Clinical Study of Aspirin and Zileuton on Biomarkers of Tobacco-Related Carcinogenesis in Current Smokers
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Study Drugs
2.3. Study Population
2.4. Study Procedure
2.5. Nasal Brushing for Gene Expression Analysis
2.6. Analysis of Urinary Biomarkers of Arachidonic Acid Metabolism
2.7. Microarray Data Acquisition and Data Preprocessing
2.8. Calculation of Gene Expression Signature Scores
2.9. Identification of Gene Expression Changes Associated with Aspirin and Zileuton Treatment
2.10. Statistical Methods
3. Results
3.1. Participant Demographics
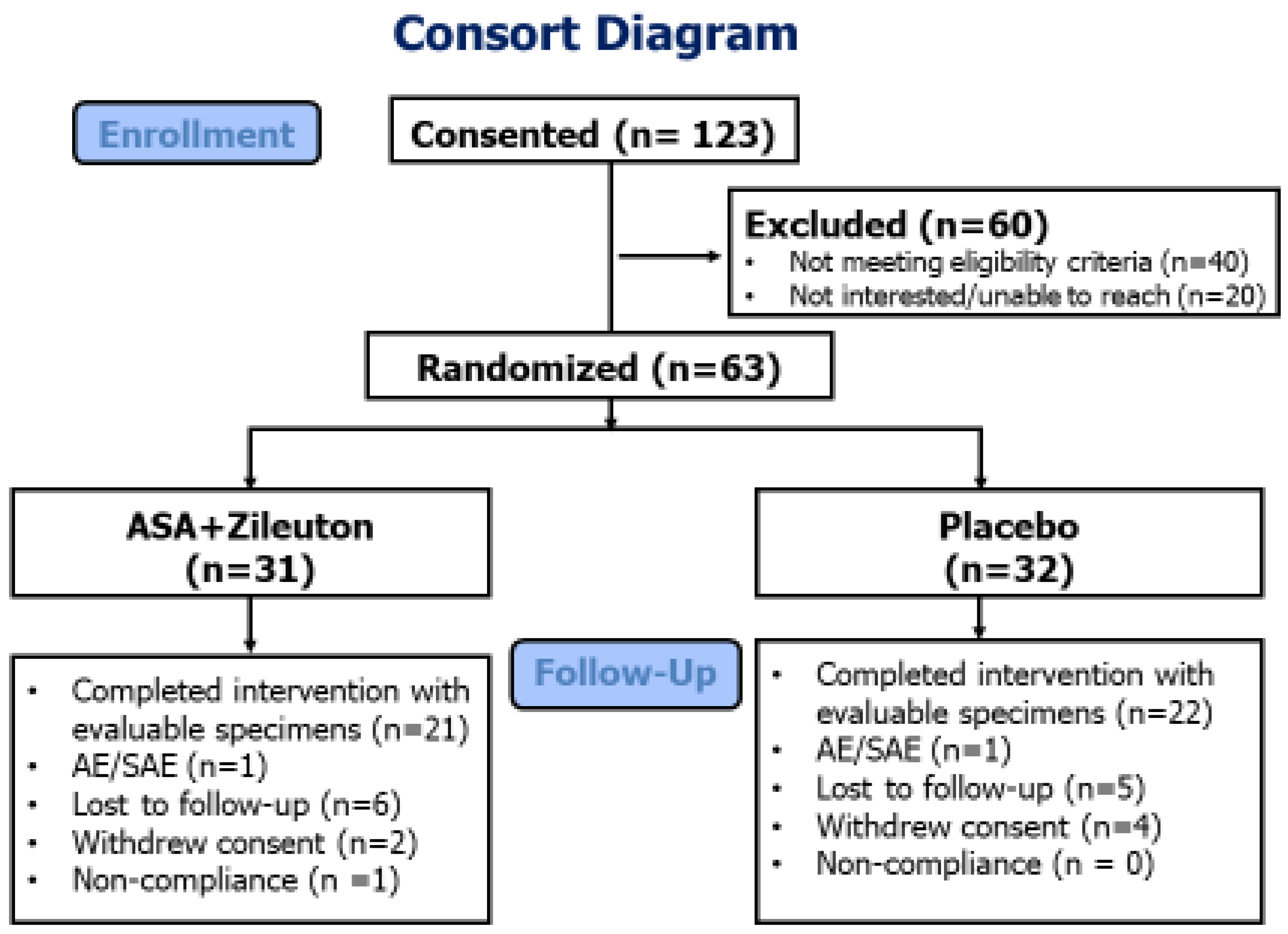
3.2. Adherence and Safety
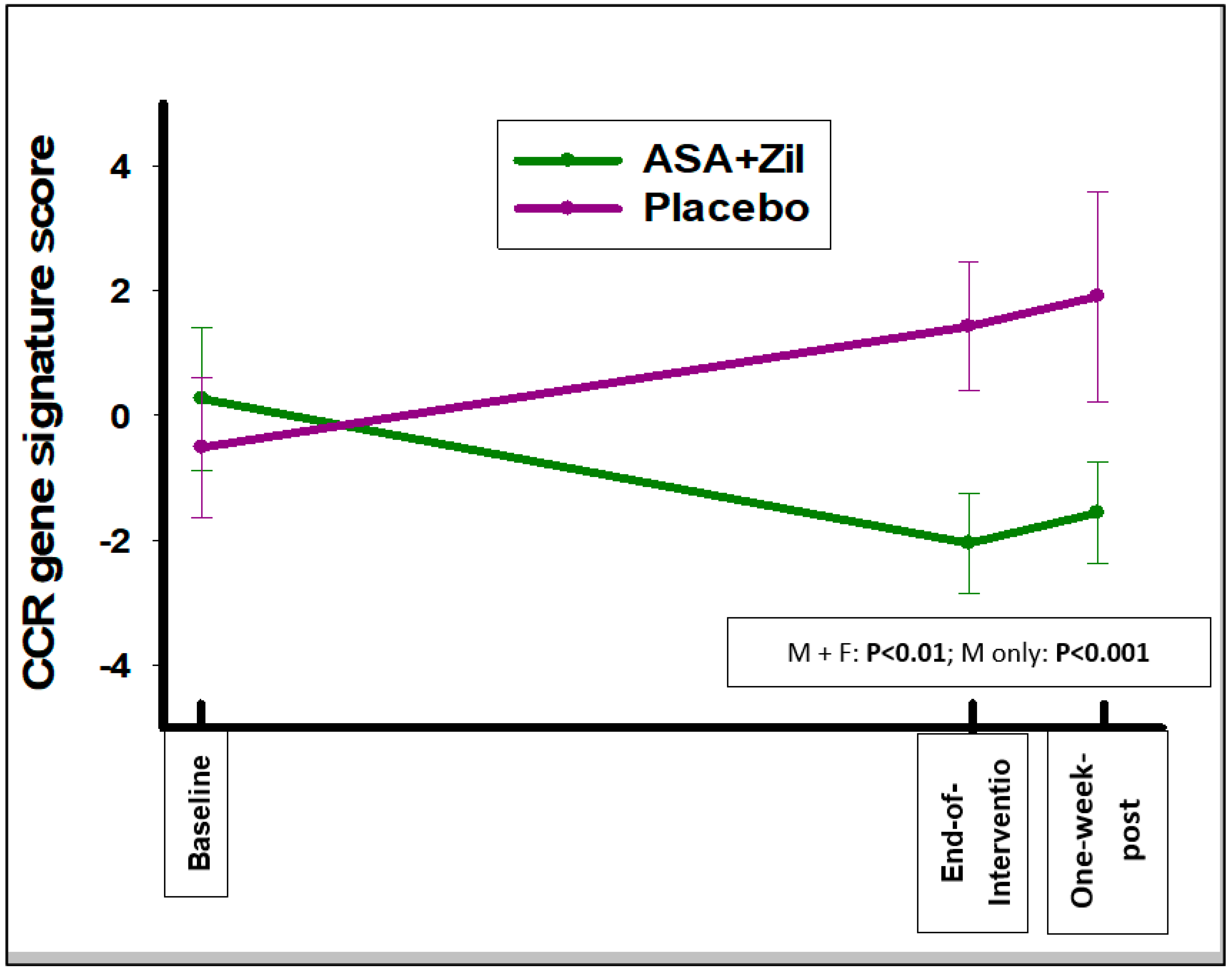
3.3. ASA and Zileuton Do Not Modulate the Smoking-Associated Gene Expression Signature; However, They Do Modulate a Bronchial Squamous Dysplasia Signature
3.4. ASA and Zileuton Suppresses the 5-LOX-Mediated AA Metabolite LTE4 but Does Not Suppress the COX-Mediated AA Metabolite PGEM
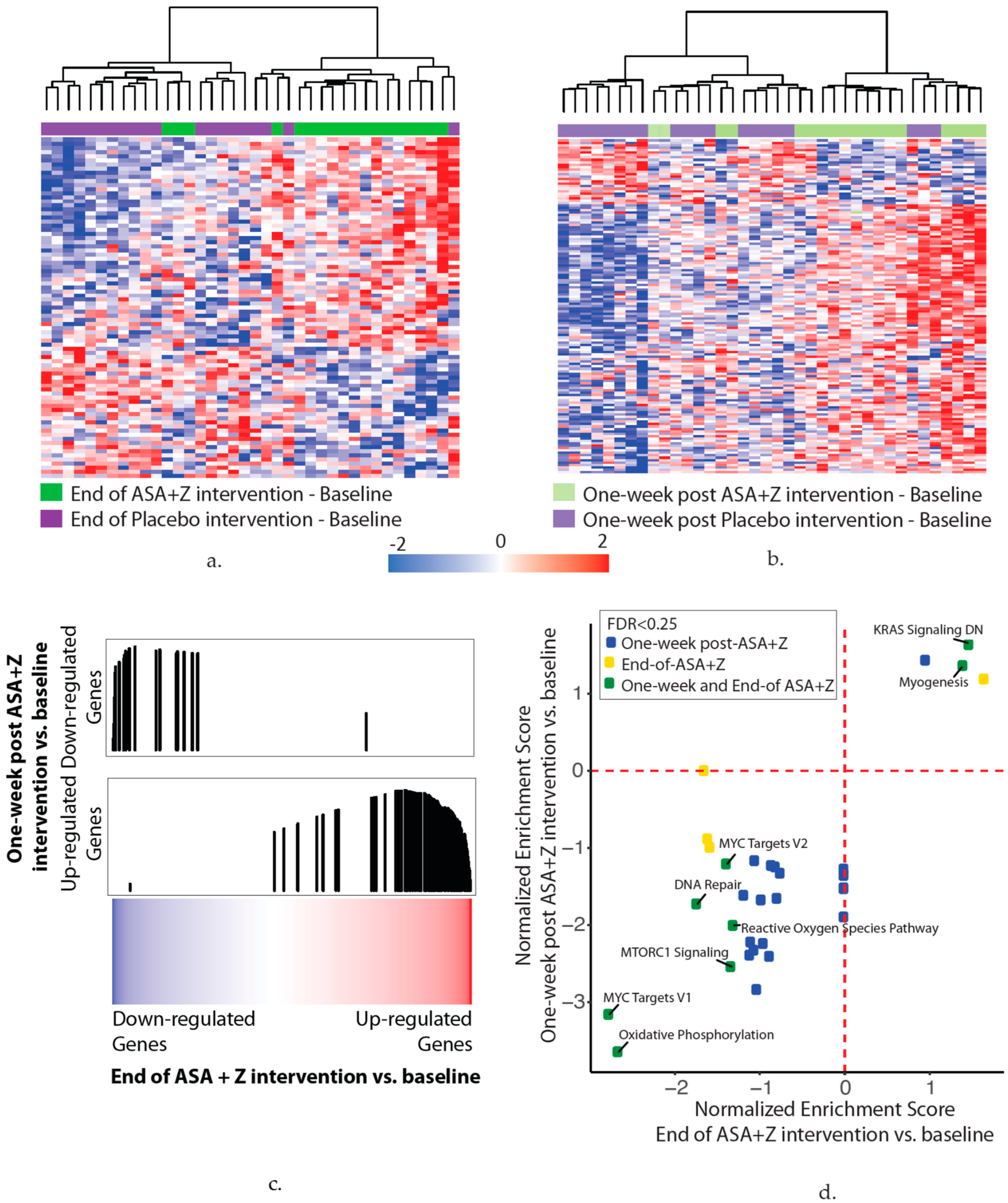
3.5. Aspirin and Zileuton Modulate Nasal Gene Expression
- (a)
- In total, 83 genes were associated with the end of the aspirin and zileuton treatment compared with baseline (p < 0.01). The heatmap shows hierarchal clustering of the z-score-normalized change in the gene expression between the end-of-treatment and baseline time points for each subject in the aspirin and zileuton (green) or placebo (purple) arms (n = 38 subjects).
- (b)
- In total, 139 genes were associated with the one-week post aspirin and zileuton treatment time point compared with baseline (p < 0.01). The heatmap shows hierarchal clustering of the z-score-normalized change in the gene expression between the one-week post-treatment and baseline time points for each subject in the aspirin and zileuton (light green) or placebo (light purple) arms (n = 38 subjects).
- (c)
- Genes that were altered at the end of the aspirin and zileuton treatment compared with baseline are concordant with the genes that were altered one week after the aspirin and zileuton treatment by GSEA (FDR < 0.05), suggesting that changes persist for at least one week post-intervention. Genes were ranked by the moderated t-statistic for the effect of the end of the aspirin and zileuton treatment versus baseline (x-axis) and the gene sets represent the genes that were up- and downregulated one week after the aspirin and zileuton treatment from (b). The black vertical lines represent the position of the genes in the ranked list (x-axis) and the height corresponds to the magnitude of the running enrichment score form GSEA (y-axis).
- (d)
- Ranked lists (genes ranked by moderated t-statistics) for the end-of-treatment and one-week post-aspirin and zileuton treatment versus baseline effects were used for pathway enrichment analysis via GSEA using the MSigDB hallmark gene set. The scatterplot shows the normalized enrichment scores for pathways enriched (FDR < 0.25) at the end of (x-axis) and one week after (y-axis) the aspirin and zileuton treatment. Positive and negative scores represent pathways enriched among upregulated and downregulated genes with the end of (yellow) and one week after (blue) or both (green) aspirin and zileuton treatment, respectively.
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer Statistics 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Mills, E.J.; Wu, P.; Alberton, M.; Kanters, S.; Lanas, A.; Lester, R. Low-dose Aspirin and Cancer Mortality: Of Randomized Trials. Am. J. Med. 2012, 125, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Algra, A.M.; Rothwell, P.M. Effects of regular aspirin on long-term cancer incidence and metastasis: A systematic evidence from observational studies versus randomised trials. Lancet Oncol. 2012, 13, 518–527. [Google Scholar] [CrossRef]
- Bosetti, C.; Rosato, V.; Gallus, S.; Cuzick, J.; La Vecchia, C. Aspirin and cancer risk: A quantitative review to 2011. Ann. Oncol. 2012, 23, 1403–1415. [Google Scholar] [CrossRef]
- Slatore, C.G.; Au, D.H.; Littman, A.J.; Satia, J.A.; White, E. Association of Nonsteroidal Anti-Inflammatory Drugs with Lung Cancer: Results from a Large Cohort Study. Cancer Epidemiol. Biomark. Prev. 2009, 18, 1203–1207. [Google Scholar] [CrossRef]
- Castonguay, A.; Rioux, N.; Duperron, C.; Jalbert, G. Inhibition of lung tumorigenesis by NSAIDS: A working hypothesis. Exp. Lung Res. 1998, 24, 605–615. [Google Scholar] [CrossRef]
- Saini, R.K.; Sanyal, S.N. Chemopreventive Effect of Nonsteroidal Anti-inflammatory Drugs on 9,10-Dimethylbenz[a]anthracene-Induced Lung Carcinogenesis in Mice. Oncol. Res. 2009, 17, 505–518. [Google Scholar] [CrossRef]
- Bell, R.L.; Young, P.R.; Albert, D.; Lanni, C.; Summers, J.B.; Brooks, D.W.; Rubin, P.; Carter, G.W. The Discovery and Development of Zileuton—An Orally Active 5-Lipoxygenase Inhibitor. Int. J. Immunopharmacol. 1992, 14, 505–510. [Google Scholar] [CrossRef]
- Carter, G.W.; Young, P.R.; Albert, D.H.; Bouska, J.; Dyer, R.; Bell, R.L.; Summers, J.B.; Brooks, D.W. 5-Lipoxygenase Inhibitory Activity of Zileuton. J. Pharmacol. Exp. Ther. 1991, 256, 929–937. [Google Scholar]
- Steele, V.E.; Holmes, C.A.; Hawk, E.T.; Kopelovich, L.; Lubet, R.A.; Crowell, J.A.; Sigman, C.C.; Kelloff, G.J. Lipoxygenase inhibitors as potential cancer chemopreventives. Cancer Epidemiol. Biomark. Prev. 1999, 8, 467–483. [Google Scholar]
- Avis, I.M.; Jett, M.; Boyle, T.; Vos, M.D.; Moody, T.; Treston, A.M.; Martínez, A.; Mulshine, J.L. Growth control of lung cancer by interruption of 5-lipoxygenase-mediated growth factor signaling. J. Clin. Investig. 1996, 97, 806–813. [Google Scholar] [CrossRef] [PubMed]
- Rioux, N.; Castonguay, A. Inhibitors of lipoxygenase: A new class of cancer chemopreventive agents. Carcinogenesis 1998, 19, 1393–1400. [Google Scholar] [CrossRef] [PubMed]
- Moody, T.W.; Leyton, J.; Martinez, A.; Hong, S.; Malkinson, A.; Mulshine, J.L. Lipoxygenase inhibitors prevent lung carcinogenesis and inhibit non-small cell lung cancer growth. Exp. Lung Res. 1998, 24, 617–628. [Google Scholar] [CrossRef]
- Gunning, W.T.; Kramer, P.M.; Steele, V.E.; Pereira, M.A. Chemoprevention by lipoxygenase and leukotriene pathway inhibitors of vinyl carbamate-induced lung tumors in mice. Cancer Res. 2002, 62, 4199–4201. [Google Scholar]
- Mohebati, A.; Milne, G.L.; Zhou, X.K.; Duffield-Lillico, A.J.; Boyle, J.O.; Knutson, A.; Bosworth, B.P.; Kingsley, P.J.; Marnett, L.J.; Brown, P.H.; et al. Effect of Zileuton and Celecoxib on Urinary LTE4 and PGE-M Levels in Smokers. Cancer Prev. Res. 2013, 6, 646–655. [Google Scholar] [CrossRef][Green Version]
- Mao, L.; Lee, J.S.; Kurie, J.M.; Fan, Y.H.; Lippman, S.M.; Lee, J.J.; Ro, J.Y.; Broxson, A.; Yu, R.; Morice, R.C.; et al. Clonal genetic alterations in the lungs of current and former smokers. J. Natl. Cancer Inst. 1997, 89, 857–862. [Google Scholar] [CrossRef]
- Harvey, B.G.; Heguy, A.; Leopold, P.L.; Carolan, B.J.; Ferris, B.; Crystal, R.G. Modification of gene expression of the small airway epithelium in response to cigarette smoking. J. Mol. Med. 2007, 85, 39–53. [Google Scholar] [CrossRef]
- Zhang, L.; Lee, J.J.; Tang, H.; Fan, Y.H.; Xiao, L.; Ren, H.; Kurie, J.; Morice, R.C.; Hong, W.K.; Mao, L. Impact of smoking cessation on global gene expression in the bronchial epithelium of chronic smokers. Cancer Prev. Res. 2008, 1, 112–118. [Google Scholar] [CrossRef]
- Hijazi, K.; Malyszko, B.; Steiling, K.; Xiao, X.; Liu, G.; Alekseyev, Y.O.; Dumas, Y.M.; Hertsgaard, L.; Jensen, J.; Hatsukami, D.; et al. Tobacco-related alterations in airway gene expression are rapidly reversed within weeks following smoking-cessation. Sci. Rep. 2019, 9, 6978. [Google Scholar] [CrossRef]
- Steiling, K.; van den Berge, M.; Hijazi, K.; Florido, R.; Campbell, J.; Liu, G.; Xiao, J.; Zhang, X.; Duclos, G.; Drizik, E.; et al. A dynamic bronchial airway gene expression signature of chronic obstructive pulmonary disease and lung function impairment. Am. J. Respir. Crit. Care Med. 2013, 187, 933–942. [Google Scholar] [CrossRef]
- Boudewijn, I.M.; Faiz, A.; Steiling, K.; van der Wiel, E.; Telenga, E.D.; Hoonhorst, S.J.M.; Ten Hacken, N.H.T.; Brandsma, C.A.; Kerstjens, H.A.M.; Timens, W.; et al. Nasal gene expression differentiates COPD from controls and overlaps bronchial gene expression. Respir. Res. 2017, 18, 213. [Google Scholar] [CrossRef] [PubMed]
- Beane, J.; Mazzilli, S.A.; Tassinari, A.M.; Liu, G.; Zhang, X.; Liu, H.; Buncio, A.D.; Dhillon, S.S.; Platero, S.J.; Lenburg, M.E.; et al. Detecting the presence and progression of premalignant lung lesions via airway gene expression. Clin. Cancer Res. 2017, 23, 5091–5100. [Google Scholar] [CrossRef] [PubMed]
- Gustafson, A.M.; Soldi, R.; Anderlind, C.; Scholand, M.B.; Qian, J.; Zhang, X.; Cooper, K.; Walker, D.; McWilliams, A.; Liu, G.; et al. Airway PI3K pathway activation is an early and reversible event in lung cancer development. Sci. Trans. Med. 2010, 2, 26ra25. [Google Scholar] [CrossRef] [PubMed]
- Beane, J.E.; Mazzilli, S.A.; Campbell, J.D.; Duclos, G.; Krysan, K.; Moy, C.; Perdomo, C.; Schaffer, M.; Liu, G.; Zhang, S.; et al. Molecular subtyping reveals immune alterations associated with progression of bronchial premalignant lesions. Nat. Commun. 2019, 10, 1856. [Google Scholar] [CrossRef]
- Beane, J.; Vick, J.; Schembri, F.; Anderlind, C.; Gower, A.; Campbell, J.; Luo, L.; Zhang, X.H.; Xiao, J.; Alekseyev, Y.O.; et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev. Res. 2011, 4, 803–817. [Google Scholar] [CrossRef]
- Sridhar, S.; Schembri, F.; Zeskind, J.; Shah, V.; Gustafson, A.M.; Steiling, K.; Liu, G.; Dumas, Y.M.; Zhang, X.; Brody, J.S.; et al. Smoking-induced gene expression changes in the bronchial airway are reflected in nasal and buccal epithelium. BMC Genom. 2008, 30, 259. [Google Scholar] [CrossRef]
- AEGIS Study Team. Shared Gene Expression Alterations in Nasal and Bronchial Epithelium for Lung Cancer Detection. J. Natl. Cancer Inst. 2017, 109, djw327. [Google Scholar]
- Garland, L.L.; Guillen-Rodriguez, J.; Hsu, C.H.; Yozwiak, M.; Zhang, H.H.; Alberts, D.S.; Davis, L.E.; Szabo, E.; Merenstein, C.; Lel, J.; et al. Effect of Intermittent Versus Continuous Low-Dose Aspirin on Nasal Epithelium Gene Expression in Current Smokers: A Randomized, Double-Blinded Trial. Cancer Prev. Res. 2019, 12, 809–820. [Google Scholar] [CrossRef]
- Zhang, X.; Sebastiani, P.; Liu, G.; Schembri, F.; Zhang, X.; Dumas, Y.M.; Langer, E.M.; Alekseyev, Y.; O’Connor, G.T.; Brooks, D.R.; et al. Similarities and differences between smoking-related gene expression in nasal and bronchial epithelium. Physiol. Genom. 2010, 41, 1–8. [Google Scholar] [CrossRef]
- Beane, J.; Sebastinani, P.; Liu, G.; Brody, J.S.; Lenburg, M.E.; Spira, A. Reversible and permanent effects of tobacco smoke exposure on airway epithelial gene expression. Genome Biol. 2007, 8, R201. [Google Scholar] [CrossRef]
- Whitney, D.H.; Elashoff, M.R.; Porta-Smith, K.; Gower, A.C.; Vachani, A.; Ferguson, J.S.; Silvestri, G.A.; Brody, J.S.; Lenburg, M.E.; Spira, A. Derivation of a bronchial genomic classifier for lung cancer in a prospective study of patients undergoing diagnostic bronchoscopy. BMC Med. Genom. 2015, 8, 18. [Google Scholar] [CrossRef] [PubMed]
- Murphey, L.J.; Williams, M.K.; Sanchez, S.C.; Byrne, L.M.; Csiki, I.; Oates, J.A.; Johnson, D.H.; Morrow, J.D. Quantification of the major urinary metabolite of PGE(2) by a liquid chromatographic/mass spectrometric assay: Determination of cyclo oxygenase-specific PGE(2) synthesis in healthy humans and those with lung cancer. Anal. Biochem. 2004, 334, 266–275. [Google Scholar] [CrossRef] [PubMed]
- Duffield-Lillico, A.J.; Boyle, J.O.; Zhou, X.K.; Ghosh, A.; Butala, G.S.; Subbaramaiah, K.; Newman, R.A.; Morrow, J.D.; Milne, G.L.; Dannenberg, A.J. Levels of Prostaglandin E Metabolite and Leukotriene E-4 Are Increased in the Urine of Smokers: Evidence that Celecoxib Shunts Arachidonic Acid into the 5-Lipoxygenase Pathway. Cancer Prev. Res. 2009, 2, 322–329. [Google Scholar] [CrossRef] [PubMed]
- McCall, M.N.; Jaffee, H.A.; Irizarry, R.A. fRMA ST: Frozen robust multiarray analysis for Affymetrix Exon and Gene ST arrays. Bioinformatics 2012, 28, 3153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Available online: http://brainarray.mbni.med.umich.edu/Brainarray/Database/CustomCDF/genomic_curated_CDF.asp (accessed on 19 April 2022).
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic. Acids. Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Chen, C.; Grennan, K.; Badner, J.; Zhang, D.; Gershon, E.; Jin, L.; Liu, C. Removing batch effects in analysis of expression microarray data: An evaluation of six batch adjustment methods. PLoS ONE 2011, 6, e17238. [Google Scholar] [CrossRef]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Liberzon, A.; Subramanian, A.; Pinchback, R.; Thorvaldsdóttir, H.; Tamayo, P.; Mesirov, J.P. Molecular signatures database (MSigDB) 3.0. Bioinformatics 2011, 27, 1739–1740. [Google Scholar] [CrossRef]
- Hänzelmann, S.; Castelo, R.; Guinney, J. GSVA: Gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013, 14, 7. [Google Scholar] [CrossRef]
- Ye, S.; Lee, M.; Lee, D.; Ha, E.H.; Chun, E.M. Association of Long-term Use of Low-Dose Aspirin as Chemoprevention With Risk of Lung Cancer. JAMA Netw. Open 2019, 2, e190185. [Google Scholar] [CrossRef]
- Rothwell, P.M.; Cook, N.R.; Gaziano, J.M.; Price, J.F.; Belch, J.; Roncaglioni, M.C.; Morimoto, T.; Mehta, Z. Effects of aspirin on risks of vascular events and cancer according to bodyweight and dose: Analysis of individual patient data from randomized trials. Lancet 2018, 392, 387–399. [Google Scholar] [CrossRef]
- La Maestra, S.; D’Agostini, F.; Izzotti, A.; Micale, R.T.; Mastracci, L.; Camoirano, A.; Balansky, R.; Trosko, J.E.; Steele, V.E.; De Flora, S. Modulation by aspirin and naproxen of nucleotide alterations and tumors in the lung of mice exposed to environmental cigarette smoke since birth. Carcinogenesis 2015, 36, 1531–1538. [Google Scholar] [CrossRef] [PubMed]
- Izzotti, A.; Balansky, R.; Ganchev, G.; Iltcheva, M.; Longobardi, M.; Pulliero, A.; Camoirano, A.; D’Agostini, F.; Geretto, M.; Micale, R.T. Early and late effects of aspirin and naproxen on microRNAs in the lung and blood of mice, either unexposed or exposed to cigarette smoke. Oncotarget 2017, 8, 85716–85748. [Google Scholar] [CrossRef] [PubMed]
- Braeckman, R.A.; Granneman, G.R.; Locke, C.S.; Machinist, J.M.; Cavannaugh, J.H.; Awni, W.M. The pharmacokinetics of zileuton in healthy young and elderly volunteers. Clin. Pharmacokinet. 1995, 29, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Pace, S.; Pergola, C.; Dehm, F.; Rossi, A.; Gerstmeier, J.; Troisi, F.; Pein, H.; Schaible, A.M.; Weinigel, C.; Rummler, S.; et al. Androgen-mediated sex bias impairs efficiency of leukotriene biosynthesis inhibitors in males. J. Clin. Investig. 2017, 127, 3167–3176. [Google Scholar] [CrossRef]
- Liu, J.Y.; Yang, J.; Inceoglu, B.; Qiu, H.; Ulu, A.; Hwang, S.H.; Chiamvimonvat, N.; Hammock, B.D. Inhibition of soluble epoxide hydrolase enhances the anti-inflammatory effects of aspirin and 5-lipoxygenase activation protein inhibitor in a murine model. Biochem. Pharmacol. 2010, 79, 880–887. [Google Scholar] [CrossRef] [PubMed]
- Celotti, F.; Durand, T. The metabolic effects of inhibitors of 5-lipoxygenase and of cyclooxygenase 1 and 2 are an advancement in the efficacy and safety of anti-inflammatory therapy. Prostaglandins Other Lipid Mediat. 2003, 71, 147–162. [Google Scholar] [CrossRef]

| Variable | All (n = 63) | ASA + Zileuton (n = 31) | Placebo (n = 32) | pb |
|---|---|---|---|---|
| Age | 51.57 ± 9.85 | 49.55 ± 8.64 | 53.53 ± 10.67 | 0.11 |
| BMI | 28.23 ± 7.02 | 28.69 ± 6.60 | 27.78 ± 7.48 | 0.61 |
| Packyears | 34.67 ± 12.42 | 32.74 ± 12.52 | 36.53 ± 12.22 | 0.23 |
| Male | 36 (57.14%) a | 18 (58.06%) | 18 (56.25%) | 1.00 |
| Female | 27 (42.86%) | 13 (41.94%) | 14 (43.75%) | 1.00 |
| White | 52 (82.54%) | 28 (90.32%) | 24 (75.00%) | 0.15 |
| Hispanic | 7 (11.11%) | 3 (9.68%) | 4 (12.50%) | 1.00 |
| Variable | All (n = 63) | ASA + Zileuton (n = 31) | Placebo (n = 32) | pb |
|---|---|---|---|---|
| Age | 51.57 ± 9.85 | 49.55 ± 8.64 | 53.53 ± 10.67 | 0.11 |
| BMI | 28.23 ± 7.02 | 28.69 ± 6.60 | 27.78 ± 7.48 | 0.61 |
| Packyears | 34.67 ± 12.42 | 32.74 ± 12.52 | 36.53 ± 12.22 | 0.23 |
| Male | 36 (57.14%) a | 18 (58.06%) | 18 (56.25%) | 1.00 |
| Female | 27 (42.86%) | 13 (41.94%) | 14 (43.75%) | 1.00 |
| White | 52 (82.54%) | 28 (90.32%) | 24 (75.00%) | 0.15 |
| Hispanic | 7 (11.11%) | 3 (9.68%) | 4 (12.50%) | 1.00 |
| Adverse Event | Aspirin + Zileuton (n = 32) | Placebo (n = 31) |
|---|---|---|
| Grades 1, 2 (%) | Grades 1, 2 (%) | |
| Blood and Lymphatic System Disorders | ||
| Anemia | 1 (3.23) | |
| Blood and lymphatic system disorders—Other-Hematocrit Low | 1 (3.23) | |
| Gastrointestinal disorders | ||
| Abdominal pain | 1 (3.23) | |
| Diarrhea | 1 (3.13) | |
| Dyspepsia | 4 (12.5) | 4 (12.9) |
| Nausea | 1 (3.13) | |
| Stomach pain | 1 (3.13) | 1 (3.23) |
| Vomiting | 1 (3.13) | |
| General disorders and administration site conditions | ||
| Fatigue | 1 (3.13) | |
| Investigations | ||
| Alkaline phosphatase increased | 1 (3.23) | |
| Blood bilirubin increased | 1 (3.13) | 1 (3.23) |
| Investigations—Other-Macrocytosis | 1 (3.13) | |
| Alanine aminotransferase increased | 1 (3.13) | |
| Aspartate aminotransferase increased | 1 (3.13) | 1 (3.23) |
| Metabolism and nutrition disorders | ||
| Dehydration | 1 (3.23) | |
| Nervous system disorders | ||
| Dizziness | 1 (3.13) | 1 (3.23) |
| Headache | 2 (6.25) | 1 (3.23) |
| Psychiatric disorders | ||
| Mania | 1 (3.13) | |
| Skin and subcutaneous tissue disorders | ||
| Rash maculo-papular | 1 (3.13) |
| All | Overall (n = 40) | ASA + Zileuton (n = 19) | Placebo (n = 21) | p a |
|---|---|---|---|---|
| Baseline | 0.31 ± 3.85 | 0.82 ± 4.00 | −0.15 ± 3.75 | 0.43 |
| Post Intervention (Int.) | 0.37 ± 3.72 | 0.67 ± 4.20 | 0.11 ± 3.30 | |
| 1-week Post Int. | −0.64 ± 4.10 | −0.03 ± 4.45 | −1.19 ± 3.79 | |
| Baseline—Post Int. | 0.06 ± 2.55 | −0.15 ± 2.89 | 0.26 ± 2.26 | 0.62 |
| Baseline-1-week Post Int. | −0.73 ± 3.31 | −0.24 ± 3.04 | −1.16 ± 3.55 | 0.40 |
| Female | n = 19 | n = 8 | n = 11 | |
| Baseline | −0.26 ± 3.05 | −1.26 ± 1.40 | 0.47 ± 3.74 | 0.18 |
| Post Int. | −0.37 ± 3.10 | −1.25 ± 3.08 | 0.27 ± 3.09 | |
| 1-week Post Int. | −1.14 ± 3.52 | −2.08 ± 3.49 | −0.45 ± 3.54 | |
| Baseline—Post Int. | −0.11 ± 2.37 | 0.01 ± 2.93 | −0.20 ± 2.01 | 0.86 |
| Baseline-1-week Post Int. | −0.87 ± 2.98 | −0.82 ± 3.57 | −0.91 ± 2.66 | 0.95 |
| Male | n = 21 | n = 11 | n = 10 | |
| Baseline | 0.83 ± 4.46 | 2.33 ± 4.62 | −0.83 ± 3.83 | 0.11 |
| Post Int. | 1.05 ± 4.16 | 2.07 ± 4.48 | −0.07 ± 3.68 | |
| 1-week Post Int. | −0.14 ± 4.66 | 1.61 ± 4.60 | −2.10 ± 4.10 | |
| Baseline—Post Int. | 0.22 ± 2.76 | −0.27 ± 2.99 | 0.76 ± 2.51 | 0.41 |
| Baseline-1-week Post Int. | −0.58 ± 3.69 | 0.22 ± 2.66 | −1.47 ± 4.58 | 0.33 |
| All | Overall (n = 40) | ASA + Zileuton (n = 19) | Placebo (n = 21) | p a |
|---|---|---|---|---|
| Baseline | −0.14 ± 5.04 a | 0.27 ± 5.01 | −0.51 ± 5.15 | 0.63 |
| Post t Intervention (Int.) | −0.22 ± 4.48 | −2.05 ± 3.50 | 1.43 ± 4.70 | |
| 1-week Post Int. | 0.27 ± 6.29 | −1.56 ± 3.56 | 1.91 ± 7.72 | |
| Baseline—Post Int. | −0.08 ± 4.66 | −2.31 ± 4.08 | 1.94 ± 4.28 | <0.01 |
| Baseline-1-week Post Int. | 0.37 ± 5.90 | −1.83 ± 5.37 | 2.35 ± 5.77 | 0.03 |
| Female | n = 19 | n = 8 | n= 11 | |
| Baseline | 0.40 ± 5.12 | 0.15 ± 5.85 | 0.58 ± 4.82 | 0.86 |
| Post Int. | −0.58 ± 4.21 | −1.33 ± 4.18 | −0.04 ± 4.35 | |
| 1-week Post Int. | 0.07 ± 6.84 | −2.68 ± 3.37 | 2.07 ± 8.11 | |
| Baseline—Post Int. | −0.98 ± 3.58 | −1.48 ± 4.82 | −0.61 ± 2.56 | 0.62 |
| Baseline-1-week Post Int. | −0.33 ± 5.96 | −2.83 ± 6.67 | 1.49 ± 4.91 | 0.12 |
| Male | n = 21 | n = 11 | n = 10 | |
| Baseline | −0.63 ± 5.04 | 0.35 ± 4.62 | −1.70 ± 5.50 | 0.36 |
| Post Int. | 0.10 ± 4.79 | −2.57 ± 3.00 | 3.05 ± 4.75 | |
| 1-week Post Int. | 0.46 ± 5.87 | −0.66 ± 3.62 | 1.71 ± 7.71 | |
| Baseline—Post Int. | 0.73 ± 5.41 | −2.93 ± 3.57 | 4.75 ± 4.09 | <0.001 |
| Baseline-1-week Post Int. | 1.06 ± 5.92 | −1.03 ± 4.27 | 3.39 ± 6.84 | 0.11 |
| All | Overall (n = 43) | ASA + Zileuton (n = 21) | Placebo (n = 22) | p b |
|---|---|---|---|---|
| Baseline | 88.17 ± 60.65 a | 89.867 ± 68.35 | 86.55 ± 53.86 | 0.86 |
| Post Int. c | 78.02 ± 90.79 | 32.25 ± 23.25 | 121.73 ± 108.97 | |
| 1-week Post Int. | 103.09 ± 97.71 | 79.57 ± 52.60 | 125.53 ± 124.01 | |
| Baseline—Post Int. | −10.14 ± 92.43 | −57.62 ± 65.56 | 35.17 ± 92.67 | <0.001 |
| Baseline-1-week Post Int. | 14.92 ± 85.67 | −10.29 ± 60.02 | 38.98 ± 100.02 | 0.06 |
| Female | n= 21 | n = 10 | n = 11 | |
| Baseline | 104.00 ± 65.78 | 114.90 ± 74.88 | 94.09 ± 58.15 | 0.48 |
| Post Int. | 104.07 ± 115.81 | 27.93 ± 22.90 | 173.28 ± 123.64 | |
| 1-week Post Int. | 112.65 ± 126.20 | 73.49 ± 32.00 | 148.24 ± 167.35 | |
| Baseline—Post Int. | 0.07 ± 125.66 | −86.97 ± 69.20 | 79.19 ± 113.19 | <0.001 |
| Baseline-1-week Post Int. | 8.65 ± 111.39 | −41.41 ± 65.73 | 54.15 ± 127.06 | <0.05 |
| Male | n = 22 | n = 11 | n = 11 | |
| Baseline | 73.06 ± 52.39 | 67.10 ± 55.66 | 79.01 ± 50.86 | 0.61 |
| Post Int. | 53.17 ± 48.85 | 36.16 ± 23.95 | 70.17 ± 61.65 | |
| 1-week Post Int. | 93.96 ± 61.20 | 85.10 ± 67.39 | 102.82 ± 56.13 | |
| Baseline—Post Int. | −19.89 ± 42.92 | −30.94 ± 51.40 | −8.84 ± 30.96 | 0.24 |
| Baseline-1-week Post Int. | 20.90 ± 52.76 | 18.00 ± 38.33 | 23.81 ± 66.01 | 0.80 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garland, L.L.; Guillen-Rodriguez, J.; Hsu, C.-H.; Davis, L.E.; Szabo, E.; Husted, C.R.; Liu, H.; LeClerc, A.; Alekseyev, Y.O.; Liu, G.; et al. Clinical Study of Aspirin and Zileuton on Biomarkers of Tobacco-Related Carcinogenesis in Current Smokers. Cancers 2022, 14, 2893. https://doi.org/10.3390/cancers14122893
Garland LL, Guillen-Rodriguez J, Hsu C-H, Davis LE, Szabo E, Husted CR, Liu H, LeClerc A, Alekseyev YO, Liu G, et al. Clinical Study of Aspirin and Zileuton on Biomarkers of Tobacco-Related Carcinogenesis in Current Smokers. Cancers. 2022; 14(12):2893. https://doi.org/10.3390/cancers14122893
Chicago/Turabian StyleGarland, Linda L., José Guillen-Rodriguez, Chiu-Hsieh Hsu, Lisa E. Davis, Eva Szabo, Christopher R. Husted, Hanqiao Liu, Ashley LeClerc, Yuriy O. Alekseyev, Gang Liu, and et al. 2022. "Clinical Study of Aspirin and Zileuton on Biomarkers of Tobacco-Related Carcinogenesis in Current Smokers" Cancers 14, no. 12: 2893. https://doi.org/10.3390/cancers14122893
APA StyleGarland, L. L., Guillen-Rodriguez, J., Hsu, C.-H., Davis, L. E., Szabo, E., Husted, C. R., Liu, H., LeClerc, A., Alekseyev, Y. O., Liu, G., Bauman, J. E., Spira, A. E., Beane, J., Wojtowicz, M., & Chow, H.-H. S. (2022). Clinical Study of Aspirin and Zileuton on Biomarkers of Tobacco-Related Carcinogenesis in Current Smokers. Cancers, 14(12), 2893. https://doi.org/10.3390/cancers14122893






