossRef]
- Senger, D.R. Vascular endothelial growth factor: Much more than an angiogenesis factor. Mol. Biol. Cell 2010, 21, 377–379. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Zhu, Y.; Xie, K.; Zhang, X.; Zhi, X.; Wang, W.; Li, Z.; Zhang, Q.; Wang, L.; Wang, J.; et al. The role of the AMOP domain in MUC4/Y-promoted tumour angiogenesis and metastasis in pancreatic cancer. J. Exp. Clin. Cancer Res. 2016, 35, 91. [Google Scholar] [CrossRef] [PubMed]
- Jang, K.; Kim, M.; Gilbert, C.A.; Simpkins, F.; Ince, T.A.; Slingerland, J.M. VEGFA activates an epigenetic pathway upregulating ovarian cancer-initiating cells. EMBO Mol. Med. 2017, 9, 304–318. [Google Scholar] [CrossRef] [PubMed]
- Hung, T.H.; Hsu, S.C.; Cheng, C.Y.; Choo, K.B.; Tseng, C.P.; Chen, T.C.; Lan, Y.W.; Huang, T.T.; Lai, H.C.; Chen, C.M.; et al. Wnt5A regulates ABCB1 expression in multidrug-resistant cancer cells through activation of the non-canonical PKA/beta-catenin pathway. Oncotarget 2014, 5, 12273–12290. [Google Scholar] [CrossRef] [PubMed]
- Rother, K.; Kirschner, R.; Sanger, K.; Bohlig, L.; Mossner, J.; Engeland, K. p53 downregulates expression of the G1/S cell cycle phosphatase Cdc25A. Oncogene 2007, 26, 1949–1953. [Google Scholar] [CrossRef] [PubMed]
- Qiu, R.; Wang, S.; Feng, X.; Chen, F.; Yang, K.; He, S. Effect of subcellular localization of P21 on proliferation and apoptosis of HepG2 cells. J. Huazhong Univ. Sci. Technol. Med. Sci. 2011, 31, 756–761. [Google Scholar] [CrossRef]
- Zhou, B.P.; Liao, Y.; Xia, W.; Spohn, B.; Lee, M.H.; Hung, M.C. Cytoplasmic localization of p21Cip1/WAF1 by Akt-induced phosphorylation in HER-2/neu-overexpressing cells. Nat. Cell Biol. 2001, 3, 245–252. [Google Scholar] [CrossRef]
- Pack, L.R.; Daigh, L.H.; Chung, M.; Meyer, T. Clinical CDK4/6 inhibitors induce selective and immediate dissociation of p21 from cyclin D-CDK4 to inhibit CDK2. Nat. Commun. 2021, 12, 3356. [Google Scholar] [CrossRef]
- Liu, Z.H.; Dai, X.M.; Du, B. Hes1: A key role in stemness, metastasis and multidrug resistance. Cancer Biol. Ther. 2015, 16, 353–359. [Google Scholar] [CrossRef]
- Wang, T.; Holt, C.M.; Xu, C.; Ridley, C.; Jones, R.P.; Baron, M.; Trump, D. Notch3 activation modulates cell growth behaviour and cross-talk to Wnt/TCF signalling pathway. Cell Signal. 2007, 19, 2458–2467. [Google Scholar] [CrossRef]
- Bali, A.; O’Brien, P.M.; Edwards, L.S.; Sutherland, R.L.; Hacker, N.F.; Henshall, S.M. Cyclin D1, p53, and p21Waf1/Cip1 expression is predictive of poor clinical outcome in serous epithelial ovarian cancer. Clin. Cancer Res. 2004, 10, 5168–5177. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mukherjee, S.; Sakpal, A.; Mehrotra, M.; Phadte, P.; Rekhi, B.; Ray, P. Homo and Heterotypic Cellular Cross-Talk in Epithelial Ovarian Cancer Impart Pro-Tumorigenic Properties through Differential Activation of the Notch3 Pathway. Cancers 2022, 14, 3365. https://doi.org/10.3390/cancers14143365
Mukherjee S, Sakpal A, Mehrotra M, Phadte P, Rekhi B, Ray P. Homo and Heterotypic Cellular Cross-Talk in Epithelial Ovarian Cancer Impart Pro-Tumorigenic Properties through Differential Activation of the Notch3 Pathway. Cancers. 2022; 14(14):3365. https://doi.org/10.3390/cancers14143365
Chicago/Turabian StyleMukherjee, Souvik, Asmita Sakpal, Megha Mehrotra, Pratham Phadte, Bharat Rekhi, and Pritha Ray. 2022. "Homo and Heterotypic Cellular Cross-Talk in Epithelial Ovarian Cancer Impart Pro-Tumorigenic Properties through Differential Activation of the Notch3 Pathway" Cancers 14, no. 14: 3365. https://doi.org/10.3390/cancers14143365
APA StyleMukherjee, S., Sakpal, A., Mehrotra, M., Phadte, P., Rekhi, B., & Ray, P. (2022). Homo and Heterotypic Cellular Cross-Talk in Epithelial Ovarian Cancer Impart Pro-Tumorigenic Properties through Differential Activation of the Notch3 Pathway. Cancers, 14(14), 3365. https://doi.org/10.3390/cancers14143365





