Circular and Fusion RNAs in Medulloblastoma Development
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Sample Information
2.2. RNA-Seq Library Preparations
2.3. RNA-Seq Data Analysis
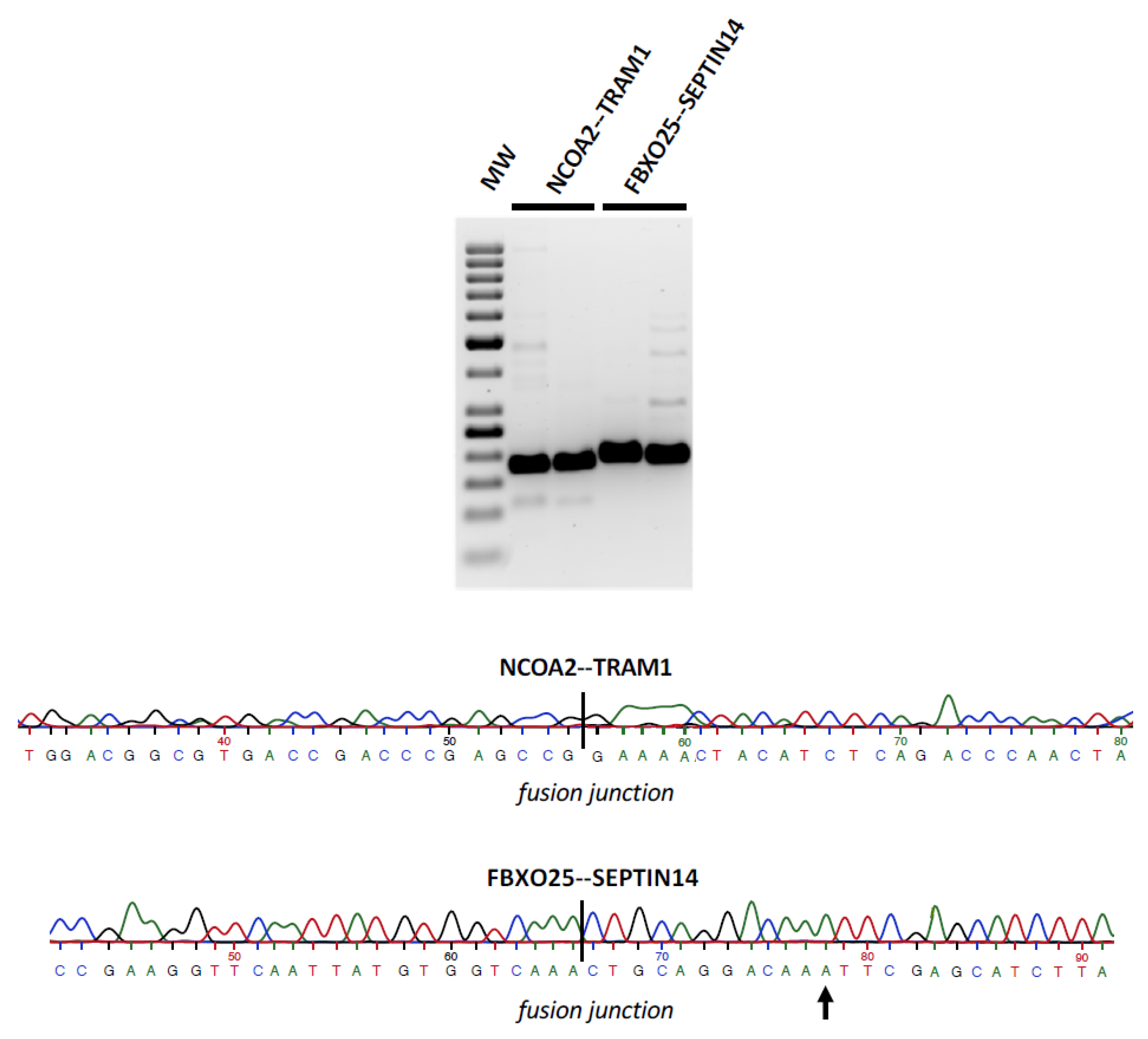
2.4. Validation of Fusion Transcripts by Sanger Sequencing
3. Results
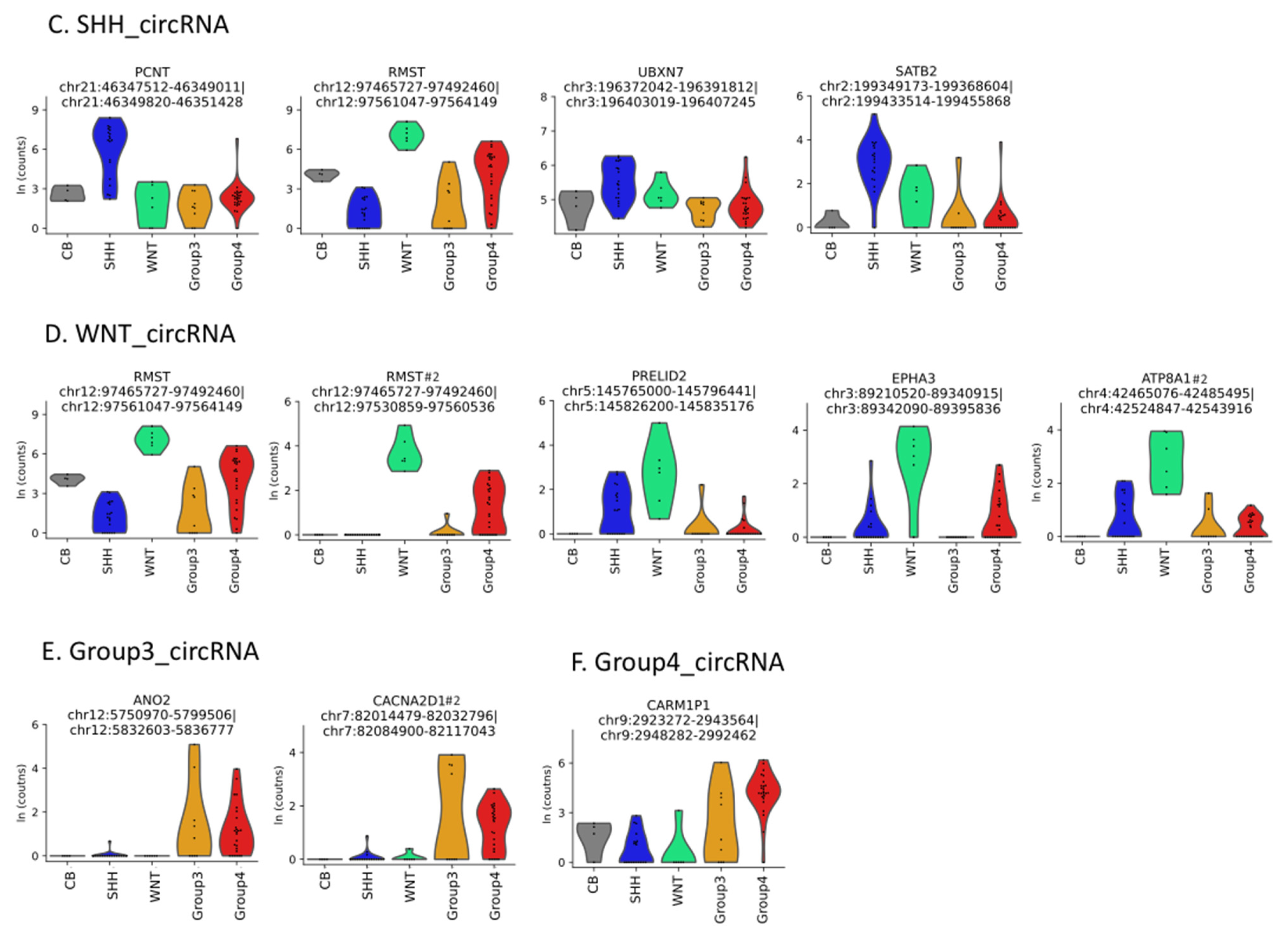
3.1. Circular RNAs
3.2. Fusion RNAs–Cerebellum
3.3. Fusion RNAs–Medulloblastoma
4. Discussion
5. Limitations
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fang, F.Y.; Rosenblum, J.S.; Ho, W.S.; Heiss, J.D. New Developments in the Pathogenesis, Therapeutic Targeting, and Treatment of Pediatric Medulloblastoma. Cancers 2022, 14, 2285. [Google Scholar] [CrossRef]
- Duc, N.M.; Huy, H.Q. Magnetic Resonance Imaging Features of Common Posterior Fossa Brain Tumors in Children: A Preliminary Vietnamese Study. Open Access Maced. J. Med. Sci. 2019, 7, 2413–2418. [Google Scholar] [CrossRef] [PubMed]
- Duc, N.M.; Huy, H.Q.; Nadarajan, C.; Keserci, B. The Role of Predictive Model Based on Quantitative Basic Magnetic Resonance Imaging in Differentiating Medulloblastoma from Ependymoma. Anticancer Res. 2020, 40, 2975–2980. [Google Scholar] [CrossRef] [PubMed]
- Girardi, F.; Allemani, C.; Coleman, M.P. Worldwide Trends in Survival from Common Childhood Brain Tumors: A Systematic Review. J. Glob. Oncol. 2019, 5, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Juraschka, K.; Taylor, M.D. Medulloblastoma in the age of molecular subgroups: A review. J. Neurosurg. Pediatr. 2019, 24, 353–363. [Google Scholar] [CrossRef]
- Salzman, J.; Gawad, C.; Wang, P.L.; Lacayo, N.; Brown, P.O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733. [Google Scholar] [CrossRef]
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.D.; Gregersen, L.H.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338. [Google Scholar] [CrossRef]
- Zaphiropoulos, P.G. Circular RNAs from transcripts of the rat cytochrome P450 2C24 gene: Correlation with exon skipping. Proc. Natl. Acad. Sci. USA 1996, 93, 6536–6541. [Google Scholar] [CrossRef]
- Nigro, J.M.; Cho, K.R.; Fearon, E.R.; Kern, S.E.; Ruppert, J.M.; Oliner, J.D.; Kinzler, K.W.; Vogelstein, B. Scrambled exons. Cell 1991, 64, 607–613. [Google Scholar] [CrossRef]
- Cocquerelle, C.; Daubersies, P.; Majerus, M.A.; Kerckaert, J.P.; Bailleul, B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 1992, 11, 1095–1098. [Google Scholar] [CrossRef]
- Piwecka, M.; Glazar, P.; Hernandez-Miranda, L.R.; Memczak, S.; Wolf, S.A.; Rybak-Wolf, A.; Filipchyk, A.; Klironomos, F.; Cerda Jara, C.A.; Fenske, P.; et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 2017, 357, eaam8526. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef] [PubMed]
- Kristensen, L.S.; Jakobsen, T.; Hager, H.; Kjems, J. The emerging roles of circRNAs in cancer and oncology. Nat. Rev. Clin. Oncol. 2021, 19, 188–206. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, C.; Du, Y.; Li, Z.; Li, M.; Hou, P.; Shen, Z.; Chu, S.; Zheng, J.; Bai, J. Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol. Cancer 2022, 21, 13. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.X.; Li, X.; Nan, F.; Jiang, S.; Gao, X.; Guo, S.K.; Xue, W.; Cui, Y.; Dong, K.; Ding, H.; et al. Structure and Degradation of Circular RNAs Regulate PKR Activation in Innate Immunity. Cell 2019, 177, 865–880.e821. [Google Scholar] [CrossRef] [PubMed]
- Vo, J.N.; Cieslik, M.; Zhang, Y.; Shukla, S.; Xiao, L.; Zhang, Y.; Wu, Y.M.; Dhanasekaran, S.M.; Engelke, C.G.; Cao, X.; et al. The Landscape of Circular RNA in Cancer. Cell 2019, 176, 869–881.e813. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Huang, V.; Xu, X.; Livingstone, J.; Soares, F.; Jeon, J.; Zeng, Y.; Hua, J.T.; Petricca, J.; Guo, H.; et al. Widespread and Functional RNA Circularization in Localized Prostate Cancer. Cell 2019, 176, 831–843.e822. [Google Scholar] [CrossRef]
- Hsiao, K.Y.; Lin, Y.C.; Gupta, S.K.; Chang, N.; Yen, L.; Sun, H.S.; Tsai, S.J. Noncoding Effects of Circular RNA CCDC66 Promote Colon Cancer Growth and Metastasis. Cancer Res. 2017, 77, 2339–2350. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.W.; Zheng, F.X.; Xiao, X.Y.; Xie, F.; Tao, D.; Huang, C.; Liu, D.; Wang, M.; Wang, L.; Zeng, F.Q.; et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017, 18, 1646–1659. [Google Scholar] [CrossRef]
- Yu, J.; Xu, Q.G.; Wang, Z.G.; Yang, Y.; Zhang, L.; Ma, J.Z.; Sun, S.H.; Yang, F.; Zhou, W.P. Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 2018, 68, 1214–1227. [Google Scholar] [CrossRef]
- Lv, T.; Miao, Y.F.; Jin, K.; Han, S.; Xu, T.Q.; Qiu, Z.L.; Zhang, X.H. Dysregulated circular RNAs in medulloblastoma regulate proliferation and growth of tumor cells via host genes. Cancer Med. 2018, 7, 6147–6157. [Google Scholar] [CrossRef] [PubMed]
- Azatyan, A.; Zhang, S.; Darabi, A.; Siesjo, P.; Wang, T.; Zaphiropoulos, P.G. Circular RNAs in Hedgehog Signaling Activation and Hedgehog-Mediated Medulloblastoma Tumors. Cancers 2021, 13, 5138. [Google Scholar] [CrossRef] [PubMed]
- Caudevilla, C.; Serra, D.; Miliar, A.; Codony, C.; Asins, G.; Bach, M.; Hegardt, F.G. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc. Natl. Acad. Sci. USA 1998, 95, 12185–12190. [Google Scholar] [CrossRef] [PubMed]
- Frantz, S.A.; Thiara, A.S.; Lodwick, D.; Ng, L.L.; Eperon, I.C.; Samani, N.J. Exon repetition in mRNA. Proc. Natl. Acad. Sci. USA 1999, 96, 5400–5405. [Google Scholar] [CrossRef]
- Guo, J.U.; Agarwal, V.; Guo, H.; Bartel, D.P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409. [Google Scholar] [CrossRef]
- Zaphiropoulos, P.G. RNA molecules containing exons originating from different members of the cytochrome P450 2C gene subfamily (CYP2C) in human epidermis and liver. Nucleic Acids Res. 1999, 27, 2585–2590. [Google Scholar] [CrossRef][Green Version]
- Finta, C.; Zaphiropoulos, P.G. The human CYP2C locus: A prototype for intergenic and exon repetition splicing events. Genomics 2000, 63, 433–438. [Google Scholar] [CrossRef]
- Finta, C.; Zaphiropoulos, P.G. Intergenic mRNA molecules resulting from trans-splicing. J. Biol. Chem. 2002, 277, 5882–5890. [Google Scholar] [CrossRef]
- Li, H.; Wang, J.; Mor, G.; Sklar, J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 2008, 321, 1357–1361. [Google Scholar] [CrossRef]
- Yuan, H.; Qin, F.; Movassagh, M.; Park, H.; Golden, W.; Xie, Z.; Zhang, P.; Sklar, J.; Li, H. A chimeric RNA characteristic of rhabdomyosarcoma in normal myogenesis process. Cancer Discov. 2013, 3, 1394–1403. [Google Scholar] [CrossRef]
- Babiceanu, M.; Qin, F.; Xie, Z.; Jia, Y.; Lopez, K.; Janus, N.; Facemire, L.; Kumar, S.; Pang, Y.; Qi, Y.; et al. Recurrent chimeric fusion RNAs in non-cancer tissues and cells. Nucleic Acids Res. 2016, 44, 2859–2872. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Qin, F.; Kumar, S.; Elfman, J.; Lin, E.; Pham, L.P.; Yang, A.; Li, H. The landscape of chimeric RNAs in non-diseased tissues and cells. Nucleic Acids Res. 2020, 48, 1764–1778. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.O.; Dong, R.; Zhang, Y.; Zhang, J.L.; Luo, Z.; Zhang, J.; Chen, L.L.; Yang, L. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 2016, 26, 1277–1287. [Google Scholar] [CrossRef] [PubMed]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Dobin, A.; Li, B.; Stransky, N.; Pochet, N.; Regev, A. Accuracy assessment of fusion transcript detection via read-mapping and de novo fusion transcript assembly-based methods. Genome Biol. 2019, 20, 213. [Google Scholar] [CrossRef]
- Liu, C.X.; Chen, L.L. Circular RNAs: Characterization, cellular roles, and applications. Cell 2022, 185, 2016–2034. [Google Scholar] [CrossRef]
- Wen, H.; Li, Y.; Malek, S.N.; Kim, Y.C.; Xu, J.; Chen, P.; Xiao, F.; Huang, X.; Zhou, X.; Xuan, Z.; et al. New fusion transcripts identified in normal karyotype acute myeloid leukemia. PLoS ONE 2012, 7, e51203. [Google Scholar] [CrossRef]
- Atak, Z.K.; Gianfelici, V.; Hulselmans, G.; De Keersmaecker, K.; Devasia, A.G.; Geerdens, E.; Mentens, N.; Chiaretti, S.; Durinck, K.; Uyttebroeck, A.; et al. Comprehensive analysis of transcriptome variation uncovers known and novel driver events in T-cell acute lymphoblastic leukemia. PLoS Genet. 2013, 9, e1003997. [Google Scholar] [CrossRef]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef]
- Cavalli, F.M.G.; Remke, M.; Rampasek, L.; Peacock, J.; Shih, D.J.H.; Luu, B.; Garzia, L.; Torchia, J.; Nor, C.; Morrissy, A.S.; et al. Intertumoral Heterogeneity within Medulloblastoma Subgroups. Cancer Cell 2017, 31, 737–754.e736. [Google Scholar] [CrossRef]
- Federico, C.; Bruno, F.; Ragusa, D.; Clements, C.S.; Brancato, D.; Henry, M.P.; Bridger, J.M.; Tosi, S.; Saccone, S. Chromosomal Rearrangements and Altered Nuclear Organization: Recent Mechanistic Models in Cancer. Cancers 2021, 13, 5860. [Google Scholar] [CrossRef] [PubMed]
- Ramsden, D.A.; Nussenzweig, A. Mechanisms driving chromosomal translocations: Lost in time and space. Oncogene 2021, 40, 4263–4270. [Google Scholar] [CrossRef]
- Jin, K.; Wang, S.; Zhang, Y.; Xia, M.; Mo, Y.; Li, X.; Li, G.; Zeng, Z.; Xiong, W.; He, Y. Long non-coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis. Cell. Mol. Life Sci. 2019, 76, 4275–4289. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Wang, Q.; Song, R.; Kong, Y.; Zhang, Z. Non-coding RNAs in depression: Promising diagnostic and therapeutic biomarkers. eBioMedicine 2021, 71, 103569. [Google Scholar] [CrossRef] [PubMed]


| Genes in Fusion | Chromosomal Position | Exon number at Fusion Junction | Relative Orientation |
|---|---|---|---|
| Cerebellum-linear | |||
| SEPTIN7P14--PSPH | Chr4-Chr7 | 1(NR_037630.1)-4(NM_004577.4) | NA |
| FBXO25--SEPTIN14 | Chr8-Chr7 | 5(NM_183420.2)-10(NM_207366.3) | NA |
| NCOA2--TRAM1 | Chr8, neighboring genes | 1(NM_006540.4)-5(NM_014294.6) | —> —> |
| PAUPAR--RCN1 | Chr11, neighboring genes | 1(NR_033971.1)-2(NM_002901.4) | —> —> |
| RPS10--NUDT3 | Chr6, neighboring genes | 5(NM_001014.5)-3(NM_006703.4) | —> —> |
| Cerebellum-circular | |||
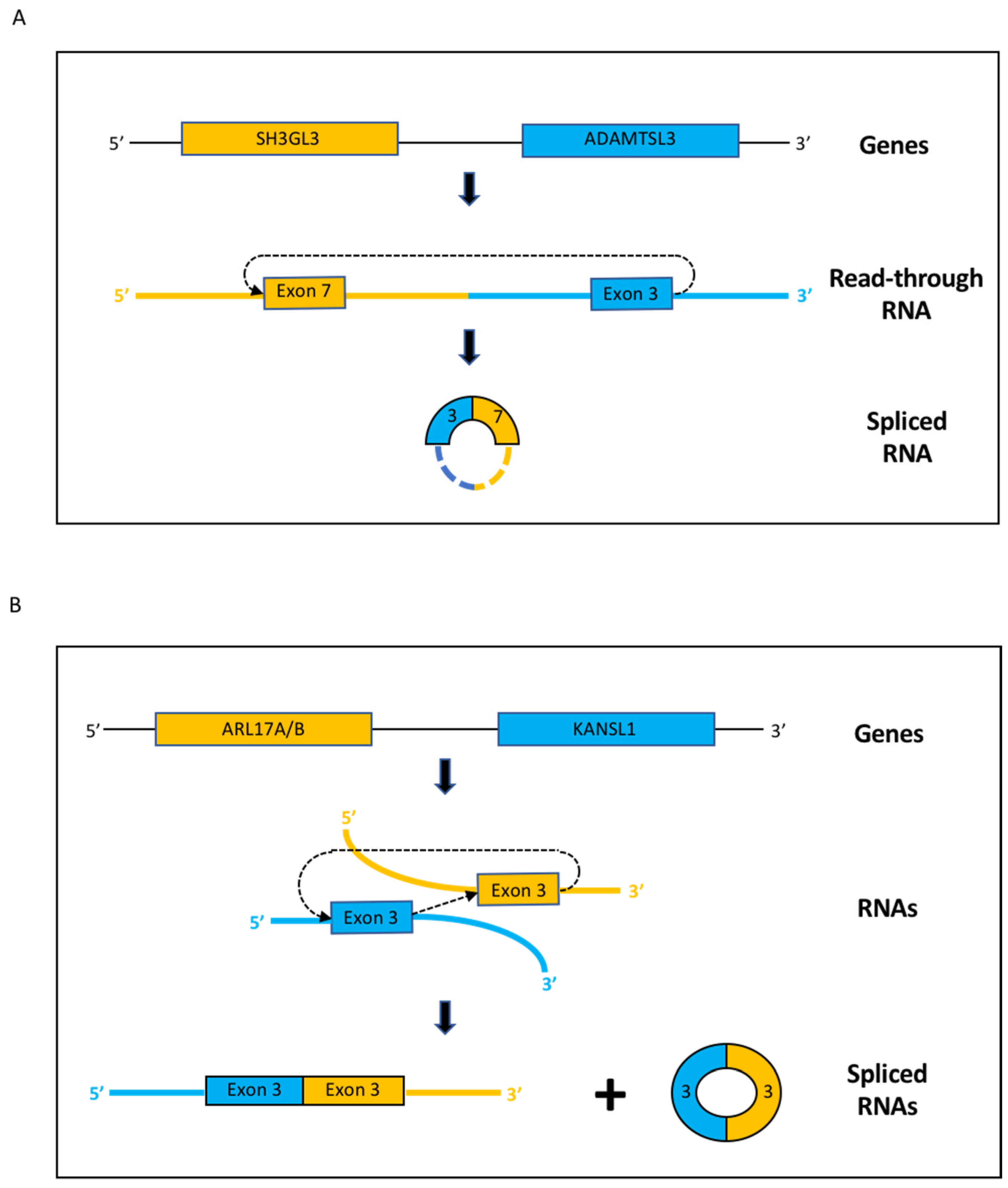
| ADAMTSL3--SH3GL3 | Chr15, neighboring genes | 3(NM_207517.3)-7(NM_003027.5) | —> —> |
| Medulloblastoma-linear | |||
| KANSL1--ARL17A | Chr17, neighboring genes | 3(NM_015443.4)-3(NM_001113738.2) | —> —> |
| TFG--ADGRG7 | Chr3, neighboring genes | 3(NM_006070.6)-2(NM_032787.3) | —> —> |
| SPSB4--PXYLP1 | Chr3, neighboring genes | 2(NM_080862.3)-2(NM_001037172.3) | —> —> |
| PVT1--CASC8 | Chr8, neighboring genes | 1(NR_003367.3)-6(NR_117100.1) | <— —> |
| Medulloblastoma-circular | |||
| ARL17A--KANSL1 | Chr17, neighboring genes | 3(NM_001113738.2)-3(NM_015443.4) | —> —> |
| KANSL1--ARL17A | Chr17, neighboring genes | 3(NM_015443.4)-3(NM_001113738.2) | —> —> |
| ARL17B--KANSL1 | Chr17, neighboring genes | 3(NM_001039083.5)-3(NM_015443.4) | —> —> |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azatyan, A.; Zaphiropoulos, P.G. Circular and Fusion RNAs in Medulloblastoma Development. Cancers 2022, 14, 3134. https://doi.org/10.3390/cancers14133134
Azatyan A, Zaphiropoulos PG. Circular and Fusion RNAs in Medulloblastoma Development. Cancers. 2022; 14(13):3134. https://doi.org/10.3390/cancers14133134
Chicago/Turabian StyleAzatyan, Ani, and Peter G. Zaphiropoulos. 2022. "Circular and Fusion RNAs in Medulloblastoma Development" Cancers 14, no. 13: 3134. https://doi.org/10.3390/cancers14133134
APA StyleAzatyan, A., & Zaphiropoulos, P. G. (2022). Circular and Fusion RNAs in Medulloblastoma Development. Cancers, 14(13), 3134. https://doi.org/10.3390/cancers14133134





