Simple Summary
Cancer remains a burden to the public health all over the world. An increasing number of studies have concentrated on the role of methyladenosine modifications on cancers. Methyladenosine modifications mainly include N6-methyladenosine (m6A), N1-methyladenosine (m1A), and 2’-O-methyladenosine (m6Am), of which dynamic changes could modulate the metabolism of RNAs in eukaryotic cells. Mounting evidence has confirmed the crucial role of methyladenosine modification in cancer, offering possibilities for cancer therapy. In this review, we discussed the regulatory role of methyladenosine modification on cancer, as well as their potential for treatment.
Abstract
Methyladenosine modifications are the most abundant RNA modifications, including N6-methyladenosine (m6A), N1-methyladenosine (m1A), and 2’-O-methyladenosine (m6Am). As reversible epigenetic modifications, methyladenosine modifications in eukaryotic RNAs are not invariable. Drastic alterations of m6A are found in a variety of diseases, including cancers. Dynamic changes of m6A modification induced by abnormal methyltransferase, demethylases, and readers can regulate cancer progression via interfering with the splicing, localization, translation, and stability of mRNAs. Meanwhile, m6A, m1A, and m6Am modifications also exert regulatory effects on noncoding RNAs in cancer progression. In this paper, we reviewed recent findings concerning the underlying biomechanism of methyladenosine modifications in oncogenesis and metastasis and discussed the therapeutic potential of methyladenosine modifications in cancer treatments.
1. Introduction
Cancer remains a considerable threat to public health worldwide. In 2022, the American Cancer Society estimates 1,918,030 new cancer cases and 609,360 cancer deaths [1]. An in-depth understanding of cancer biology is essential in identifying effective therapeutics strategies.
Cancer cells, similar to other eukaryotic cells, perform most of their functions through translated proteins by mRNAs. There are increasing numbers of chemical modifications observed on the bases of mRNA nucleosides. Methyladenosine modification refers to an additional methyl group inserted onto adenosine. mRNA maturation, exportation, degradation, and translation are largely regulated by methyladenosine modifications [2,3,4], including N6-methyladenosine (m6A), N1-methyladenosine (m1A), and 2’-O-methyladenosine (m6Am), among which the m6A modification is the most abundant [5,6]. In addition to mRNAs, methyladenosine modification also regulates noncoding RNAs (ncRNAs) [7] during oncogenesis, cancer metastasis, and therapeutic intervention [8]. The number of studies on RNA methylation in cancer has exploded since 2012. The role of methyladenosine modification has expanded from tumor genesis and metastasis to the regulation of tumor microenvironment, immunotherapy, drug resistance, etc. However, current reviews found in the literature do not cover the latest research findings. We present this review to fill this gap and to offer our perspective on the research direction in this field. We describe the structure of m6A, m1A, and m6Am in eukaryotic cells; explicitly state how corresponding enzymes including methyltransferases (writers), demethylases (erasers), and readers modify RNAs in a dynamic manner, with focus on m6A-, m1A-, and m6Am regulated RNAs in the oncogenesis and metastasis of cancer; and present our perspective on their therapeutic potential in cancer (Figure 1).
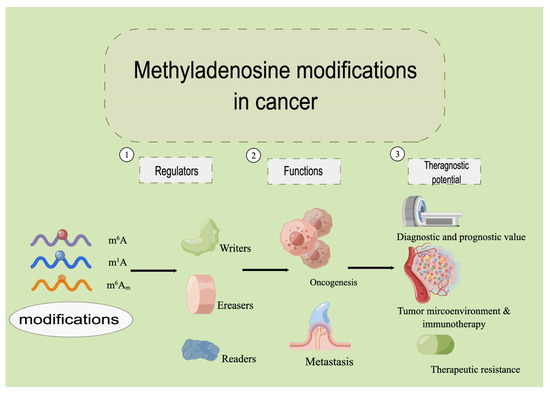
Figure 1.
The overview of the methyladenosine modifications in cancer.
2. Methyladenosine Modifications in Eukaryotic RNAs
Nucleotides are the basic unit of nucleic acid. Since the 1950s, more and more chemical modifications of nucleotides on RNAs have been identified, enriching our knowledge about the structure, biogenesis, and function of RNAs to a great extent [9]. More than 60% of RNA chemical modifications are methylation modifications, which play a vital role in the RNA family [10,11]. In eukaryotic messenger RNAs (mRNAs), m6A is the most abundant type of chemical modification, affecting the structural stability, progress of translation, and regulation of the secondary structure of mRNAs [11,12]. While fifty years have passed since m6A was initially discovered in eukaryotic mRNA [13,14], it was not until the past decade that considerable progress has been made in understanding this modification.
In 2012, employing a combination of m6A-specific antibodies and gene sequencing techniques, transcriptome-wide analysis of m6A revealed the first glimpse of its landscape [10,15]. m6A is widely distributed in the mammalian transcriptome of more than 7000 mRNAs and 300 ncRNAs. However, the distribution of m6A modification is not random in most transcripts. The level of m6A modification at a certain site is determined by local sequence, followed by the secondary structure, and the distance from a “potential selected site” to the gene terminal area [16]. There is a fairly specific consensus motif, D(G, A, or U)R(G or A)ACH(A, C, or U) [9], at the m6A modification site, which is enriched in the 3′ untranslated region (3′UTR) as well as coding sequence (CDS) region, especially inside the long internal exons and around the area of stop codon [10,15] (Figure 2A). The 5’ end of introns was also found significantly enriched in m6A modification [12]. m6A could cause alterations to the mRNA structure, facilitating the binding of nuclear RNA-binding protein heterogeneous nuclear ribonucleoprotein C (HNRNPC) to intervene in the processing of precursor mRNA (pre-mRNA), which is a mechanism termed “m6A switch’’ [17]. Although the modification sites are evolutionarily conserved [10,15,16], there are some differences among different cells [12]. Besides sequence-specific sites, m6A modification seems more likely to be present in regions with secondary structures [12], although this remains controversial. m6A modifications are found in virtually all types of RNAs, including rRNAs, tRNAs [18], mRNAs, ncRNAs (microRNAs (miRNAs), circular RNAs (circRNAs), and long noncoding RNAs (lncRNAs)) [9], long intergenic noncoding (lincRNAs) [10], and small nucleolar RNAs (snoRNAs) [19] in various pathophysiological processes, such as hematopoiesis [12], germ cell genesis [20], and virus infection [21].

Figure 2.
Dynamic changes in methyladenosine modifications on mRNA. (A) Distribution of m6A, m1A, and m6Am modifications on mRNA; (B) methylases, demethylases, and readers of m6A modification; (C) methylases, demethylases, and readers of m6Am modification; and (D) methylases, demethylases, and readers of m1A modification.
Another common base methylation modification is N1-methyladenosine (m1A). Dominissini et al., found that m1A modification is quite abundant in mammalian mRNAs around the starting codon, and its methyl donor is S-adenosylmethionine (SAM) [22]. m1A modification is evolutionarily conserved too, and positively correlates with translation efficiency and protein-level transcripts. m1A is mainly located in rRNAs and tRNAs in cytoplasm [6]. Unlike m6A, m1A can disturb the Watson–Crick base pairing [23], possessing the ability to inhibit the activity of translation by affecting ribosome scanning [6].
3. Writers, Erasers, and Readers in the Coregulation of the Dynamic Adjustment of Methyladenosine Modification
m6A is the most common type of reversible methyladenosine modification in mRNA [24] and has been a hot topic in epigenetic research for decades. Methylation or demethylation is realized with the help of methyltransferases (“Writers”) or demethylases (“Erasers”). Moreover, in the nucleus, these modifications could be bound by numerous nucleus-specific “Reader” proteins to affect the splicing and transport of mRNAs, and so on. When the transcript is transported to the cytoplasm, the m6A modification site will be bound to the cytoplasm-specific reader proteins, exerting its effect on a series of processes, such as mRNA metabolism and translation [25] (Figure 2B–D). The function of methyladenosine modification-related regulators was summarized in Table 1.

Table 1.
The functions of m6A-related regulators.
3.1. Writers
In mammalian cells, four distinct methyltransferases are known to be responsible for methylating m6A in different types of RNAs. Since most of the m6A sites in mRNAs are catalyzed by the methyltransferase-like 3 (METTL3)/METTL14 complex, this heterodimer is primarily described here [25].
The labeling of m6A in mRNAs depends mainly on a kind of methyltransferase complex (MTX) containing multiple components including the METTL3-METTL14 heterodimer (the core element), and several regulatory factors, for example, Wilm’s tumor 1-associated protein (WTAP) and KIAA1429, which are necessary for the integrity of enzyme function [28,29]. MTX is distributed in the nucleus rather than the cytoplasm, being selective to the sequences of the substrate, with no obvious preference for the structure of RNA [28]. In this heterodimer, METTL3 seems to be the only subunit comprising an active center with methyltransferase activity where there is a SAM binding site. METTL14 is not primarily responsible for catalyzing methyltranslation but provides an RNA-binding scaffold that increases enzyme activity by stabilizing the conformation of METTL3. In addition, this subunit seems to play a role in the recognition of bases, METTL3 cannot exercise catalytic activity alone in absence of METTL14. However, METTL14 may exhibit catalytic activity under certain circumstances, and the catalytic sites of METTL14 homologs vary among different species [26,27]. Although MTX has been well studied, there is a lack of research on its catalytic efficiency, which provides a direction for future research.
Labeling of m1A in mRNAs is known through MTX of TRMT6/TRMT61A complex and TRMT10C [30]. Phosphorylated C-terminal domain-interacting factor 1 (PCIF1), a cap-specific adenosine-N6-methyltransferase, catalyzes the formation of m6Am on the first adenosine adjacent to m7G cap of mRNAs [30].
3.2. Erasers
A set of enzymes can demethylate RNA m6A, participating in the dynamic regulation of RNA activity. Fat-mass- and obesity-associated protein (FTO) was discovered as the first human m6A demethylase, existing in the nucleoplasm in a form of dot-like structure [24]. FTO is a homolog of AlkB family dioxygenases, catalyzes the reaction of oxidative demethylation of m6A in single-stranded RNAs (ssRNAs) instead of double-stranded RNAs (dsRNAs) in a Fe (II)- and alpha-ketoglutarate(α-KG)-dependent manner [31]. The activity of FTO is affected by pH in the environment with the optimum pH value being around 6.0. FTO also mediates the demethylation of m6Am and shows the same enzyme activity for m6Am and m6A on identical RNA sequences. It was reported that m6A is the most suitable substrate for FTO [31]. However, this is controversial because certain researchers consider FTO to mainly demethylate m6Am on small nuclear RNAs (snRNAs), instead of demethylating m6A on mRNA, due to the cross-reaction of m6A antibodies with m6Am [16,38]. Several antibody-independent testing techniques have been proposed for further research in this aspect [12,16,38]. In addition, m1A is a physiological substrate of FTO. FTO can demethylate m1A in tRNAs and RNAs with loop structure, lacking activity on linear ssRNAs and ssDNAs [31].
AlkB homolog 5 (ALKBH5), a member of the AlkB homolog (ALKBH) family, is the second identified RNA demethylase in mammals with a strong preference for ssRNAs, on which m6A modification is the main physiological substrate. ALKBH5 is diffusely dispersed in the nucleoplasm with an obvious granular aggregation, co-located well with some mRNA processing factors, such as phosphorylated serine/arginine-rich splicing factor 2 (SC35-PI), Smith antigen (SM), and alternative splicing factor/splicing factor 2(ASF/SF2) in nuclear speckles. Various cellular processes are under the control of ALKBH5, including assembly and modification of mRNA processing factors, translocation of transcripts, and the metabolism of RNAs [20]. Unlike FTO, ALKBH5 seems to be more specific in demethylating m6A rather than acting on m6Am [32].
3.3. Readers
The first three proteins found to be m6A-binding proteins were two YT521-B homology (YTH) family proteins, namely YTH-domain-containing family protein 2 (YTHDF2), YTHDF3, and ELAVL1 (also known as HUR) [15]. Five kinds of YTH proteins are distributed in mammalian cells, divided into three classes: YTH-domain-containing protein (YTHDC)1, YTHDC2, and YTHDF protein family (YTHDF1-3). These five proteins contain the YTH domain that selectively binds to m6A [15,25,33].
Cytoplasm is the main repository of YTHDF1, YTHDF2, and YTHDF3. Earlier studies suggested that YTHDF1 could accelerate the translation process by promoting ribosome binding to target transcripts and increasing translation efficiency, whereas YTHDF2 plays a role in mRNA degradation by transporting mRNA with m6A modification to processing bodies (P bodies) and other RNA decay sites. YTHDF3 has both of the aforementioned effects; it promotes protein translation in coordination with YTHDF1, while also accelerating mRNA degradation together with YTHDF2 [33,34,35]. At the moment, there is a dispute over whether each YTHDF member performs totally distinct functions [25]. Given the high similarity of amino acid sequences among YTHDF proteins, it is not clear how they regulate diverse functions, and further research is necessary [12].
In contrast to its counterparts, YTHDC2 can be found in both the cytoplasm and nucleus. In addition to its DEAH-box domain and R3H core domain, the protein possesses two ankyrin repeat domains. YTHDC2 binds to 5′-3′ exonuclease by recognizing ankyrin repeat domains, which are independent of RNAs, indicating the possibility that it is involved in the regulation of RNA stability. YTHDC2 might also function in the translation process by liaising m6A-containing transcripts and ribosomes [36].
Similarly, YTHDC proteins could act as readers to combine with m1A sites, for which the affinity is weaker than that of m6A. m1A is bound directly by YTHDF-binding proteins 1–3 as well as the YTH domain of YTHDC1, but not YTHDC2. In addition, YTHDF1 seems to promote the translation of transcripts with an m1A modification while YTHDF2 accelerates the degradation of modified mRNAs [37].
4. Methyladenosine Modification: An Inseparable Part of Cancers
A growing number of studies have demonstrated the participation of methyladenosine modification in the formation (oncogenesis) and metastatic progression of cancer (Table 2). Overall, most m6A-related proteins show cancer-promoting properties. The exception is that METTL14 is predominantly cancer-inhibiting.

Table 2.
The emerging roles of methyladenosine modification in cancer.
4.1. Writers in Oncogenesis
Research into oncogenesis, or tumorigenesis, has been employed for a long time to describe genetic changes contributing to the transformation of normal cells into cancer cells [98]. Incessant efforts have identified a series of oncogenes or tumor suppressor genes in leading to the genesis of cancers when aberrant [99]. By increasing the modification of m6A, METTL3 plays a crucial role in the oncogenesis of several cancers. Choe et al., indicated that METTL3 promotes the oncogenic transformation of human lung cancer cells by increasing the translation of oncogene-bromodomain-containing protein 4 (BRD4) in a eukaryotic initiation factor 3 (eIF3h)-dependent way [40]. METTL3 has been found to guarantee the stability of hexokinase 2 (HK2) and solute carrier family 2 member 1 (SLC2A1) via an m6A-insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2)/3-dependent pathway, contributing to the tumorigenesis of colorectal cancer (CRC) [41]. Approximately 80% of colorectal cancer cases are likely to be affected by numerical and structural aneuploidy, especially broad copy number variations (BCNAs). The analyses conducted by Condorelli et al., revealed that the transcript levels of eIF3h (Chr8, q24.11) is 4–5-fold higher in Chr8q-, Chr13q-, and Chr20q-gained cancer samples, which might play a significant role in the recurrence of BCNAs in CRC [100,101]. Combining these together, it is reasonable to speculate that methyladenosine modification could impact CRC by modulating BCNAs in an eIF3h-dependent way. Further, METTL3 accelerates the oncogenesis of CRC by reducing the expression of yippee-like 5 (YPEL5) epigenetically in an m6A-YTHDF2-dependent way [42]. In bladder cancer, METTL3 reduces the expression of PTEN via expediting the maturity of pri-miR221/222, resulting in the malignant proliferation of cancer cells [43]. Moreover, the regulatory effect of METTL3 is prevalent in other types of cancers, including glioma [45,46], gastric cancer [47], hepatocellular carcinoma (HCC) [48], breast cancer (BRCA) [49], and acute myeloid leukemia (AML) [50].
METTL3 and METTL14 constitute a heterodimer MTX, in which METTL14 functions as the stabilizer for METTL3 to play the catalyzing reaction [102]. In previous studies, METTL14 has been regarded as a contributor to the oncogenesis of many cancers. For example, METTL14 plays an oncogenic role in AML by controlling leukemia stem cell self-renewal [51]. lnc942 upregulates the METTL14-mediated stability of mRNA including C-X-C motif chemokine receptor 4 (CXCR4) and Cytochrome P450 family 1 subfamily B member 1 (CYP1B1) in BRCA cells and thus promotes cell proliferation [52]. However, a growing number of studies have shown that METTL14 may have a predominantly negative regulatory role in oncogenesis. Yang et al., found that METTL14 suppresses the proliferation of CRC cells by inhibiting the transcription of oncogenic lncRNA X-inactive specific transcript (XIST) [53]. Similarly, the growth of CRC cells can be inhibited by METTL14 through changing the m6A modifications of miR-375/Yes-associated protein 1 (YAP1) pathway [54]. In gastric cancer, METTL14 was reported to be able to modulate the m6A modifications on circORC5 and thus abate the cell growth by the miR-30c-2-3p/AKT1 substrate 1 (AKT1S1) axis [55]. METTL14 was verified to restrain the skin oncogenesis induced by ultraviolet B (UVB) radiation via facilitating damage-specific DNA-binding protein 2 (DDB2)-mediated global genome repair in an m6A-dependent manner [56].
Recently, a study showed that WTAP upregulates the proliferation capability of HCC cells by suppressing the expression of ETS proto-oncogene 1 (ETS1) in a Hu-Antigen R (HuR)/p21/p27-dependent manner [57]. By mediating the stability of diaphanous-related formin 1 antisense RNA 1 (DIAPH1-AS1) in an m6A-dependent manner, WTAP promotes the formation of MTDH-LASP1 complex and the expression of LASP1, facilitating the cell growth of nasopharyngeal carcinoma [58].
Moreover, m1A methylation in a subset of tRNA is upregulated by TRMT6/TRMT61A, resulting in an increment of peroxisome-proliferator-activated receptor delta (PPARδ) translation, which in turn stimulates cholesterol synthesis and activates hedgehog signaling to promote the oncogenesis of HCC [59]. In bladder cancer, m1A upregulated by TRMT6/TRMT61A dysregulates the targetome of tRNA fragments, leading to the unfolded protein response and silencing of tumor suppressor genes [60].
4.2. Erasers in Oncogenesis
As an “eraser” of m6A, FTO removes the m6A modifications on RRACH motifs in mRNAs or lncRNAs in a Fe (II)/α-KG-dependent manner [103]. Oncogenesis is proven to be impacted by FTO. FTO is reported to demethylate m6A modifications in subtypes of AML, exerting their effect on the leukemogenesis [61,62]. In lung cancer, proliferation of cancer cells could be promoted by FTO-mediated upregulation of KRAS or MZF1 signaling [63,64]. FTO acts as a key factor in the tumorigenesis of oral squamous cell carcinoma by demethylating the translation initiation factor eIF4G1 [65]. Niu et al., reported that FTO lessens the pro-apoptosis effect of BCL2-interacting protein 3 (BNIP3) in an m6A-YTHDF2-dependent manner, contributing to the growth of breast cancer [66]. Furthermore, FTO was also deemed to promote oncogenesis by demethylating lincRNAs. For example, FTO upregulates LINC00022 epigenetically to promote esophageal squamous cell carcinoma [67]. However, FTO also exerts inhibitory effects on certain types of cancer [104]. FTO suppresses the oncogenesis of pancreatic cancer by inhibiting Wnt signaling as well as demethylating PJA2 [68]. In papillary thyroid cancer (PTC), Huang et al., found that FTO restrains the stability of Apolipoprotein E (APOE), repressing the glycolysis and growth of PTC in an m6A-IGF2BP2-mediated manner [69]. Another type of demethylase, ALKBH5, was found to promote oncogenesis of AML selectively in an m6A-dependent manner [70]. In addition, it is reported that ALKBH5 activates period circadian clock 1 (PER1), serving as a suppressor for pancreatic cancer in an m6A-YTHDF2 dependent way [71].
4.3. Readers in Oncogenesis
As mentioned above, m6A readers (YTHDF2, IGF2BP2, etc.) can work synergistically with m6A writers and erasers during the oncogenesis of various cancers. Some of them might call forth oncogenesis individually as well. A recent study showed that YTHDF1 increases eIF3C translation, which amplifies ovarian cancer oncogenesis [72]. In gastric cancer, YTHDF1 facilitates the expression of Wnt receptor frizzled 7 (FZD7), resulting in the upregulation of Wnt/β-catenin-related cell proliferation [73]. YTHDF2 is able to suppress the expression of AXIN1, which is an inhibitor of Wnt/β-catenin, resulting in the tumorigenesis of lung cancer [74]. Paternally expressed 10 (PEG10) is associated with proliferation, differentiation, and apoptosis of certain types of cancers, Zhang et al., reported that IGF2BP1 contributes to the progression of endometrial cancer by stabilizing paternally PEG10 mRNA in an m6A-dependent manner [75]. IGF2BP1 also promotes the expression of serum response factor (SRF, a transcriptional regulator) and stabilizes the expression of 35 kinds of SRF-mediated oncogenic genes including PDZ, LIM domain 7 (PDLIM7), and Forkhead box K1 (FOXK1), leading to the growth and invasion of ovarian, liver, and lung cancer [76].
4.4. Methyladenosine Modifications in Metastasis
An increasing number of studies have investigated the methyladenosine–tumor invasion–metastasis relationship. Yue et al., unveiled that METTL3 modulates the expression of zinc finger MYM-type containing 1 (ZMYM1) in an m6A-mediated manner, thus contributing to the metastasis of gastric cancer by recruiting the CtBP/LSD1/CoREST complex [77]. In prostate cancer, METTL3 improves the YTHDF2–HNRNPD-recognized degradation of ubiquitin-specific peptidase 4 (USP4) by mediating m6A modification on A2696 and then maintains the ubiquitin of ELAV-like RNA-binding protein 1 (ELAVL1), giving rise to the migration and invasion of cancer cells [78]. METTL3 enhances the maturation of pri-miR-1246 in an m6A-dependent manner, contributing to the metastasis of CRC via sprouty-related EVH1-domain-containing 2 (SPRED2)/MAPK signaling pathway [79]. In addition, the maturation of pri-miR-1246 mediated by METTL3 facilitates the metastasis of ovarian cancer via repressing the cyclin G2 (CCNG2) pathway [80]. Matrix metalloproteinase (MMPs) are proteolytic enzymes with the capability of disintegrating the membrane of extracellular band basement, potentiating the invasion of certain cancers [105]. According to Dahal et al., METTL3 reinforces the expression of MMP2, leading to an augmented invasion capacity of melanoma cells [81].
As for METTL14, most studies have identified its inhibitory effect on the metastasis of cancers. In CRC, METTL14 promotes the degradation of SRY-related high-mobility group box 4 (SOX4) in an m6A-dependent way, triggering the inhibition of metastasis of CRC via phosphatidylinositol 3-kinase (PI3K)/Akt signal [82]. During miRNA processing, METTL14 modulates the maturity of pri-miR-126 by interacting with DGCR8, repressing the metastasis of HCC [83]. Chen et al., reported that exon skipping of METTL14 induced by Cdc2-like kinases 1 (CLK1)/SR-like splicing factors5 (SRSF5) pathway tends to give rise to the metastasis of pancreatic cancer [84]. However, METTL14 was found to act as a fortifier for the metastasis of pancreatic cancer because it facilitates the turnover of p53 apoptosis effector related to PMP22 (PERP) mRNA [85]. Given the contradiction, the role of METTL14 in metastasis of cancers needs to be confirmed by further studies. Another methylase writer, WTAP, contributes to the migration and invasion of pancreatic cancer via stabilizing Fak mRNA and activating Fak-related pathways [86].
Erasers have been shown to have dual roles in the metastasis of cancers. In BRCA, FTO facilitates tumor cell migration and invasion by upregulating ADP ribosylation factor-like GTPase 5B (ARL5B) through demethylating the miR-181b-3p [87]. By decreasing the m6A modification level on integrin β1(ITGB1), FTO enables the metastasis of gastric cancer [88]. Conversely, FTO erases m6A modifications on metastasis-associated protein 1 (MTA1) mRNA and lowers the stability of MTA1 transcripts mediated by IGF2BP2, leading to the suppression of metastasis in CRC [89]. There have been numerous studies demonstrating that ALKBH5 inhibits cancer metastasis. For example, ALKBH5 serves as a suppressor for the metastasis of gastric cancer by downregulating the expression of protein kinase and membrane-associated tyrosine/threonine 1 (PKMYT1) in an IGF2BP3-m6A-mediated manner [90]. In addition, the suppressive effect of ALKBH5 was observed in prostate cancer, CRC, and non-small-cell lung cancers [91,92,93].
The regulatory role of methylated modification readers on cancer metastasis was also reported. Su et al., indicated that YTHDF1 binds with m6A modifications on EGFR, promoting EGFR expression to stimulate the metastasis of HCC after insufficient radiofrequency ablation [94]. YTHDC1 was found to be able to bind to m6A modifications on metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), contributing to the expression of certain key oncogenes by maintaining the genomic binding sites of nuclear speckles, thus promoting metastasis [95]. According to Chang et al., YTHDF3 promotes breast cancer brain metastasis by inducing the upregulation of ST6 N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (ST6GALNAC5), gap junction protein (GJA1), and EGFR, whose transcripts are enriched in m6A modifications [96]. The inhibitory effect of YTHDF2 on the metastasis of lung adenocarcinoma was also investigated [97].
5. Theragnostic Potential of Methyladenosine Modifications in Cancer
The previous sections presented the non-negligible role of methyladenosine modifications in cancer oncogenesis and metastasis. Given the wide range of essential roles, many researchers have focused on their theragnostic potential on cancer, divided into three parts: acting as diagnostic markers and prognostic predictors, conditioning tumor microenvironment (TME) and immunotherapy, and modulating therapeutic resistance and self-renewal of cancer.
5.1. Acting as Diagnostic Markers and Prognostic Predictors
Early diagnostic and accurate prognostic biomarkers are both meaningful in helping clinicians make rational decisions in cancer treatment. The occurrence of cancer is often accompanied by alterations in the profiles of m6A expression. Alterations of m6A in tissues or in blood provide a possibility for early diagnosis of cancer. Huang et al., conducted liquid chromatography–electrospray ionization tandem mass spectrometry (LC-ESI-MS/MS) on nucleosides extracted from captured circulating tumor cells (CTC) and found an elevation of m6A modifications on mRNA that demonstrated the possibility of a noninvasive method for diagnosing cancer [106]. Significant upregulated m6A modification was detected in peripheral blood RNA in patients with breast cancer and was closely associated with the disease stages [107]. Receiver operating characteristic curve (ROC) analysis showed that the area under the curve (AUC) value of m6A is 0.887, larger than that of carbohydrate antigen 153 (CA153) as well as carcinoembryonic antigen (CEA) in the cohort. Ge et al., found that the m6A level has an AUC value of 0.929 in distinguishing gastric cancer from benign gastric disease, greater than that for CEA and carbohydrate antigen 199 (CA199) [108]. In lung cancer, the m6A level in leukocytes has an ascendant sensitivity and specificity in discriminating lung adenocarcinoma, lung squamous cell carcinoma, and non-small-cell lung cancer from healthy adults [109]. Through a support vector machine algorithm, Zhang et al., constructed a serum m6A-miRNA diagnostic signature to detect cancer ulteriorly [110]. They obtained 18 candidate miRNAs and tested them on 14,965 serum samples including 12 cancer types via training, internal validation, and external validation cohort. The results showed that this m6A-miRNA signature has a satisfactory diagnostic performance in identifying cancers, especially lung cancer, gastric cancer, and HCC [110]. However, current studies on the diagnostic potential of m6A tend to focus on overall expression profiles rather than on tumor-specific expression profiles. This creates a problem, even when changes in m6A modifications are detected in the blood, it is still challenging to localize specific tumors. Hence, it is imperative to find specific m6A-tagged markers for different cancers.
Prognosis prediction is important for cancer treatment as it can be used to guide treatment methods in order to achieve better therapeutic effects. Plenty of studies have focused on the prognostic value of methyladenosine modification. In gastric cancer, Wang et al., found that the expression level of METTL3, remarkably elevated in tumor tissue, is closely associated with poor prognosis [111]. Conversely, downregulation of METTL14 in rectal cancer is correlated with poor outcome in patients [112]. When it comes to lower-grade glioma, lung adenocarcinoma, breast cancer, and bladder cancer, risk models including m6A-mediated lncRNAs were built, revealing an effective value in predicting the prognoses [113,114,115,116]. More recently, Shen et al., conducted comprehensive analyses for the predictive value of 23 m6A regulators and 83 mRNAs and noncoding RNAs among 9804 pan-cancer samples [117]. The m6A signature they constructed exhibited a favorable performance in predicting the overall survival rate in 24 types of cancers. Zheng et al., found that m1A-regulating genes are correlated with advanced clinical stages and poor prognosis of pancreatic cancer, implying the possibility of m1A being an effective prognostic predictor [118]. Furthermore, Zhao et al., established an m1A signature model, by which the prognosis of HCC can be easily forecasted via evaluating the condition of tumor microenvironment (TME) [119].
These data implicate that methyladenosine modification could be implemented in the diagnosis and prediction of various cancers (Figure 3). Further basic and clinical research is needed to validate their effectiveness, which will pave the way for a better early-stage diagnosis as well as a more specific treatment for cancers.

Figure 3.
Methyladenosine modifications could act as diagnostic markers and prognostic predictors.
5.2. Conditioning Tumor Microenvironment and Immunotherapy
TME refers to the complex ecosystem comprising non-cancer host cells and non-cellular components, including endothelial cells, fibroblasts, immune cells, extracellular matrix, growth factors, cytokines, and so on [120]. As a breeding ground for neoplasms, TME potentiates the initiation, proliferation, invasion, and therapeutic resistance of cancer [121]. TME is involved in the complicated regulatory network of methyladenosine modifications, which, in turn, influences tumor development, progression, and efficacy of therapy.
Hypoxia is a characteristic of TME that could activate the progression of cancers through augmenting hypoxia-inducible factors (HIFs) [122]. So far, studies have confirmed that there is an interactive relationship between TME-related hypoxia and changes in m6A modification in tumors. For instance, breast cancer is associated with transcriptional activity of ALKBH5 mediated by HIF-1α and HIF-2α, leading to a decrease in m6A modification on NANOG mRNA and initiation of the tumor in the hypoxic TME [123]. In glioblastoma multiforme, Dong et al., found that demethylase ALKBH5 is associated with the recruitment of hypoxia-induced tumor-associated macrophages (TAMs); ALKBH5 deficiency suppresses the expression and secretion of CXCL8/IL8 [124]. Another study demonstrated that YTHDF1 facilitates the hypoxic adaption of non-small-cell lung cancer via Keap1-nuclear respiratory factor 2 (Nrf2)-Aldo-keto reductase family 1 member C1 (AKR1C1) axis [125]. In stomach cancer, m6A reader IGF2BP3 regulates the hypoxia-induced metastasis by increasing the expression of HIF-1α in an m6A-dependent manner.
The Warburg Effect refers to abnormally elevated glycolysis activation in cancer cells and the TME, a hallmark of tumor progression [126]. This metabolic dysfunction of cancer cells induces a hypoglycemic and acid condition of TME and further accelerates the progression of cancer [127]. METTL3 was found to upregulate glycolysis in CRC by stabilizing HK2 and SLC2A1 in an m6A-IGF2BP2/3-dependent manner, leading to cancer progression [41]. Conversely, FTO represses the glycolysis of PTC by destabilizing APOE, thus restraining the growth of cancer [69]. By modulating hypoxia and glycolysis in the TME, altering m6A modifications may serve as a therapeutic strategy in treating cancers, though more in-depth research is required.
There is bidirectional crosstalk between the immune system and bone microenvironment, exerting a non-negligible effect on cancer [128]. Among them, the receptor activator of NF-kB (RANK)/RANK ligand (RANKL)/osteoprotegerin (OPG) pathway contributes to the interactions between cancer immunity and osteocytes [129,130]. For instance, the disturbance of RANKL/OPG ratio and osteoclastogenesis induced by cancer cells could facilitate the disruption of bone and implantation of metastases via downregulating the immune system pathway in a vicious circle [128]. If the bidirectional interaction between bone microenvironment and immune system can be effectively regulated during metastasis, it will be helpful for cancer treatment. More recently, Fang et al., reported that YTHDF2 alleviates osteoclast formation, bone resorption, and secretion of inflammatory cytokines in RANKL-primed osteoclast precursors by mediating NF-kB and MAPK signaling [131]. Further, m6A modification on circ_0008542 extracted from osteoblast exosomes could promote bone resorption by mediating the increasing RANK in osteoclast [132]. These findings implied the potential of m6A on the bidirectional crosstalk between the immune system and bone microenvironment. However, these current studies are limited to non-cancerous fields and more direct evidence is needed to demonstrate its effect on cancer.
TME has a characteristic of restraining immunoreaction and harboring immune-suppressive cells and cytokines, as well as immune checkpoint inhibitor receptors [133,134,135]. Further, TME accelerates the exhaustion of tumor killer cells, including CD8+ T cells and antigen-presenting cells (APCs) [136,137]. Numerous studies have indicated the regulatory role of m6A modification in the immunosuppression of TME. m6A regulates immune repression in the tumor microenvironment in two main ways: one is to regulate the infiltration of immune cells, and the other is to affect immune checkpoints inhibitors (ICIs). Xiong et al., found that METTL3 improves the m6A modification on Jak1 mRNA in tumor-infiltrating myeloid cells (TIMs) under the influence of lactate, promoting the immunosuppressive effect of TIMs [138]. Dong et al., reported that knockout of METTL14 in TAMs promotes the dysfunction of CD8+, imposing an immunosuppressive effect on the progression of CRC [139]. In intra-hepatic cholangiocarcinoma, ALKBH5 has been regarded to orchestra the expression of PD-L1 in TME in an m6A-mediated manner [140]. The modulating effect of m6A on immunosuppression of TME is expected to provide a new anchor for immunotherapy. Recently, METTL3 and 14 were found to weaken the potency of anti-PD-1 therapy by interfering with interferon-γ-signal transducer and activator of transcription 1 (STAT1)- interferon regulatory factor 1 (IRF-1) signaling [141]. This finding implies that knockdown of METTL3 and 14 would strengthen the immune response of anti-PD-1 immunotherapy. In breast cancer, Wan et al., found that METTL3/IGF2BP3 upregulates the expression of PD-L1 epigenetically, leading to a PD-L1-dependent T cell exhaustion and infiltration [142], suggesting METTL3/IGF2BP3-mediated m6A modification might be a novel immunotherapeutic target. In addition, depletion of FTO improves the sensitivity of melanoma to the anti-PD-1 blockade immunotherapy [143]. Deletion of ALKBH5 was also found to enhance the efficiency of anti-PD-1 immunotherapy via decreasing lactate and the infiltration of suppressive immune cells in TME [144]. Li et al., found that circNDUFB2 acts as an antitumor immune modulator via destabilizing IGF2BPs and activating RIG-I-MAVS signaling to facilitate the recruitment of immune cells in non-small-cell lung cancer (NSCLC) [145]. Furthermore, m6A scores were constructed in prostate cancer and pancreatic cancer to predict the prognosis and response to ICI therapy [146]. Meanwhile, Gao et al., provided an m1A score to evaluate the recruitment of immune cells in TME in CRC [147]. These findings together suggest that modulation of m6A/m1A could assist ICIs and forecast their curative effect, paving the way for an improvement of immunotherapy (Figure 4). Of note, m6A-associated proteins such as METTL3 tend to function globally instead of having a characteristic of tissue or RNA specificity. It is difficult to ensure that the altered methyladenosine modifications in specific RNA can be regulated without changing modifications in other RNAs. Therefore, how to specify the regulation of methyladenosine modifications is a complex problem for its application in cancer therapy.

Figure 4.
Methyladenosine modifications are able to condition tumor microenvironment and immunotherapy.
5.3. Modulating Therapeutic Resistance and Self-Renewal of Cancers
Therapeutic resistance, such as radioresistance and chemoresistance, is one of the major challenges in cancer management [148]. A growing number of studies confirmed the role of m6A in therapeutic resistance (chemoresistance, drug resistance, and radioresistance), pointing to a new direction for future cancer therapy. Through targeting the myelocytomatosis oncogene (MYC)-miR-155/23a-Cluster-MXI1 pathway, the inhibition of demethylase FTO was able to potentiate the therapeutic effect of temozolomide in glioma [149]. In lung adenocarcinoma, METTL7B was found to mediate the resistance to EGFR-tyrosine kinase inhibitors (EGFR-TKIs) via methylating GPX4, HMOX1, and SOD1 mRNA. Jin et al., found that METTL3 induces the drug resistance of NSCLC via activating the MALAT1-miR-1914-3p-YAP axis in an m6A-dependent manner [150]. Meanwhile, Liu et al., unveiled that METTL3-mediated autophagy reduces the resistance of NSCLC cells to gefitinib via β-elemene [151]. In MCF-7 breast cancer xenograft model, METTL3 contributes to the adriamycin resistance by promoting the maturity of pri-microRNA-221-3p in an m6A-dependent manner [152]. WTAP promotes the chemoresistance to gemcitabine in pancreatic cancer by fortifying the stability of Fak mRNA [86]. Poly-(ADP-ribose) polymerase (PARP) inhibitor (PARPi) resistance remains an obstacle to the treatment of BRCA-mutated epithelial ovarian cancer (EOC). Fukumoto et al., found that m6A modification on FZD10 mRNA is correlated with the PARPi resistance via Wnt/β-catenin pathway [153]. Interestingly, PARP1 and Wnt/β-catenin pathways were found to be enriched in BRCA bearing the specific chromosomal aberrations (Chr1q gains and Chr16q losses) [154]. More studies are needed to investigate the possibility of methyladenosine modification modulating therapeutic resistance by regulating aberrations of chromosomes in cancers. ALKBH5-HOXA10 demethylates m6A modification on JAK2 mRNA, resulting in cisplatin resistance of EOC. Depletion of METTL3 enhances the sorafenib resistance of HCC through modifying the hypoxia TME via FOXO3-mediated autophagy [155]. In addition to the aforementioned drug resistance, m6A modification also exerts its effects on radioresistance. Wu et al., found that the stabilization of circCUX1 induced by m6A modification augments the radioresistance of hypopharyngeal squamous cell carcinoma via inhibiting the caspase-1 pathway [156]. These findings not only pinpoint the role of m6A modification on therapeutic resistance but also provide a new prospect for drug and radiation therapy for cancers.
In tumors, cancer stem cells are capable of sustaining pluripotency, promoting tumor progression, facilitating self-renewal, and proving resistance to drugs. Numerous studies have indicated that m6A modification acts as a modulator for the maintenance and self-renewal of cancer stem cells, providing a novel target for remedying tumors and eliminating the therapeutic resistance of cancer. Cui et al., demonstrated that the self-renewal and tumorigenesis of glioblastoma stem cells are largely dependent on m6A modification [157]. Dixit et al., also found that m6A reader YTHDF2 is a dependency of glioblastoma stem cells via stabilizing mRNAs of oncogene MYC and VEGFA [158]. In leukemia, inhibition of FTO attenuates the self-renewal of leukemia stem cells and suppresses immune checkpoint receptors, especially leukocyte immunoglobulin-like receptor B4 (LILRB4) [159]. ALKBH5 plays a crucial role in self-renewal and maintenance of leukemia stem cells and initiating cells in m6A-dependent ways [70]. In CRC cells, demethylation of cytoplasmic m6Am by FTO enhances colorectal cancer stem-like phenotypes, potentiating the drug resistance of CRC [160]. What is more, FTO suppresses the self-renewal of cancer stem cells in ovarian cancer by inhibiting 3’, 5’-cyclic adenosine monophosphate (cAMP) signaling [161] (Figure 5). The above findings suggest that m6A can positively impact cancer therapy by regulating the self-renewal ability and drug resistance of tumor cells. For clinical applications, research on its specific effects needs to be strengthened.
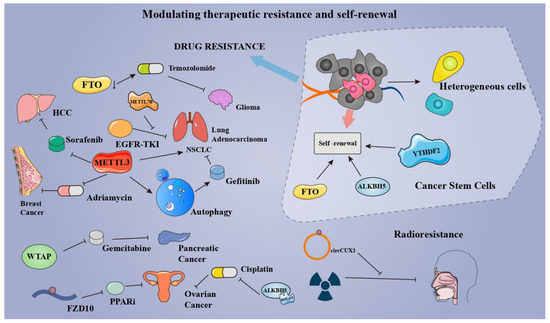
Figure 5.
By modulating therapeutic resistance and self-renewal of cancers, methyladenosine modifications have the potential to mediate the efficacy of cancer therapy.
6. Conclusions and Perspectives
Understanding methyladenosine modification in the development, regulation, and biological processes of diseases is a hot topic in recent years. Mounting evidence shows the important function of methyladenosine modification in controlling the fate of RNA. m6A modification is able to control the splice, exportation, stabilization, and translation of mRNA, ensuring normal physiological processes. Meanwhile, aberrant m6A modification on mRNAs leads to certain diseases, especially cancers. m6A writer, eraser, and reader proteins play a critical role in the oncogenesis and metastasis of glioma, breast cancer, HCC, gastric cancer, and CRC, etc. Thus, controlling the inordinate expression of m6A-related genes would be a novel strategy for the treatment of cancers. m6A modifications are also regarded as secondary structure modifications of ncRNAs. lncRNAs, circRNAs, and miRNAs themselves play vital roles in cancers [162]. Regulation of m6A modification to cancer can be achieved indirectly through their regulatory roles in ncRNAs. These characteristics render m6A an indispensable target in cancer research. This review systematically presents the role of methyladenosine modification in cancer and classifies its theragnostic effects into three categories, providing a new perspective for future research.
In view of the aforementioned properties, methyladenosine modification has the potential to be effective in cancer treatment. Considerable efforts have been made to implement the modulation of m6A in the remedy of cancer. On account of the maturation of methylated RNA immunoprecipitation sequencing (MeRIP-seq), the alteration profiles of a wide range of cancers have been screened. Among them, the upregulation of m6A modification in the peripheral blood RNA or leukocyte is able to diagnose certain types of cancer. In addition, the aberrantly modified m6A modification has the potency to predict the outcome of cancer. Regulating the m6A modification is supposed to control the TME of solid tumors, exerting an impact on the immunotherapy of tumors. Lastly, m6A modification adjusts the self-renewal of cancer stem cells and regulation of m6A is expected to have a unique value in the therapeutic resistance of tumors.
Clinical application of targeting m6A in cancer requires further research. First, exploring suitable small-molecule inhibitors for the writers, erasers, and readers of m6A modification is desirable. Treatment might be improved through selective inhibition of m6A-related proteins in tumor tissues. More recently, Yankova et al., have identified a selective METTL3 catalytic inhibitor, STM2457, which could suppress the growth of AML [163]. Lee et al., reported that eltrombopag selectively inhibits the catalytic form of the METTL3-14 complex that can decrease the m6A modification on RNA in AML cells [164]. Meanwhile, two FTO inhibitors (FB23 and FB23-2) have been designed by Huang et al., exhibiting a selective inhibitory effect on the proliferation of AML [165]. Sabnis applied a novel small molecule ALKBH5 inhibitor with the activity of suppressing demethylase [166]. These advances pave the way for further development of cancer treatment. Second, as lack of specificity is a concern in epigenetic therapeutics, inhibition of methylases or demethylases may affect m6A modification in normal cells and interfere with their physiological functions. At the same time, the spectrum of m6A expression in different types of cancer may be extremely heterogeneous, and a unique m6A signature needs to be established for each type or subtype of cancer. Finally, an m6A-centered post-transcriptional editing needs to be investigated as an effective supplement to the existing gene editing therapy in cancer.
Author Contributions
X.Q., D.R. and H.Z. conceived the theme; X.Q. and X.S. conducted the writing of the manuscript; X.Q., Y.Z. and D.R. drew the figures and table; D.R., H.Z. and S.T.C.W. reviewed, edited, and finalized this manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by NIH R01CA238727 to H.Z. and S.T.W., NIH U01CA253553, T.T. and W.F. Chao Foundation and John S. Dunn Research Foundation to S.T.W., Houston Methodist Cornerstone Award to H.Z. and Changzheng Hospital Pyramid Talent Project (2020) of Second Military Medical University and PLA Navy No. 905 Hospital Management Project (2019 and 2021) to D.R.
Conflicts of Interest
All the authors declare they have no conflict of interest.
Abbreviations
Methyltransferase-like 3—METTL 3; Wilm’s tumor 1-associated protein—WTAP; fat-mass- and obesity-associated protein—FTO; single-stranded RNAs—ssRNAs; double-stranded RNAs—dsRNAs; AlkB homolog 5—ALKBH5; S-adenosylmethionine—SAM; eukaryotic initiation factor 3—eIF3; metastasis-associated lung adenocarcinoma transcript 1—MALAT1; inactive X-specific transcripts—XIST; insulin-like growth factor 2 mRNA-binding protein 2—IGF2BP2; liquid chromatography–electrospray ionization tandem mass spectrometry—LC-ESI-MS/MS; signal transducer and activator of transcription 1—STAT1; interferon regulatory factor 1—IRF-1; non-small-cell lung cancer—NSCLC; immune checkpoint inhibitors—ICIs; myelocytomatosis oncogene—MYC; poly-(ADP-ribose) polymerase—PARP; methylated RNA immunoprecipitation sequencing—MeRIP-seq.
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Q.; Hou, J.; Zhou, Y.; Li, Z.; Cao, X. The RNA helicase DDX46 inhibits innate immunity by entrapping mA-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 2017, 18, 1094–1103. [Google Scholar] [CrossRef] [PubMed]
- Bartosovic, M.; Molares, H.C.; Gregorova, P.; Hrossova, D.; Kudla, G.; Vanacova, S. N6-methyladenosine demethylase FTO targets pre-mRNAs and regulates alternative splicing and 3’-end processing. Nucleic Acids Res. 2017, 45, 11356–11370. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef]
- Oerum, S.; Meynier, V.; Catala, M.; Tisné, C. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res. 2021, 49, 7239–7255. [Google Scholar] [CrossRef]
- Safra, M.; Sas-Chen, A.; Nir, R.; Winkler, R.; Nachshon, A.; Bar-Yaacov, D.; Matthias, E.; Walter, R.; Noam, S.-G.; Schraga, S. The m1A landscape on cytosolic and mitochondrial mRNA at single-base resolution. Nature 2017, 551, 251–255. [Google Scholar] [CrossRef]
- Rong, D.; Sun, G.; Wu, F.; Cheng, Y.; Sun, G.; Jiang, W.; Li, X.; Zhong, Y.; Wu, L.; Zhang, C.; et al. Epigenetics: Roles and therapeutic implications of non-coding RNA modifications in human cancers. Mol. Ther. Nucleic Acids 2021, 25, 67–82. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, B.; Nie, Z.; Duan, L.; Xiong, Q.; Jin, Z.; Yang, C.; Chen, Y. The role of m6A modification in the biological functions and diseases. Signal Transduct. Target. Ther. 2021, 6, 74. [Google Scholar] [CrossRef]
- Huang, H.; Weng, H.; Chen, J. mA Modification in Coding and Non-coding RNAs: Roles and Therapeutic Implications in Cancer. Cancer Cell 2020, 37, 270–288. [Google Scholar] [CrossRef]
- Meyer, K.D.; Saletore, Y.; Zumbo, P.; Elemento, O.; Mason, C.E.; Jaffrey, S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3’ UTRs and near stop codons. Cell 2012, 149, 1635–1646. [Google Scholar] [CrossRef]
- Zhang, Y.; Geng, X.; Li, Q.; Xu, J.; Tan, Y.; Xiao, M.; Song, J.; Liu, F.; Fang, C.; Wang, H. m6A modification in RNA: Biogenesis, functions and roles in gliomas. J. Exp. Clin. Cancer Res. 2020, 39, 192. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S.; Peng, Y.; Ge, R.; Su, R.; Senevirathne, C.; Harada, B.T.; Dai, Q.; Wei, J.; Zhang, L.; et al. m6A RNA modifications are measured at single-base resolution across the mammalian transcriptome. Nat. Biotechnol. 2022, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Perry, R.P.; Kelley, D.E. Existence of methylated messenger RNA in mouse L cells. Cell 1974, 1, 37–42. [Google Scholar] [CrossRef]
- Desrosiers, R.; Friderici, K.; Rottman, F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. USA 1974, 71, 3971–3975. [Google Scholar] [CrossRef] [PubMed]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Campos, M.A.; Edelheit, S.; Toth, U.; Safra, M.; Shachar, R.; Viukov, S.; Winkler, R.; Nir, R.; Lasman, L.; Brandis, A.; et al. Deciphering the "mA Code" via Antibody-Independent Quantitative Profiling. Cell 2019, 178, 731–747. [Google Scholar] [CrossRef]
- Liu, N.; Dai, Q.; Zheng, G.; He, C.; Parisien, M.; Pan, T. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature 2015, 518, 560–564. [Google Scholar] [CrossRef]
- Schmidt, W.; Arnold, H.H.; Kersten, H. Biosynthetic pathway of ribothymidine in B. subtilis and M. lysodeikticus involving different coenzymes for transfer RNA and ribosomal RNA. Nucleic Acids Res. 1975, 2, 1043–1051. [Google Scholar] [CrossRef][Green Version]
- Linder, B.; Grozhik, A.V.; Olarerin-George, A.O.; Meydan, C.; Mason, C.E.; Jaffrey, S.R. Single-nucleotide-resolution mapping of m6A and m6Am throughout the transcriptome. Nat. Methods 2015, 12, 767–772. [Google Scholar] [CrossRef]
- Zheng, G.; Dahl, J.A.; Niu, Y.; Fedorcsak, P.; Huang, C.-M.; Li, C.J.; Vågbø, C.B.; Shi, Y.; Wang, W.-L.; Song, S.-H.; et al. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell 2013, 49, 18–29. [Google Scholar] [CrossRef]
- Kim, G.-W.; Siddiqui, A. N6-methyladenosine modification of HCV RNA genome regulates cap-independent IRES-mediated translation via YTHDC2 recognition. Proc. Natl. Acad. Sci. USA 2021, 118, e2022024118. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Li, X.; Yi, C. N-methyladenosine methylome in messenger RNA and non-coding RNA. Curr. Opin. Chem. Biol. 2018, 45, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Yi, C.; Jian, X.; Zheng, G.; He, C. Structure determination of DNA methylation lesions N1-meA and N3-meC in duplex DNA using a cross-linked protein-DNA system. Nucleic Acids Res. 2010, 38, 4415–4425. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Fu, Y.; Zhao, X.; Dai, Q.; Zheng, G.; Yang, Y.; Yi, C.; Lindahl, T.; Pan, T.; Yang, Y.-G.; et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011, 7, 885–887. [Google Scholar] [CrossRef]
- Zaccara, S.; Ries, R.J.; Jaffrey, S.R. Reading, writing and erasing mRNA methylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 608–624. [Google Scholar] [CrossRef]
- Wang, P.; Doxtader, K.A.; Nam, Y. Structural Basis for Cooperative Function of Mettl3 and Mettl14 Methyltransferases. Mol. Cell 2016, 63, 306–317. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Xue, Y.; Guan, Z.; Zhang, D.; Liu, Z.; Gong, Z.; Huang, J.; Tang, C.; Zou, T.; et al. Structural basis of N(6)-adenosine methylation by the METTL3-METTL14 complex. Nature 2016, 534, 575–578. [Google Scholar] [CrossRef]
- Liu, J.; Yue, Y.; Han, D.; Wang, X.; Fu, Y.; Zhang, L.; Jia, G.; Yu, M.; Lu, Z.; Deng, X.; et al. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat. Chem. Biol. 2014, 10, 93–95. [Google Scholar] [CrossRef]
- Schwartz, S.; Mumbach, M.R.; Jovanovic, M.; Wang, T.; Maciag, K.; Bushkin, G.G.; Mertins, P.; Ter-Ovanesyan, D.; Habib, N.; Cacchiarelli, D.; et al. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5’ sites. Cell Rep. 2014, 8, 284–296. [Google Scholar] [CrossRef]
- Akichika, S.; Hirano, S.; Shichino, Y.; Suzuki, T.; Nishimasu, H.; Ishitani, R.; Sugita, A.; Hirose, Y.; Iwasaki, S.; Nureki, O.; et al. Cap-specific terminal -methylation of RNA by an RNA polymerase II-associated methyltransferase. Science 2019, 363. [Google Scholar] [CrossRef]
- Zhang, X.; Wei, L.-H.; Wang, Y.; Xiao, Y.; Liu, J.; Zhang, W.; Yan, N.; Amu, G.; Tang, X.; Zhang, L.; et al. Structural insights into FTO’s catalytic mechanism for the demethylation of multiple RNA substrates. Proc. Natl. Acad. Sci. USA 2019, 116, 2919–2924. [Google Scholar] [CrossRef] [PubMed]
- Mauer, J.; Luo, X.; Blanjoie, A.; Jiao, X.; Grozhik, A.V.; Patil, D.P.; Linder, B.; Pickering, B.F.; Vasseur, J.-J.; Chen, Q.; et al. Reversible methylation of mA in the 5’ cap controls mRNA stability. Nature 2017, 541, 371–375. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, B.; Roundtree, I.A.; Lu, Z.; Han, D.; Ma, H.; Weng, X.; Chen, K.; Shi, H.; He, C. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015, 161, 1388–1399. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lu, Z.; Gomez, A.; Hon, G.C.; Yue, Y.; Han, D.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef]
- Shi, H.; Wang, X.; Lu, Z.; Zhao, B.S.; Ma, H.; Hsu, P.J.; Liu, C.; He, C. YTHDF3 facilitates translation and decay of N-methyladenosine-modified RNA. Cell Res. 2017, 27, 315–328. [Google Scholar] [CrossRef]
- Kretschmer, J.; Rao, H.; Hackert, P.; Sloan, K.E.; Höbartner, C.; Bohnsack, M.T. The mA reader protein YTHDC2 interacts with the small ribosomal subunit and the 5′-3′ exoribonuclease XRN1. RNA 2018, 24, 1339–1350. [Google Scholar] [CrossRef]
- Dai, X.; Wang, T.; Gonzalez, G.; Wang, Y. Identification of YTH Domain-Containing Proteins as the Readers for N1-Methyladenosine in RNA. Anal. Chem. 2018, 90, 6380–6384. [Google Scholar] [CrossRef]
- Meyer, K.D. DART-seq: An antibody-free method for global mA detection. Nat. Methods 2019, 16, 1275–1280. [Google Scholar] [CrossRef]
- Lin, S.; Choe, J.; Du, P.; Triboulet, R.; Gregory, R.I. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol. Cell 2016, 62, 335–345. [Google Scholar] [CrossRef]
- Choe, J.; Lin, S.; Zhang, W.; Liu, Q.; Wang, L.; Ramírez-Moya, J.; Du, P.; Kim, W.; Tang, S.; Sliz, P.; et al. mRNA circularization by METTL3-eIF3h enhances translation and promotes oncogenesis. Nature 2018, 561, 556–560. [Google Scholar] [CrossRef]
- Shen, C.; Xuan, B.; Yan, T.; Ma, Y.; Xu, P.; Tian, X.; Zhang, X.; Cao, Y.; Ma, D.; Zhu, X.; et al. mA-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer 2020, 19, 72. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Tang, W.; Xu, Y.; Xu, Y.; Xu, B.; Fu, S.; Wang, Y.; Chen, F.; Chen, Y.; Han, Y.; et al. METTL3/YTHDF2 m6A axis accelerates colorectal carcinogenesis through epigenetically suppressing YPEL5. Mol. Oncol. 2021, 15, 2172–2184. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Wang, J.-Z.; Yang, X.; Yu, H.; Zhou, R.; Lu, H.-C.; Yuan, W.-B.; Lu, J.-C.; Zhou, Z.-J.; Lu, Q.; et al. METTL3 promote tumor proliferation of bladder cancer by accelerating pri-miR221/222 maturation in m6A-dependent manner. Mol. Cancer 2019, 18, 110. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Wang, H.; Zhao, W.; Fu, S.; Li, Y.; Ni, W.; Xin, Y.; Li, W.; Yang, C.; Bai, Y.; et al. SUMO1 modification of methyltransferase-like 3 promotes tumor progression via regulating Snail mRNA homeostasis in hepatocellular carcinoma. Theranostics 2020, 10, 5671–5686. [Google Scholar] [CrossRef] [PubMed]
- Tassinari, V.; Cesarini, V.; Tomaselli, S.; Ianniello, Z.; Silvestris, D.A.; Ginistrelli, L.C.; Martini, M.; De Angelis, B.; De Luca, G.; Vitiani, L.R.; et al. ADAR1 is a new target of METTL3 and plays a pro-oncogenic role in glioblastoma by an editing-independent mechanism. Genome Biol. 2021, 22, 51. [Google Scholar] [CrossRef]
- Chai, R.-C.; Wu, F.; Wang, Q.-X.; Zhang, S.; Zhang, K.-N.; Liu, Y.-Q.; Zhao, Z.; Jiang, T.; Wang, Y.-Z.; Kang, C.-S. mA RNA methylation regulators contribute to malignant progression and have clinical prognostic impact in gliomas. Aging 2019, 11, 1204–1225. [Google Scholar] [CrossRef]
- Huo, F.-C.; Zhu, Z.-M.; Zhu, W.-T.; Du, Q.-Y.; Liang, J.; Mou, J. METTL3-mediated mA methylation of SPHK2 promotes gastric cancer progression by targeting KLF2. Oncogene 2021, 40, 2968–2981. [Google Scholar] [CrossRef]
- Yang, N.; Wang, T.; Li, Q.; Han, F.; Wang, Z.; Zhu, R.; Zhou, J. HBXIP drives metabolic reprogramming in hepatocellular carcinoma cells via METTL3-mediated m6A modification of HIF-1α. J. Cell Physiol. 2021, 236, 3863–3880. [Google Scholar] [CrossRef]
- Wang, H.; Xu, B.; Shi, J. N6-methyladenosine METTL3 promotes the breast cancer progression via targeting Bcl-2. Gene 2020, 722, 144076. [Google Scholar] [CrossRef]
- Vu, L.P.; Pickering, B.F.; Cheng, Y.; Zaccara, S.; Nguyen, D.; Minuesa, G.; Chou, T.; Chow, A.; Saletore, Y.; MacKay, M.; et al. The N-methyladenosine (mA)-forming enzyme METTL3 controls myeloid differentiation of normal hematopoietic and leukemia cells. Nat. Med. 2017, 23, 1369–1376. [Google Scholar] [CrossRef]
- Weng, H.; Huang, H.; Wu, H.; Qin, X.; Zhao, B.S.; Dong, L.; Shi, H.; Skibbe, J.; Shen, C.; Hu, C.; et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA mA Modification. Cell Stem. Cell 2018, 22, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Wu, Z.; Wang, X.; Wang, Y.; Hu, X.; Qin, W.; Lu, S.; Xu, D.; Wu, Y.; Chen, Q.; et al. LNC942 promoting METTL14-mediated mA methylation in breast cancer cell proliferation and progression. Oncogene 2020, 39, 5358–5372. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, S.; He, C.; Xue, P.; Zhang, L.; He, Z.; Zang, L.; Feng, B.; Sun, J.; Zheng, M. METTL14 suppresses proliferation and metastasis of colorectal cancer by down-regulating oncogenic long non-coding RNA XIST. Mol. Cancer 2020, 19, 46. [Google Scholar] [CrossRef]
- Chen, X.; Xu, M.; Xu, X.; Zeng, K.; Liu, X.; Sun, L.; Pan, B.; He, B.; Pan, Y.; Sun, H.; et al. METTL14 Suppresses CRC Progression via Regulating N6-Methyladenosine-Dependent Primary miR-375 Processing. Mol. Ther. 2020, 28, 599–612. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.-N.; Chen, Z.-Y.; Chen, X.-Y.; Chen, M.; Yi, Y.-C.; Zhu, J.-S.; Zhang, J. METTL14-mediated mA modification of circORC5 suppresses gastric cancer progression by regulating miR-30c-2-3p/AKT1S1 axis. Mol. Cancer 2022, 21, 51. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Yang, S.; Cui, Y.-H.; Wei, J.; Shah, P.; Park, G.; Cui, X.; He, C.; He, Y.-Y. METTL14 facilitates global genome repair and suppresses skin tumorigenesis. Proc. Natl. Acad. Sci. USA 2021, 118, e2025948118. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, C.; Chen, J.; Chen, D.; Yang, B.; He, B.; Hu, W.; Zhang, Y.; Liu, H.; Dai, L.; et al. WTAP facilitates progression of hepatocellular carcinoma via m6A-HuR-dependent epigenetic silencing of ETS1. Mol. Cancer 2019, 18, 127. [Google Scholar] [CrossRef]
- Li, Z.-X.; Zheng, Z.-Q.; Yang, P.-Y.; Lin, L.; Zhou, G.-Q.; Lv, J.-W.; Zhang, L.-L.; Chen, F.; Li, Y.-Q.; Wu, C.-F.; et al. WTAP-mediated m6A modification of lncRNA DIAPH1-AS1 enhances its stability to facilitate nasopharyngeal carcinoma growth and metastasis. Cell Death Differ. 2022, 29, 1137–1151. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Li, X.; Xiong, X.; Wang, J.; Zhou, Z.; Zhu, X.; Gu, Y.; Dominissini, D.; He, L.; et al. N-methyladenosine methylation in tRNA drives liver tumourigenesis by regulating cholesterol metabolism. Nat. Commun. 2021, 12, 6314. [Google Scholar] [CrossRef]
- Su, Z.; Monshaugen, I.; Wilson, B.; Wang, F.; Klungland, A.; Ougland, R.; Dutta, A. TRMT6/61A-dependent base methylation of tRNA-derived fragments regulates gene-silencing activity and the unfolded protein response in bladder cancer. Nat. Commun. 2022, 13, 2165. [Google Scholar] [CrossRef]
- Li, Z.; Weng, H.; Su, R.; Weng, X.; Zuo, Z.; Li, C.; Huang, H.; Nachtergaele, S.; Dong, L.; Hu, C.; et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N-Methyladenosine RNA Demethylase. Cancer Cell 2017, 31, 127–141. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Su, R.; Deng, X.; Chen, Y.; Chen, J. FTO in cancer: Functions, molecular mechanisms, and therapeutic implications. Trends Cancer 2022, 8, 598–614. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Zhao, J.; Han, L.; Xu, M.; Wang, K.; Shi, J.; Dong, Z. Retrospective study of gene signatures and prognostic value of m6A regulatory factor in non-small cell lung cancer using TCGA database and the verification of FTO. Aging 2020, 12, 17022–17037. [Google Scholar] [CrossRef]
- Liu, J.; Ren, D.; Du, Z.; Wang, H.; Zhang, H.; Jin, Y. mA demethylase FTO facilitates tumor progression in lung squamous cell carcinoma by regulating MZF1 expression. Biochem. Biophys. Res. Commun. 2018, 502, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liao, Y.; Zhang, M.; Zhu, Y.; Wang, W.; Cai, H.; Liang, J.; Song, F.; Hou, C.; Huang, S.; et al. N6-methyladenosine demethyltransferase FTO-mediated autophagy in malignant development of oral squamous cell carcinoma. Oncogene 2021, 40, 3885–3898. [Google Scholar] [CrossRef] [PubMed]
- Niu, Y.; Lin, Z.; Wan, A.; Chen, H.; Liang, H.; Sun, L.; Wang, Y.; Li, X.; Xiong, X.-F.; Wei, B.; et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol. Cancer 2019, 18, 46. [Google Scholar] [CrossRef]
- Cui, Y.; Zhang, C.; Ma, S.; Li, Z.; Wang, W.; Li, Y.; Ma, Y.; Fang, J.; Wang, Y.; Cao, W.; et al. RNA m6A demethylase FTO-mediated epigenetic up-regulation of LINC00022 promotes tumorigenesis in esophageal squamous cell carcinoma. J. Exp. Clin. Cancer Res. 2021, 40, 294. [Google Scholar] [CrossRef]
- Zeng, J.; Zhang, H.; Tan, Y.; Wang, Z.; Li, Y.; Yang, X. m6A demethylase FTO suppresses pancreatic cancer tumorigenesis by demethylating and inhibiting Wnt signaling. Mol. Ther. Nucleic Acids 2021, 25, 277–292. [Google Scholar] [CrossRef]
- Huang, J.; Sun, W.; Wang, Z.; Lv, C.; Zhang, T.; Zhang, D.; Dong, W.; Shao, L.; He, L.; Ji, X.; et al. FTO suppresses glycolysis and growth of papillary thyroid cancer via decreasing stability of APOE mRNA in an N6-methyladenosine-dependent manner. J. Exp. Clin. Cancer Res. 2022, 41, 42. [Google Scholar] [CrossRef]
- Shen, C.; Sheng, Y.; Zhu, A.C.; Robinson, S.; Jiang, X.; Dong, L.; Chen, H.; Su, R.; Yin, Z.; Li, W.; et al. RNA Demethylase ALKBH5 Selectively Promotes Tumorigenesis and Cancer Stem Cell Self-Renewal in Acute Myeloid Leukemia. Cell Stem. Cell 2020, 27, 64–80.e9. [Google Scholar] [CrossRef]
- Guo, X.; Li, K.; Jiang, W.; Hu, Y.; Xiao, W.; Huang, Y.; Feng, Y.; Pan, Q.; Wan, R. RNA demethylase ALKBH5 prevents pancreatic cancer progression by posttranscriptional activation of PER1 in an m6A-YTHDF2-dependent manner. Mol. Cancer 2020, 19, 91. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wei, Q.; Jin, J.; Luo, Q.; Liu, Y.; Yang, Y.; Cheng, C.; Li, L.; Pi, J.; Si, Y.; et al. The m6A reader YTHDF1 promotes ovarian cancer progression via augmenting EIF3C translation. Nucleic Acids Res. 2020, 48, 3816–3831. [Google Scholar] [CrossRef] [PubMed]
- Pi, J.; Wang, W.; Ji, M.; Wang, X.; Wei, X.; Jin, J.; Liu, T.; Qiang, J.; Qi, Z.; Li, F.; et al. YTHDF1 Promotes Gastric Carcinogenesis by Controlling Translation of. Cancer Res. 2021, 81, 2651–2665. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Sheng, H.; Ma, F.; Wu, Q.; Huang, J.; Chen, Q.; Sheng, L.; Zhu, X.; Zhu, X.; Xu, M. RNA mA reader YTHDF2 facilitates lung adenocarcinoma cell proliferation and metastasis by targeting the AXIN1/Wnt/β-catenin signaling. Cell Death Dis. 2021, 12, 479. [Google Scholar] [CrossRef]
- Zhang, L.; Wan, Y.; Zhang, Z.; Jiang, Y.; Gu, Z.; Ma, X.; Nie, S.; Yang, J.; Lang, J.; Cheng, W.; et al. IGF2BP1 overexpression stabilizes PEG10 mRNA in an m6A-dependent manner and promotes endometrial cancer progression. Theranostics 2021, 11, 1100–1114. [Google Scholar] [CrossRef]
- Müller, S.; Glaß, M.; Singh, A.K.; Haase, J.; Bley, N.; Fuchs, T.; Lederer, M.; Dahl, A.; Huang, H.; Chen, J.; et al. IGF2BP1 promotes SRF-dependent transcription in cancer in a m6A- and miRNA-dependent manner. Nucleic Acids Res. 2019, 47, 375–390. [Google Scholar] [CrossRef] [PubMed]
- Yue, B.; Song, C.; Yang, L.; Cui, R.; Cheng, X.; Zhang, Z.; Zhao, G. METTL3-mediated N6-methyladenosine modification is critical for epithelial-mesenchymal transition and metastasis of gastric cancer. Mol. Cancer 2019, 18, 142. [Google Scholar] [CrossRef]
- Chen, Y.; Pan, C.; Wang, X.; Xu, D.; Ma, Y.; Hu, J.; Chen, P.; Xiang, Z.; Rao, Q.; Han, X. Silencing of METTL3 effectively hinders invasion and metastasis of prostate cancer cells. Theranostics 2021, 11, 7640–7657. [Google Scholar] [CrossRef]
- Peng, W.; Li, J.; Chen, R.; Gu, Q.; Yang, P.; Qian, W.; Ji, D.; Wang, Q.; Zhang, Z.; Tang, J.; et al. Upregulated METTL3 promotes metastasis of colorectal Cancer via miR-1246/SPRED2/MAPK signaling pathway. J. Exp. Clin. Cancer Res. 2019, 38, 393. [Google Scholar] [CrossRef]
- Bi, X.; Lv, X.; Liu, D.; Guo, H.; Yao, G.; Wang, L.; Liang, X.; Yang, Y. METTL3 promotes the initiation and metastasis of ovarian cancer by inhibiting CCNG2 expression via promoting the maturation of pri-microRNA-1246. Cell Death Discov. 2021, 7, 237. [Google Scholar] [CrossRef]
- Dahal, U.; Le, K.; Gupta, M. RNA m6A methyltransferase METTL3 regulates invasiveness of melanoma cells by matrix metallopeptidase 2. Melanoma Res. 2019, 29, 382–389. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Xu, M.; Xu, X.; Zeng, K.; Liu, X.; Pan, B.; Li, C.; Sun, L.; Qin, J.; Xu, T.; et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol. Cancer 2020, 19, 106. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.-Z.; Yang, F.; Zhou, C.-C.; Liu, F.; Yuan, J.-H.; Wang, F.; Wang, T.-T.; Xu, Q.-G.; Zhou, W.-P.; Sun, S.-H. METTL14 suppresses the metastatic potential of hepatocellular carcinoma by modulating N -methyladenosine-dependent primary MicroRNA processing. Hepatology 2017, 65, 529–543. [Google Scholar] [CrossRef]
- Chen, S.; Yang, C.; Wang, Z.-W.; Hu, J.-F.; Pan, J.-J.; Liao, C.-Y.; Zhang, J.-Q.; Chen, J.-Z.; Huang, Y.; Huang, L.; et al. CLK1/SRSF5 pathway induces aberrant exon skipping of METTL14 and Cyclin L2 and promotes growth and metastasis of pancreatic cancer. J. Hematol. Oncol. 2021, 14, 60. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Liu, J.; Zhao, Y.; He, R.; Xu, X.; Guo, X.; Li, X.; Xu, S.; Miao, J.; Guo, J.; et al. Upregulation of METTL14 mediates the elevation of PERP mRNA N adenosine methylation promoting the growth and metastasis of pancreatic cancer. Mol. Cancer 2020, 19, 130. [Google Scholar] [CrossRef]
- Li, B.-Q.; Liang, Z.-Y.; Seery, S.; Liu, Q.-F.; You, L.; Zhang, T.-P.; Guo, J.-C.; Zhao, Y.-P. WT1 associated protein promotes metastasis and chemo-resistance to gemcitabine by stabilizing Fak mRNA in pancreatic cancer. Cancer Lett. 2019, 451, 48–57. [Google Scholar] [CrossRef]
- Xu, Y.; Ye, S.; Zhang, N.; Zheng, S.; Liu, H.; Zhou, K.; Wang, L.; Cao, Y.; Sun, P.; Wang, T. The FTO/miR-181b-3p/ARL5B signaling pathway regulates cell migration and invasion in breast cancer. Cancer Commun. 2020, 40, 484–500. [Google Scholar] [CrossRef]
- Wang, D.; Qu, X.; Lu, W.; Wang, Y.; Jin, Y.; Hou, K.; Yang, B.; Li, C.; Qi, J.; Xiao, J.; et al. N-Methyladenosine RNA Demethylase FTO Promotes Gastric Cancer Metastasis by Down-Regulating the m6A Methylation of ITGB1. Front. Oncol. 2021, 11, 681280. [Google Scholar] [CrossRef]
- Ruan, D.-Y.; Li, T.; Wang, Y.-N.; Meng, Q.; Li, Y.; Yu, K.; Wang, M.; Lin, J.-F.; Luo, L.-Z.; Wang, D.-S.; et al. FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis. Oncogene 2021, 40, 5168–5181. [Google Scholar] [CrossRef]
- Hu, Y.; Gong, C.; Li, Z.; Liu, J.; Chen, Y.; Huang, Y.; Luo, Q.; Wang, S.; Hou, Y.; Yang, S.; et al. Demethylase ALKBH5 suppresses invasion of gastric cancer via PKMYT1 m6A modification. Mol. Cancer 2022, 21, 34. [Google Scholar] [CrossRef]
- Jin, D.; Guo, J.; Wu, Y.; Yang, L.; Wang, X.; Du, J.; Dai, J.; Chen, W.; Gong, K.; Miao, S.; et al. mA demethylase ALKBH5 inhibits tumor growth and metastasis by reducing YTHDFs-mediated YAP expression and inhibiting miR-107/LATS2-mediated YAP activity in NSCLC. Mol. Cancer 2020, 19, 40. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Dai, M.; Li, J.; Cai, J.; Zuo, Z.; Ni, S.; Zhang, Q.; Zhou, Z. m(6)A demethylase ALKBH5 inhibits cell proliferation and the metastasis of colorectal cancer by regulating the FOXO3/miR-21/SPRY2 axis. Am. J. Transl. Res. 2021, 13, 11209–11222. [Google Scholar] [PubMed]
- Zhu, K.; Li, Y.; Xu, Y. The FTO mA demethylase inhibits the invasion and migration of prostate cancer cells by regulating total mA levels. Life Sci. 2021, 271, 119180. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Huang, M.; Liao, J.; Lin, S.; Yu, P.; Yang, J.; Cai, Y.; Zhu, S.; Xu, L.; Peng, Z.; et al. Insufficient Radiofrequency Ablation Promotes Hepatocellular Carcinoma Metastasis Through N6-Methyladenosine mRNA Methylation-Dependent Mechanism. Hepatology 2021, 74, 1339–1356. [Google Scholar] [CrossRef]
- Wang, X.; Liu, C.; Zhang, S.; Yan, H.; Zhang, L.; Jiang, A.; Liu, Y.; Feng, Y.; Li, D.; Guo, Y.; et al. N-methyladenosine modification of MALAT1 promotes metastasis via reshaping nuclear speckles. Dev. Cell 2021, 56, 702–715.e8. [Google Scholar] [CrossRef]
- Chang, G.; Shi, L.; Ye, Y.; Shi, H.; Zeng, L.; Tiwary, S.; Huse, J.T.; Huo, L.; Ma, L.; Ma, Y.; et al. YTHDF3 Induces the Translation of mA-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020, 38, 857–871. [Google Scholar] [CrossRef]
- Zhao, T.; Wang, M.; Zhao, X.; Weng, S.; Qian, K.; Shi, K.; Gu, Y.; Ying, W.; Qian, X.; Zhang, Y. YTHDF2 Inhibits the Migration and Invasion of Lung Adenocarcinoma by Negatively Regulating the FAM83D-TGFβ1-SMAD2/3 Pathway. Front. Oncol. 2022, 12, 763341. [Google Scholar] [CrossRef]
- Bergers, G.; Benjamin, L.E. Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer 2003, 3, 401–410. [Google Scholar] [CrossRef]
- Lee, E.Y.H.P.; Muller, W.J. Oncogenes and tumor suppressor genes. Cold Spring Harb. Perspect. Biol. 2010, 2, a003236. [Google Scholar] [CrossRef]
- Condorelli, D.F.; Privitera, A.P.; Barresi, V. Chromosomal Density of Cancer Up-Regulated Genes, Aberrant Enhancer Activity and Cancer Fitness Genes Are Associated with Transcriptional Cis-Effects of Broad Copy Number Gains in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4652. [Google Scholar] [CrossRef]
- Condorelli, D.F.; Spampinato, G.; Valenti, G.; Musso, N.; Castorina, S.; Barresi, V. Positive Caricature Transcriptomic Effects Associated with Broad Genomic Aberrations in Colorectal Cancer. Sci. Rep. 2018, 8, 14826. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Li, L.; Huang, Y.; Ma, J.; Min, J. Readers, writers and erasers of N-methylated adenosine modification. Curr. Opin. Struct. Biol. 2017, 47, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Gerken, T.; Girard, C.A.; Tung, Y.-C.L.; Webby, C.J.; Saudek, V.; Hewitson, K.S.; Yeo, G.S.H.; McDonough, M.A.; Cunliffe, S.; McNeill, L.A.; et al. The obesity-associated FTO gene encodes a 2-oxoglutarate-dependent nucleic acid demethylase. Science 2007, 318, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Zuidhof, H.R.; Calkhoven, C.F. Oncogenic and tumor-suppressive functions of the RNA demethylase FTO. Cancer Res. 2022, 82, 2201–2212. [Google Scholar] [CrossRef] [PubMed]
- Roomi, M.W.; Monterrey, J.C.; Kalinovsky, T.; Rath, M.; Niedzwiecki, A. In vitro modulation of MMP-2 and MMP-9 in human cervical and ovarian cancer cell lines by cytokines, inducers and inhibitors. Oncol. Rep. 2010, 23, 605–614. [Google Scholar]
- Huang, W.; Qi, C.-B.; Lv, S.-W.; Xie, M.; Feng, Y.-Q.; Yuan, B.-F. Determination of DNA and RNA Methylation in Circulating Tumor Cells by Mass Spectrometry. Anal. Chem. 2016, 88, 1378–1384. [Google Scholar] [CrossRef]
- Xiao, H.; Fan, X.; Zhang, R.; Wu, G. Upregulated N6-Methyladenosine RNA in Peripheral Blood: Potential Diagnostic Biomarker for Breast Cancer. Cancer Res. Treat. 2021, 53, 399–408. [Google Scholar] [CrossRef]
- Ge, L.; Zhang, N.; Chen, Z.; Song, J.; Wu, Y.; Li, Z.; Chen, F.; Wu, J.; Li, D.; Li, J.; et al. Level of N6-Methyladenosine in Peripheral Blood RNA: A Novel Predictive Biomarker for Gastric Cancer. Clin. Chem. 2020, 66, 342–351. [Google Scholar] [CrossRef]
- Pei, Y.; Lou, X.; Li, K.; Xu, X.; Guo, Y.; Xu, D.; Yang, Z.; Xu, D.; Cui, W.; Zhang, D. Peripheral Blood Leukocyte N6-methyladenosine is a Noninvasive Biomarker for Non-small-cell Lung Carcinoma. Onco Targets Ther. 2020, 13, 11913–11921. [Google Scholar] [CrossRef]
- Zhang, B.; Chen, Z.; Tao, B.; Yi, C.; Lin, Z.; Li, Y.; Shao, W.; Lin, J.; Chen, J. mA target microRNAs in serum for cancer detection. Mol. Cancer 2021, 20, 170. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated mA modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.; Long, J.; Huang, Q.; Han, Y.; Peng, Y.; Guo, C.; Liu, S.; Chen, Y.; Shen, E.; Long, K.; et al. M6A “Writer” Gene: A Favorable Prognostic Biomarker and Correlated With Immune Infiltrates in Rectal Cancer. Front. Oncol. 2021, 11, 615296. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Huang, X.; Li, Y.; Chen, Y.; Lin, L. mA-related lncRNAs are potential biomarkers for predicting prognoses and immune responses in patients with LUAD. Mol. Ther. Nucleic Acids 2021, 24, 780–791. [Google Scholar] [CrossRef]
- Tu, Z.; Wu, L.; Wang, P.; Hu, Q.; Tao, C.; Li, K.; Huang, K.; Zhu, X. N6-Methylandenosine-Related lncRNAs Are Potential Biomarkers for Predicting the Overall Survival of Lower-Grade Glioma Patients. Front. Cell Dev. Biol. 2020, 8, 642. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Meng, Q.; Ma, B. Characterization of the Prognostic m6A-Related lncRNA Signature in Gastric Cancer. Front. Oncol. 2021, 11, 630260. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, B.; He, M.; Cai, Y.; Ying, X.; Jiang, C.; Ji, W.; Zeng, J. N6-Methylandenosine-Related lncRNAs Predict Prognosis and Immunotherapy Response in Bladder Cancer. Front. Oncol. 2021, 11, 710767. [Google Scholar] [CrossRef] [PubMed]
- Shen, S.; Zhang, R.; Jiang, Y.; Li, Y.; Lin, L.; Liu, Z.; Zhao, Y.; Shen, H.; Hu, Z.; Wei, Y.; et al. Comprehensive analyses of m6A regulators and interactive coding and non-coding RNAs across 32 cancer types. Mol. Cancer 2021, 20, 67. [Google Scholar] [CrossRef]
- Zheng, Q.; Yu, X.; Zhang, Q.; He, Y.; Guo, W. Genetic characteristics and prognostic implications of m1A regulators in pancreatic cancer. Biosci. Rep. 2021, 41. [Google Scholar] [CrossRef]
- Zhao, M.; Shen, S.; Xue, C. A Novel m1A-Score Model Correlated With the Immune Microenvironment Predicts Prognosis in Hepatocellular Carcinoma. Front. Immunol. 2022, 13, 805967. [Google Scholar] [CrossRef]
- Xiao, Y.; Yu, D. Tumor microenvironment as a therapeutic target in cancer. Pharmacol. Ther. 2021, 221, 107753. [Google Scholar] [CrossRef]
- Gu, Y.; Wu, X.; Zhang, J.; Fang, Y.; Pan, Y.; Shu, Y.; Ma, P. The evolving landscape of N-methyladenosine modification in the tumor microenvironment. Mol. Ther. 2021, 29, 1703–1715. [Google Scholar] [CrossRef] [PubMed]
- Subarsky, P.; Hill, R.P. The hypoxic tumour microenvironment and metastatic progression. Clin. Exp. Metastasis 2003, 20, 237–250. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Samanta, D.; Lu, H.; Bullen, J.W.; Zhang, H.; Chen, I.; He, X.; Semenza, G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m⁶A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA 2016, 113, E2047–E2056. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Qin, X.; Wang, B.; Li, Q.; Hu, J.; Cheng, X.; Guo, D.; Cheng, F.-L.; Fang, C.; Tan, Y.; et al. ALKBH5 Facilitates Hypoxia-Induced Paraspeckle Assembly and IL8 Secretion to Generate an Immunosuppressive Tumor Microenvironment. Cancer Res. 2021, 81, 5876–5888. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Fan, S.; Wu, M.; Zuo, Z.; Li, X.; Jiang, L.; Shen, Q.; Xu, P.; Zeng, L.; Zhou, Y.; et al. YTHDF1 links hypoxia adaptation and non-small cell lung cancer progression. Nat. Commun. 2019, 10, 4892. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Zhang, T.; Suo, C.; Zheng, C.; Zhang, H. Hypoxia and Metabolism in Metastasis. Adv. Exp. Med. Biol. 2019, 1136, 87–95. [Google Scholar]
- Gnoni, A.; Brunetti, O.; Longo, V.; Calabrese, A.; Argentiero, A.-l.; Calbi, R.; Solimando Antonio, G.; Licchetta, A. Immune system and bone microenvironment: Rationale for targeted cancer therapies. Oncotarget 2020, 11, 480–487. [Google Scholar]
- Morony, S.; Capparelli, C.; Sarosi, I.; Lacey, D.L.; Dunstan, C.R.; Kostenuik, P.J. Osteoprotegerin inhibits osteolysis and decreases skeletal tumor burden in syngeneic and nude mouse models of experimental bone metastasis. Cancer Res. 2001, 61, 4432–4436. [Google Scholar]
- Wong, S.K.; Mohamad, N.-V.; Giaze, T.R.; Chin, K.-Y.; Mohamed, N.; Ima-Nirwana, S. Prostate Cancer and Bone Metastases: The Underlying Mechanisms. Int. J. Mol. Sci. 2019, 20, 2587. [Google Scholar] [CrossRef]
- Fang, C.; He, M.; Li, D.; Xu, Q. YTHDF2 mediates LPS-induced osteoclastogenesis and inflammatory response via the NF-κB and MAPK signaling pathways. Cell Signal 2021, 85, 110060. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Qiao, S.-C.; Wu, X.-B.; Sun, B.; Yang, J.-G.; Li, X.; Zhang, X.; Qian, S.-J.; Gu, Y.-X.; Lai, H.-C. Circ_0008542 in osteoblast exosomes promotes osteoclast-induced bone resorption through m6A methylation. Cell Death Dis. 2021, 12, 628. [Google Scholar] [CrossRef] [PubMed]
- Previte, D.M.; Martins, C.P.; O’Connor, E.C.; Marre, M.L.; Coudriet, G.M.; Beck, N.W.; Menk, A.V.; Wright, R.H.; Tse, H.M.; Delgoffe, G.M.; et al. Lymphocyte Activation Gene-3 Maintains Mitochondrial and Metabolic Quiescence in Naive CD4 T Cells. Cell Rep. 2019, 27, 129–141.e4. [Google Scholar] [CrossRef] [PubMed]
- Bengsch, B.; Johnson, A.L.; Kurachi, M.; Odorizzi, P.M.; Pauken, K.E.; Attanasio, J.; Stelekati, E.; McLane, L.M.; Paley, M.A.; Delgoffe, G.M.; et al. Bioenergetic Insufficiencies Due to Metabolic Alterations Regulated by the Inhibitory Receptor PD-1 Are an Early Driver of CD8(+) T Cell Exhaustion. Immunity 2016, 45, 358–373. [Google Scholar] [CrossRef]
- Liu, Y.; Cao, X. Immunosuppressive cells in tumor immune escape and metastasis. J. Mol. Med. 2016, 94, 509–522. [Google Scholar] [CrossRef]
- Li, M.; Zha, X.; Wang, S. The role of N6-methyladenosine mRNA in the tumor microenvironment. Biochim. Biophys. Acta Rev. Cancer 2021, 1875, 188522. [Google Scholar] [CrossRef]
- Patsoukis, N.; Bardhan, K.; Chatterjee, P.; Sari, D.; Liu, B.; Bell, L.N.; Karoly, E.D.; Freeman, G.J.; Petkova, V.; Seth, P.; et al. PD-1 alters T-cell metabolic reprogramming by inhibiting glycolysis and promoting lipolysis and fatty acid oxidation. Nat. Commun. 2015, 6, 6692. [Google Scholar] [CrossRef]
- Xiong, J.; He, J.; Zhu, J.; Pan, J.; Liao, W.; Ye, H.; Wang, H.; Song, Y.; Du, Y.; Cui, B.; et al. Lactylation-driven METTL3-mediated RNA mA modification promotes immunosuppression of tumor-infiltrating myeloid cells. Mol. Cell 2022, 82, 1660–1677. [Google Scholar] [CrossRef]
- Dong, L.; Chen, C.; Zhang, Y.; Guo, P.; Wang, Z.; Li, J.; Liu, Y.; Liu, J.; Chang, R.; Li, Y.; et al. The loss of RNA N-adenosine methyltransferase Mettl14 in tumor-associated macrophages promotes CD8 T cell dysfunction and tumor growth. Cancer Cell 2021, 39, 945–957.e10. [Google Scholar] [CrossRef]
- Qiu, X.; Yang, S.; Wang, S.; Wu, J.; Zheng, B.; Wang, K.; Shen, S.; Jeong, S.; Li, Z.; Zhu, Y.; et al. MA Demethylase ALKBH5 Regulates PD-L1 Expression and Tumor Immunoenvironment in Intrahepatic Cholangiocarcinoma. Cancer Res. 2021, 81, 4778–4793. [Google Scholar] [CrossRef]
- Wang, L.; Hui, H.; Agrawal, K.; Kang, Y.; Li, N.; Tang, R.; Yuan, J.; Rana, T.M. m A RNA methyltransferases METTL3/14 regulate immune responses to anti-PD-1 therapy. EMBO J. 2020, 39, e104514. [Google Scholar] [CrossRef] [PubMed]
- Wan, W.; Ao, X.; Chen, Q.; Yu, Y.; Ao, L.; Xing, W.; Guo, W.; Wu, X.; Pu, C.; Hu, X.; et al. METTL3/IGF2BP3 axis inhibits tumor immune surveillance by upregulating N-methyladenosine modification of PD-L1 mRNA in breast cancer. Mol. Cancer 2022, 21, 60. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wei, J.; Cui, Y.-H.; Park, G.; Shah, P.; Deng, Y.; Aplin, A.E.; Lu, Z.; Hwang, S.; He, C.; et al. mA mRNA demethylase FTO regulates melanoma tumorigenicity and response to anti-PD-1 blockade. Nat. Commun. 2019, 10, 2782. [Google Scholar] [CrossRef]
- Li, N.; Kang, Y.; Wang, L.; Huff, S.; Tang, R.; Hui, H.; Agrawal, K.; Gonzalez, G.M.; Wang, Y.; Patel, S.P.; et al. ALKBH5 regulates anti-PD-1 therapy response by modulating lactate and suppressive immune cell accumulation in tumor microenvironment. Proc. Natl. Acad. Sci. USA 2020, 117, 20159–20170. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Zhu, L.; Lu, C.; Wang, C.; Wang, H.; Jin, H.; Ma, X.; Cheng, Z.; Yu, C.; Wang, S.; et al. circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nat. Commun. 2021, 12, 295. [Google Scholar] [CrossRef]
- Liu, Z.; Zhong, J.; Zeng, J.; Duan, X.; Lu, J.; Sun, X.; Liu, Q.; Liang, Y.; Lin, Z.; Zhong, W.; et al. Characterization of the m6A-Associated Tumor Immune Microenvironment in Prostate Cancer to Aid Immunotherapy. Front. Immunol. 2021, 12, 735170. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Wang, H.; Li, H.; Ye, X.; Xia, Y.; Yuan, S.; Lu, J.; Xie, X.; Wang, L.; Zhang, J. Integrated analyses of mA regulator-mediated modification patterns in tumor microenvironment-infiltrating immune cells in colon cancer. Oncoimmunology 2021, 10, 1936758. [Google Scholar] [CrossRef]
- Shriwas, O.; Mohapatra, P.; Mohanty, S.; Dash, R. The Impact of m6A RNA Modification in Therapy Resistance of Cancer: Implication in Chemotherapy, Radiotherapy, and Immunotherapy. Front. Oncol. 2020, 10, 612337. [Google Scholar] [CrossRef]
- Xiao, L.; Li, X.; Mu, Z.; Zhou, J.; Zhou, P.; Xie, C.; Jiang, S. FTO Inhibition Enhances the Antitumor Effect of Temozolomide by Targeting MYC-miR-155/23a Cluster-MXI1 Feedback Circuit in Glioma. Cancer Res. 2020, 80, 3945–3958. [Google Scholar] [CrossRef]
- Jin, D.; Guo, J.; Wu, Y.; Du, J.; Yang, L.; Wang, X.; Di, W.; Hu, B.; An, J.; Kong, L.; et al. mA mRNA methylation initiated by METTL3 directly promotes YAP translation and increases YAP activity by regulating the MALAT1-miR-1914-3p-YAP axis to induce NSCLC drug resistance and metastasis. J. Hematol. Oncol. 2019, 12, 135. [Google Scholar] [CrossRef]
- Liu, S.; Li, Q.; Li, G.; Zhang, Q.; Zhuo, L.; Han, X.; Zhang, M.; Chen, X.; Pan, T.; Yan, L.; et al. The mechanism of mA methyltransferase METTL3-mediated autophagy in reversing gefitinib resistance in NSCLC cells by β-elemene. Cell Death Dis. 2020, 11, 969. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.; Hong, X.; Li, S.; Meng, P.; Xiao, F. METTL3 promotes adriamycin resistance in MCF-7 breast cancer cells by accelerating pri-microRNA-221-3p maturation in a m6A-dependent manner. Exp. Mol. Med. 2021, 53, 91–102. [Google Scholar] [CrossRef] [PubMed]
- Fukumoto, T.; Zhu, H.; Nacarelli, T.; Karakashev, S.; Fatkhutdinov, N.; Wu, S.; Liu, P.; Kossenkov, A.V.; Showe, L.C.; Jean, S.; et al. N-Methylation of Adenosine of mRNA Contributes to PARP Inhibitor Resistance. Cancer Res. 2019, 79, 2812–2820. [Google Scholar] [CrossRef] [PubMed]
- Privitera, A.P.; Barresi, V.; Condorelli, D.F. Aberrations of Chromosomes 1 and 16 in Breast Cancer: A Framework for Cooperation of Transcriptionally Dysregulated Genes. Cancers 2021, 13, 1585. [Google Scholar] [CrossRef]
- Lin, Z.; Niu, Y.; Wan, A.; Chen, D.; Liang, H.; Chen, X.; Sun, L.; Zhan, S.; Chen, L.; Cheng, C.; et al. RNA m A methylation regulates sorafenib resistance in liver cancer through FOXO3-mediated autophagy. EMBO J. 2020, 39, e103181. [Google Scholar] [CrossRef]
- Wu, P.; Fang, X.; Liu, Y.; Tang, Y.; Wang, W.; Li, X.; Fan, Y. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021, 12, 298. [Google Scholar] [CrossRef]
- Cui, Q.; Shi, H.; Ye, P.; Li, L.; Qu, Q.; Sun, G.; Sun, G.; Lu, Z.; Huang, Y.; Yang, C.-G.; et al. mA RNA Methylation Regulates the Self-Renewal and Tumorigenesis of Glioblastoma Stem Cells. Cell Rep. 2017, 18, 2622–2634. [Google Scholar] [CrossRef]
- Dixit, D.; Prager, B.C.; Gimple, R.C.; Poh, H.X.; Wang, Y.; Wu, Q.; Qiu, Z.; Kidwell, R.L.; Kim, L.J.Y.; Xie, Q.; et al. The RNA m6A Reader YTHDF2 Maintains Oncogene Expression and Is a Targetable Dependency in Glioblastoma Stem Cells. Cancer Discov. 2021, 11, 480–499. [Google Scholar] [CrossRef]
- Su, R.; Dong, L.; Li, Y.; Gao, M.; Han, L.; Wunderlich, M.; Deng, X.; Li, H.; Huang, Y.; Gao, L.; et al. Targeting FTO Suppresses Cancer Stem Cell Maintenance and Immune Evasion. Cancer Cell 2020, 38, 79–96. [Google Scholar] [CrossRef]
- Relier, S.; Ripoll, J.; Guillorit, H.; Amalric, A.; Achour, C.; Boissière, F.; Vialaret, J.; Attina, A.; Debart, F.; Choquet, A.; et al. FTO-mediated cytoplasmic mA demethylation adjusts stem-like properties in colorectal cancer cell. Nat. Commun. 2021, 12, 1716. [Google Scholar] [CrossRef]
- Huang, H.; Wang, Y.; Kandpal, M.; Zhao, G.; Cardenas, H.; Ji, Y.; Chaparala, A.; Tanner, E.J.; Chen, J.; Davuluri, R.V.; et al. FTO-Dependent -Methyladenosine Modifications Inhibit Ovarian Cancer Stem Cell Self-Renewal by Blocking cAMP Signaling. Cancer Res. 2020, 80, 3200–3214. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Yang, H.; Yang, C.; Bao, Y.-L.; Yang, S.-M.; Liu, J.; Xiao, Y.-F. Translation of noncoding RNAs and cancer. Cancer Lett. 2021, 497, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Yankova, E.; Blackaby, W.; Albertella, M.; Rak, J.; De Braekeleer, E.; Tsagkogeorga, G.; Pilka, E.S.; Aspris, D.; Leggate, D.; Hendrick, A.G.; et al. Small-molecule inhibition of METTL3 as a strategy against myeloid leukaemia. Nature 2021, 593, 597–601. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-H.; Choi, N.; Kim, S.; Jin, M.S.; Shen, H.; Kim, Y.-C. Eltrombopag as an Allosteric Inhibitor of the METTL3-14 Complex Affecting the mA Methylation of RNA in Acute Myeloid Leukemia Cells. Pharmaceuticals 2022, 15, 440. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Su, R.; Sheng, Y.; Dong, L.; Dong, Z.; Xu, H.; Ni, T.; Zhang, Z.S.; Zhang, T.; Li, C.; et al. Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia. Cancer Cell 2019, 35, 677–691. [Google Scholar] [CrossRef]
- Sabnis, R.W. Novel Small Molecule RNA m6A Demethylase AlkBH5 Inhibitors for Treating Cancer. ACS Med. Chem. Lett. 2021, 12, 856–857. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).