Current and Emerging Methods for Ovarian Cancer Screening and Diagnostics: A Comprehensive Review
Simple Summary
Abstract
1. Introduction
2. Prologue—Early Screening and Risk Assessment Considerations
Cost-Effectiveness of Ovarian Cancer Screening
3. Current Screening Options
CA125 and Transvaginal Ultrasound
4. Protein Diagnostics
4.1. Protein Screening and Diagnostic Assays for Asymptomatic Women
4.2. Emerging Protein Diagnostics
5. Imaging Diagnostics
5.1. Internal Organ Scans
5.2. Hysteroscopy Imaging
5.3. Emerging Imaging Techniques
6. Preoperative Diagnostics
6.1. Multivariate Index Assays
6.2. The Ovarian Risk of Malignancy Algorithm
6.3. The Risk of Malignancy Index
6.4. The Copenhagen Index
7. Genetic Diagnostics
8. Epigenetic Diagnostics
9. Pan-Cancer Screening
9.1. FDA Approved Pan-Cancer Screening
9.2. Pan-Cancer Tests in Development
10. Emerging Diagnostic Approaches
10.1. MicroRNA
10.2. Extracellular Vesicles
10.3. Autoantibodies
10.4. Microbiome
10.5. Metabolomics
11. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Genetic Risk Factors
Appendix A.2. Reproductive Risk Factors
Appendix A.3. Dietary and Lifestyle Risk Factors
References
- Shih, I.M.; Wang, Y.; Wang, T.L. The Origin of Ovarian Cancer Species and Precancerous Landscape. Am. J. Pathol. 2021, 191, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Rendi, M.H. Epithelial carcinoma of the ovary, fallopian tube, and peritoneum: Histopathology. In UpToDate; Chakrabarti, A., Ed.; UpToDate: Waltham, MA, USA, 2021. [Google Scholar]
- Bowtell, D.D.; Böhm, S.; Ahmed, A.A.; Aspuria, P.J.; Bast, R.C.; Beral, V.; Berek, J.S.; Birrer, M.J.; Blagden, S.; Bookman, M.A.; et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer 2015, 15, 668–679. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics, 2022. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Berek, J.S.; Kehoe, S.T.; Kumar, L.; Friedlander, M. Cancer of the ovary, fallopian tube, and peritoneum. Int. J. Gynecol. Obstet. 2018, 143, 59–78. [Google Scholar] [CrossRef] [PubMed]
- Goff, B.A.; Mandel, L.S.; Drescher, C.W.; Urban, N.; Gough, S.; Schurman, K.M.; Patras, J.; Mahony, B.S.; Robyn Andersen, M. Development of an ovarian cancer symptom index. Cancer 2007, 109, 221–227. [Google Scholar] [CrossRef]
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef]
- Howlader, N.; Noon, A.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Rhul, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. Cancer Statistics Review, 1975–2018—SEER Statistics; National Cancer Institute: Bethesda, MD, USA, 2021. [Google Scholar]
- Torre, L.A.; Trabert, B.; DeSantis, C.E.; Miller, K.D.; Samimi, G.; Runowicz, C.D.; Gaudet, M.M.; Jemal, A.; Siegel, R.L. Ovarian cancer statistics, 2018. CA Cancer J. Clin. 2018, 68, 284–296. [Google Scholar] [CrossRef]
- Safaeian, M.; Solomon, D.; Castle, P.E. Cervical Cancer Prevention—Cervical Screening: Science in Evolution. Obstet. Gynecol. Clin. N. Am. 2007, 34, 739. [Google Scholar] [CrossRef]
- Doubeni, C.A.; Corley, D.A.; Quinn, V.P.; Jensen, C.D.; Zauber, A.G.; Goodman, M.; Johnson, J.R.; Mehta, S.J.; Becerra, T.A.; Zhao, W.K.; et al. Effectiveness of screening colonoscopy in reducing the risk of death from right and left colon cancer: A large community-based study. Gut 2018, 67, 291–298. [Google Scholar] [CrossRef]
- Matteson, K.A.; Gunderson, C.; Richardson, D.L. Committee Opinion No. 716 Summary: The Role of the Obstetrician–Gynecologist in the Early Detection of Epithelial Ovarian Cancer in Women at Average Risk. Obstet. Gynecol. 2017, 130, 664–665. [Google Scholar] [CrossRef]
- Grossman, D.C.; Curry, S.J.; Owens, D.K.; Barry, M.J.; Davidson, K.W.; Doubeni, C.A.; Epling, J.W.; Kemper, A.R.; Krist, A.H.; Kurth, A.E.; et al. Screening for Ovarian Cancer: US Preventive Services Task Force Recommendation Statement. JAMA 2018, 319, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Barrett, J.; Jenkins, V.; Farewell, V.; Menon, U.; Jacobs, I.; Kilkerr, J.; Ryan, A.; Langridge, C.; Fallowfield, L. Psychological morbidity associated with ovarian cancer screening: Results from more than 23,000 women in the randomised trial of ovarian cancer screening (UKCTOCS). BJOG 2014, 121, 1071–1079. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Berek, J.S. Epithelial carcinoma of the ovary, fallopian tube, and peritoneum: Clinical features and diagnosis. In UpToDate; Chakrabarti, A., Ed.; UpToDate: Waltham, MA, USA, 2021. [Google Scholar]
- Kurman, R.J.; Shih, I.M. Molecular pathogenesis and extraovarian origin of epithelial ovarian cancer—Shifting the paradigm. Hum. Pathol. 2011, 42, 918–931. [Google Scholar] [CrossRef] [PubMed]
- Koshiyama, M.; Matsumura, N.; Konishi, I. Subtypes of Ovarian Cancer and Ovarian Cancer Screening. Diagnostics 2017, 7, 12. [Google Scholar] [CrossRef]
- Ilenkovan, N.; Gourley, C. Pathogenesis, Genetics, and Genomics of Non–High Grade Serous Ovarian Cancers. Hematol. Oncol. Clin. N. Am. 2018, 32, 929–942. [Google Scholar] [CrossRef]
- Gockley, A.; Melamed, A.; Bregar, A.J.; Clemmer, J.T.; Birrer, M.; Schorge, J.O.; Del Carmen, M.G.; Alejandro Rauh-Hain, J. Outcomes of women with high-grade and low-grade advanced-stage serous epithelial ovarian cancer. Obstet. Gynecol. 2017, 129, 439–447. [Google Scholar] [CrossRef]
- Bell, D.; Berchuck, A.; Birrer, M.; Chien, J.; Cramer, D.W.; Dao, F.; Dhir, R.; Disaia, P.; Gabra, H.; Glenn, P.; et al. Integrated genomic analyses of ovarian carcinoma. Nature 2011, 474, 609–615. [Google Scholar] [CrossRef]
- Differences between Screening and Diagnostic Tests and Case Finding. Available online: https://www.healthknowledge.org.uk/public-health-textbook/disease-causation-diagnostic/2c-diagnosis-screening/screening-diagnostic-case-finding (accessed on 22 February 2022).
- Katki, H.A.; Kinney, W.K.; Fetterman, B.; Lorey, T.; Poitras, N.E.; Cheung, L.; Demuth, F.; Schiffman, M.; Wacholder, S.; Castle, P.E. Cervical cancer risk for women undergoing concurrent testing for human papillomavirus and cervical cytology: A population-based study in routine clinical practice. Lancet Oncol. 2011, 12, 663–672. [Google Scholar] [CrossRef]
- Haghighi, F.; Ghanbarzadeh, N.; Ataee, M.; Sharifzadeh, G.; Mojarrad, J.S.; Najafi-Semnani, F. A comparison of liquid-based cytology with conventional Papanicolaou smears in cervical dysplasia diagnosis. Adv. Biomed. Res. 2016, 5, 162. [Google Scholar] [CrossRef]
- Renshaw, A.A.; Davey, D.D.; Birdsong, G.G.; Walsh, M.; Styer, P.E.; Mody, D.R.; Colgan, T.J. Precision in Gynecologic Cytologic Interpretation: A Study From the College of American Pathologists Interlaboratory Comparison Program in Cervicovaginal Cytology. Arch. Pathol. Lab. Med. 2003, 127, 1413–1420. [Google Scholar] [CrossRef]
- Fairman, A.; Tan, J.; Quinn, M. Women with low-grade abnormalities on Pap smear should be referred for colposcopy. Aust. N. Z. J. Obstet. Gynaecol. 2004, 44, 252–255. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.Y.H.; Ladame, S. Diagnostic, prognostic, and predictive biomarkers for cancer. In Bioengineering Innovative Solutions for Cancer; Academic Press: Cambridge, MA, USA, 2020; pp. 3–21. [Google Scholar] [CrossRef]
- Sassu, C.M.; Palaia, I.; Boccia, S.M.; Caruso, G.; Perniola, G.; Tomao, F.; Di Donato, V.; Musella, A.; Muzii, L. Role of Circulating Biomarkers in Platinum-Resistant Ovarian Cancer. Int. J. Mol. Sci. 2021, 22, 13650. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.; Khan, S.; Sun, Y.; Hess, K.; Shmulevich, I.; Sood, A.K.; Zhang, W. Association of BRCA1 and BRCA2 mutations with survival, chemotherapy sensitivity, and gene mutator phenotype in patients with ovarian cancer. JAMA 2011, 306, 1557–1565. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Lee, M.; Kim, H.S.; Chung, H.H.; Kim, J.W.; Park, N.H.; Song, Y.S. Effect of BRCA mutational status on survival outcome in advanced-stage high-grade serous ovarian cancer. J. Ovarian Res. 2019, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Helleday, T. The underlying mechanism for the PARP and BRCA synthetic lethality: Clearing up the misunderstandings. Mol. Oncol. 2011, 5, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Dziadkowiec, K.N.; Gasiorowska, E.; Nowak-Markwitz, E.; Jankowska, A. PARP inhibitors: Review of mechanisms of action and BRCA1/2 mutation targeting. Menopause Rev. 2016, 15, 215. [Google Scholar] [CrossRef]
- Lee, J.M.; Ledermann, J.A.; Kohn, E.C. PARP Inhibitors for BRCA1/2 mutation-associated and BRCA-like malignancies. Ann. Oncol. 2014, 25, 32–40. [Google Scholar] [CrossRef]
- Elias, K.M.; Guo, J.; Bast, R.C. Early Detection of Ovarian Cancer. Hematol. Oncol. Clin. N. Am. 2018, 32, 903–914. [Google Scholar] [CrossRef]
- Greene, M.H.; Feng, Z.; Gail, M.H. The importance of test positive predictive value in ovarian cancer screening. Clin. Cancer Res. 2008, 14, 7574. [Google Scholar] [CrossRef]
- Ranganathan, P.; Aggarwal, R. Common pitfalls in statistical analysis: Understanding the properties of diagnostic tests—Part 1. Perspect. Clin. Res. 2018, 9, 40. [Google Scholar] [CrossRef]
- Wilson, J.M.G.; Jungner, G. Principles and Practice of Screening for Disease; Wold Health Organization (WHO): Geneva, Switzerland, 1968; Volume 34. [Google Scholar]
- Dobrow, M.J.; Hagens, V.; Chafe, R.; Sullivan, T.; Rabeneck, L. Consolidated principles for screening based on a systematic review and consensus process. CMAJ 2018, 190, E422–E429. [Google Scholar] [CrossRef] [PubMed]
- Moss, H.A.; Berchuck, A.; Neely, M.L.; Myers, E.R.; Havrilesky, L.J. Estimating Cost-effectiveness of a Multimodal Ovarian Cancer Screening Program in the United States: Secondary Analysis of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS). JAMA Oncol. 2018, 4, 190–195. [Google Scholar] [CrossRef] [PubMed]
- Kearns, B.; Chilcott, J.; Whyte, S.; Preston, L.; Sadler, S. Cost-effectiveness of screening for ovarian cancer amongst postmenopausal women: A model-based economic evaluation. BMC Med. 2016, 14, 200. [Google Scholar] [CrossRef] [PubMed]
- Havrilesky, L.J.; Sanders, G.D.; Kulasingam, S.; Myers, E.R. Reducing ovarian cancer mortality through screening: Is it possible, and can we afford it? Gynecol. Oncol. 2008, 111, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Kaijser, J.; Bourne, T.; Valentin, L.; Sayasneh, A.; Van Holsbeke, C.; Vergote, I.; Testa, A.C.; Franchi, D.; Van Calster, B.; Timmerman, D. Improving strategies for diagnosing ovarian cancer: A summary of the International Ovarian Tumor Analysis (IOTA) studies. Ultrasound Obstet. Gynecol. 2013, 41, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Zurawski, V.R.; Orjaseter, H.; Andersen, A.; Jellum, E. Elevated serum CA 125 levels prior to diagnosis of ovarian neoplasia: Relevance for early detection of ovarian cancer. Int. J. Cancer 1988, 42, 677–680. [Google Scholar] [CrossRef]
- Bast, R.C.; Klug, T.L.; John, E.S.; Jenison, E.; Niloff, J.M.; Lazarus, H.; Berkowitz, R.S.; Leavitt, T.; Griffiths, C.T.; Parker, L.; et al. A radioimmunoassay using a monoclonal antibody to monitor the course of epithelial ovarian cancer. N. Engl. J. Med. 1983, 309, 883–887. [Google Scholar] [CrossRef]
- Bast, R.C.; Feeney, M.; Lazarus, H.; Nadler, L.M.; Colvin, R.B.; Knapp, R.C. Reactivity of a monoclonal antibody with human ovarian carcinoma. J. Clin. Investig. 1981, 68, 1331–1337. [Google Scholar] [CrossRef]
- Timmerman, D.; Van Calster, B.; Jurkovic, D.; Valentin, L.; Testa, A.C.; Bernard, J.P.; Van Holsbeke, C.; Van Huffel, S.; Vergote, I.; Bourne, T. Inclusion of CA-125 does not improve mathematical models developed to distinguish between benign and malignant adnexal tumors. J. Clin. Oncol. 2007, 25, 4194–4200. [Google Scholar] [CrossRef]
- Buys, S.S.; Partridge, E.; Greene, M.H.; Prorok, P.C.; Reding, D.; Riley, T.L.; Hartge, P.; Fagerstrom, R.M.; Ragard, L.R.; Chia, D.; et al. Ovarian cancer screening in the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial: Findings from the initial screen of a randomized trial. Am. J. Obstet. Gynecol. 2005, 193, 1630–1639. [Google Scholar] [CrossRef]
- Pinsky, P.F.; Yu, K.; Kramer, B.S.; Black, A.; Buys, S.S.; Partridge, E.; Gohagan, J.; Berg, C.D.; Prorok, P.C. Extended mortality results for ovarian cancer screening in the PLCO trial with median 15years follow-up. Gynecol. Oncol. 2016, 143, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, I.J.; Menon, U.; Ryan, A.; Gentry-Maharaj, A.; Burnell, M.; Kalsi, J.K.; Amso, N.N.; Apostolidou, S.; Benjamin, E.; Cruickshank, D.; et al. Ovarian cancer screening and mortality in the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS): A randomised controlled trial. Lancet 2016, 387, 945–956. [Google Scholar] [CrossRef]
- Jacobs, I.; Bast, R.C. The CA 125 tumour-associated antigen: A review of the literature. Hum. Reprod. 1989, 4, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Høgdall, E.V.S.; Christensen, L.; Kjaer, S.K.; Blaakaer, J.; Kjærbye-Thygesen, A.; Gayther, S.; Jacobs, I.J.; Høgdall, C.K. CA125 expression pattern, prognosis and correlation with serum CA125 in ovarian tumor patients: From The Danish “MALOVA” Ovarian Cancer Study. Gynecol. Oncol. 2007, 104, 508–515. [Google Scholar] [CrossRef]
- Johnson, C.C.; Kessel, B.; Riley, T.L.; Ragard, L.R.; Williams, C.R.; Xu, J.L.; Buys, S.S. The epidemiology of CA-125 in women without evidence of ovarian cancer in the Prostate, Lung, Colorectal and Ovarian Cancer (PLCO) Screening Trial. Gynecol. Oncol. 2008, 110, 383–389. [Google Scholar] [CrossRef]
- Pauler, D.; Menon, U.; McIntosh, M.; Symecko, H.L.; Skates, S.J.; Jacobs, I.J. Factors Influencing Serum CA125II Levels in Healthy Postmenopausal Women. Cancer Epidemiol. Biomark. Prev. 2001, 10, 489–493. [Google Scholar]
- Guo, N.; Peng, Z. Does serum CA125 have clinical value for follow-up monitoring of postoperative patients with epithelial ovarian cancer? Results of a 12-year study. J. Ovarian Res. 2017, 10, 14. [Google Scholar] [CrossRef]
- Piatek, S.; Panek, G.; Lewandowski, Z.; Piatek, D.; Kosinski, P.; Bidzinski, M. Nadir CA-125 has prognostic value for recurrence, but not for survival in patients with ovarian cancer. Sci. Rep. 2021, 11, 18190. [Google Scholar] [CrossRef]
- Ruggeri, G.; Bandiera, E.; Zanotti, L.; Belloli, S.; Ravaggi, A.; Romani, C.; Bignotti, E.; Tassi, R.A.; Tognon, G.; Galli, C.; et al. HE4 and epithelial ovarian cancer: Comparison and clinical evaluation of two immunoassays and a combination algorithm. Clin. Chim. Acta 2011, 412, 1447–1453. [Google Scholar] [CrossRef]
- Drapkin, R.; Von Horsten, H.H.; Lin, Y.; Mok, S.C.; Crum, C.P.; Welch, W.R.; Hecht, J.L. Human epididymis protein 4 (HE4) is a secreted glycoprotein that is overexpressed by serous and endometrioid ovarian carcinomas. Cancer Res. 2005, 65, 2162–2169. [Google Scholar] [CrossRef]
- Abbink, K.; Zusterzeel, P.L.M.; Geurts-Moespot, A.J.; Herwaarden, A.E.V.; Pijnenborg, J.M.A.; Sweep, F.C.G.J.; Massuger, L.F.A.G. HE4 is superior to CA125 in the detection of recurrent disease in high-risk endometrial cancer patients. Tumour Biol. 2018, 40, 1010428318757103. [Google Scholar] [CrossRef] [PubMed]
- Anastasi, E.; Giovanna Marchei, G.; Viggiani, V.; Gennarini, G.; Frati, L.; Reale, M.G. HE4: A new potential early biomarker for the recurrence of ovarian cancer. Tumor Biol. 2010, 31, 113–119. [Google Scholar] [CrossRef] [PubMed]
- 510(k) Premarket Notification K112624, Food and Drug Administration. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?ID=K112624 (accessed on 6 June 2022).
- 510(k) Premarket Notification K093957, Food and Drug Administration. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpmn/pmn.cfm?id=K093957 (accessed on 6 June 2022).
- Pavai, S.; Yap, S.F. The Clinical Significance of Elevated Levels of Serum CA 19·9. Med. J. Malaysia 2003, 8, 667–672. [Google Scholar]
- Motoyama, T.; Watanabe, H.; Takeuchi, S.; Watanabe, T.; Gotoh, S.; Okazaki, E. Cancer antigen 125, carcinoembryonic antigen, and carbohydrate determinant 19-9 in ovarian tumors—PubMed. Cancer 1990, 66, 2628–2635. [Google Scholar] [CrossRef]
- Kelly, P.J.; Archbold, P.; Price, J.H.; Cardwell, C.; McCluggage, W.G. Serum CA19.9 levels are commonly elevated in primary ovarian mucinous tumours but cannot be used to predict the histological subtype. J. Clin. Pathol. 2010, 63, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Cho, H.Y.; Kyung, M.S. Serum CA19-9 as a Predictor of Malignancy in Primary Ovarian Mucinous Tumors: A Matched Case-Control Study. Med. Sci. Monit. 2014, 20, 1339. [Google Scholar] [CrossRef]
- Lertkhachonsuk, A.-A.; Buranawongtrakoon, S.; Lekskul, N.; Rermluk, N.; Wee-Stekly, W.-W.; Charakorn, C. Serum CA19-9, CA-125 and CEA as tumor markers for mucinous ovarian tumors. J. Obstet. Gynaecol. Res. 2020, 46, 2287–2291. [Google Scholar] [CrossRef]
- Tingulstad, S.; Hagen, B.; Skjeldestad, F.E.; Halvorsen, T.; Nustad, K.; Onsrud, M. The risk-of-malignancy index to evaluate potential ovarian cancers in local hospitals. Obstet. Gynecol. 1999, 93, 448–452. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Yamada, R.; Oguri, H.; Maeda, N.; Fukaya, T. Comparison of four malignancy risk indices in the preoperative evaluation of patients with pelvic masses. Eur. J. Obstet. Gynecol. Reprod. Biol. 2009, 144, 163–167. [Google Scholar] [CrossRef]
- Karlsen, M.A.; Høgdall, E.V.S.; Christensen, I.J.; Borgfeldt, C.; Kalapotharakos, G.; Zdrazilova-Dubska, L.; Chovanec, J.; Lok, C.A.R.; Stiekema, A.; Mutz-Dehbalaie, I.; et al. A novel diagnostic index combining HE4, CA125 and age may improve triage of women with suspected ovarian cancer—An international multicenter study in women with an ovarian mass. Gynecol. Oncol. 2015, 138, 640–646. [Google Scholar] [CrossRef]
- Moore, R.G.; Miller, M.C.; Disilvestro, P.; Landrum, L.M.; Gajewski, W.; Ball, J.J.; Skates, S.J. Evaluation of the diagnostic accuracy of the risk of ovarian malignancy algorithm in women with a pelvic mass. Obstet. Gynecol. 2011, 118, 280–288. [Google Scholar] [CrossRef] [PubMed]
- Watrowski, R.; Obermayr, E.; Wallisch, C.; Aust, S.; Concin, N.; Braicu, E.I.; Vn Gorp, T.; Hasenburg, A.; Sehouli, J.; Vergote, I.; et al. Biomarker-Based Models for Preoperative Assessment of Adnexal Mass: A Multicenter Validation Study. Cancers 2022, 14, 1780. [Google Scholar] [CrossRef] [PubMed]
- Carreras-Dieguez, N.; Glickman, A.; Munmany, M.; Casanovas, G.; Agustí, N.; Díaz-Feijoo, B.; Saco, A.; Sánchez, B.; Gaba, L.; Angeles, M.A.; et al. Comparison of HE4, CA125, ROMA and CPH-I for Preoperative Assessment of Adnexal Tumors. Diagnostics 2022, 12, 226. [Google Scholar] [CrossRef] [PubMed]
- Moore, R.G.; McMeekin, D.S.; Brown, A.K.; DiSilvestro, P.; Miller, M.C.; Allard, W.J.; Gajewski, W.; Kurman, R.; Bast, R.C.; Skates, S.J. A novel multiple marker bioassay utilizing HE4 and CA125 for the prediction of ovarian cancer in patients with a pelvic mass. Gynecol. Oncol. 2009, 112, 40–46. [Google Scholar] [CrossRef]
- Minar, L.; Felsinger, M.; Cermakova, Z.; Zlamal, F.; Bienertova-Vasku, J. Comparison of the Copenhagen Index versus ROMA for the preoperative assessment of women with ovarian tumors. Int. J. Gynecol. Obstet. 2018, 140, 241–246. [Google Scholar] [CrossRef]
- Romagnolo, C.; Leon, A.E.; Fabricio, A.S.C.; Taborelli, M.; Polesel, J.; Del Pup, L.; Steffan, A.; Cervo, S.; Ravaggi, A.; Zanotti, L.; et al. HE4, CA125 and risk of ovarian malignancy algorithm (ROMA) as diagnostic tools for ovarian cancer in patients with a pelvic mass: An Italian multicenter study. Gynecol. Oncol. 2016, 141, 303–311. [Google Scholar] [CrossRef]
- Meys, E.M.J.; Kaijser, J.; Kruitwagen, R.F.P.M.; Slangen, B.F.M.; Van Calster, B.; Aertgeerts, B.; Verbakel, J.Y.; Timmerman, D.; Van Gorp, T. Subjective assessment versus ultrasound models to diagnose ovarian cancer: A systematic review and meta-analysis. Eur. J. Cancer 2016, 58, 17–29. [Google Scholar] [CrossRef]
- Tran, D.T.; Khoa Vo, V.; Le, M.T.; Chuang, L.; Quoc, V.; Nguyen, H. Copenhagen Index versus ROMA in preoperative ovarian malignancy risk stratification: Result from the first Vietnamese prospective cohort study. Gynecol. Oncol. 2021, 162, 113–119. [Google Scholar] [CrossRef]
- Yoshida, A.; Derchain, S.F.; Pitta, D.R.; Andrade, L.A.L.D.A.; Sarian, L.O. Comparing the Copenhagen Index (CPH-I) and Risk of Ovarian Malignancy Algorithm (ROMA): Two equivalent ways to differentiate malignant from benign ovarian tumors before surgery? Gynecol. Oncol. 2016, 140, 481–485. [Google Scholar] [CrossRef]
- Li, F.; Tie, R.; Chang, K.; Wang, F.; Deng, S.; Lu, W.; Yu, L.; Chen, M. Does risk for ovarian malignancy algorithm excel human epididymis protein 4 and CA125 in predicting epithelial ovarian cancer: A meta-analysis. BMC Cancer 2012, 12, 258. [Google Scholar] [CrossRef]
- Wang, Z.; Tao, X.; Ying, C. CPH-I and HE4 Are More Favorable Than CA125 in Differentiating Borderline Ovarian Tumors from Epithelial Ovarian Cancer at Early Stages. Dis. Markers 2019, 2019, 6241743. [Google Scholar] [CrossRef] [PubMed]
- Ueland, F.R.; Desimone, C.P.; Seamon, L.G.; Miller, R.A.; Goodrich, S.; Podzielinski, I.; Sokoll, L.; Smith, A.; Van Nagell, J.R.; Zhang, Z. Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors. Obstet. Gynecol. 2011, 117, 1289–1297. [Google Scholar] [CrossRef] [PubMed]
- Bristow, R.E.; Smith, A.; Zhang, Z.; Chan, D.W.; Crutcher, G.; Fung, E.T.; Munroe, D.G. Ovarian malignancy risk stratification of the adnexal mass using a multivariate index assay. Gynecol. Oncol. 2013, 128, 252–259. [Google Scholar] [CrossRef] [PubMed]
- Bristow, R.E.; Hodeib, M.; Smith, A.; Chan, D.W.; Zhang, Z.; Fung, E.T.; Tewari, K.S.; Munroe, D.G.; Ueland, F.R. Impact of a multivariate index assay on referral patterns for surgical management of an adnexal mass. Am. J. Obstet. Gynecol. 2013, 209, 581.e1–581.e8. [Google Scholar] [CrossRef] [PubMed]
- Longoria, T.C.; Ueland, F.R.; Zhang, Z.; Chan, D.W.; Smith, A.; Fung, E.T.; Munroe, D.G.; Bristow, R.E. Clinical performance of a multivariate index assay for detecting early-stage ovarian cancer. Am. J. Obstet. Gynecol. 2014, 210, 78.e1–78.e9. [Google Scholar] [CrossRef] [PubMed]
- Ware Miller, R.; Smith, A.; DeSimone, C.P.; Seamon, L.; Goodrich, S.; Podzielinski, I.; Sokoll, L.; van Nagell, J.R.J.; Zhang, Z.; Ueland, F.R. Performance of the American College of Obstetricians and Gynecologists’ ovarian tumor referral guidelines with a multivariate index assay. Obstet. Gynecol. 2011, 117, 1298–1306. [Google Scholar] [CrossRef]
- Grenache, D.G.; Heichman, K.A.; Werner, T.L.; Vucetic, Z. Clinical performance of two multi-marker blood tests for predicting malignancy in women with an adnexal mass. Clin. Chim. Acta 2015, 438, 358–363. [Google Scholar] [CrossRef]
- Shulman, L.P.; Francis, M.; Bullock, R.; Pappas, T. Clinical Performance Comparison of Two In-Vitro Diagnostic Multivariate Index Assays (IVDMIAs) for Presurgical Assessment for Ovarian Cancer Risk. Adv. Ther. 2019, 36, 2402–2413. [Google Scholar] [CrossRef]
- Coleman, R.L.; Herzog, T.J.; Chan, D.W.; Munroe, D.G.; Pappas, T.C.; Smith, A.; Zhang, Z.; Wolf, J. Validation of a second-generation multivariate index assay for malignancy risk of adnexal masses. Am. J. Obstet. Gynecol. 2016, 215, 82.e1–82.e11. [Google Scholar] [CrossRef]
- Skates, S.J.; Horick, N.; Yu, Y.; Xu, F.J.; Berchuck, A.; Havrilesky, L.J.; De Bruijn, H.W.A.; Van Der Zee, A.G.J.; Woolas, R.P.; Jacobs, I.J.; et al. Preoperative sensitivity and specificity for early-stage ovarian cancer when combining cancer antigen CA-125II, CA 15-3, CA 72-4, and macrophage colony-stimulating factor using mixtures of multivariate normal distributions. J. Clin. Oncol. 2004, 22, 4059–4066. [Google Scholar] [CrossRef]
- Zhang, Z.; Yu, Y.; Xu, F.; Berchuck, A.; van Haaften-Day, C.; Havrilesky, L.J.; de Bruijn, H.W.A.; van der Zee, A.G.J.; Woolas, R.P.; Jacobs, I.J.; et al. Combining multiple serum tumor markers improves detection of stage I epithelial ovarian cancer. Gynecol. Oncol. 2007, 107, 526–531. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Li, L.; Douville, C.; Cohen, J.D.; Yen, T.T.; Kinde, I.; Sundfelt, K.; Kjær, S.K.; Hruban, R.H.; Shih, I.M.; et al. Evaluation of liquid from the Papanicolaou test and other liquid biopsies for the detection of endometrial and ovarian cancers. Sci. Transl. Med. 2018, 10, eaap8793. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Yu, L.; Luo, X.; Huang, L.; Li, Q.S.; Shao, X.S.; Liu, Y.; Fan, Y.; Yang, G.Z. Detection of OPCML methylation, a possible epigenetic marker, from free serum circulating DNA to improve the diagnosis of early-stage ovarian epithelial cancer. Oncol. Lett. 2017, 14, 217–223. [Google Scholar] [CrossRef] [PubMed]
- Shao, Y.; Kong, J.; Xu, H.; Wu, X.; Cao, Y.P.; Li, W.; Han, J.; Li, D.; Xie, K.; Wu, J. OPCML Methylation and the Risk of Ovarian Cancer: A Meta and Bioinformatics Analysis. Front. Cell Dev. Biol. 2021, 9, 570898. [Google Scholar] [CrossRef]
- Widschwendter, M.; Zikan, M.; Wahl, B.; Lempiäinen, H.; Paprotka, T.; Evans, I.; Jones, A.; Ghazali, S.; Reisel, D.; Eichner, J.; et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med. 2017, 9, 116. [Google Scholar] [CrossRef]
- Sánchez-Vega, F.; Gotea, V.; Petrykowska, H.M.; Margolin, G.; Krivak, T.C.; DeLoia, J.A.; Bell, D.W.; Elnitski, L. Recurrent patterns of DNA methylation in the ZNF154, CASP8, and VHL promoters across a wide spectrum of human solid epithelial tumors and cancer cell lines. Epigenetics 2013, 8, 1355. [Google Scholar] [CrossRef]
- Miller, B.F.; Petrykowska, H.M.; Elnitski, L. Assessing ZNF154 methylation in patient plasma as a multicancer marker in liquid biopsies from colon, liver, ovarian and pancreatic cancer patients. Sci. Rep. 2021, 11, 221. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef]
- Elias, K.M.; Fendler, W.; Stawiski, K.; Fiascone, S.J.; Vitonis, A.F.; Berkowitz, R.S.; Frendl, G.; Konstantinopoulos, P.; Crum, C.P.; Kedzierska, M.; et al. Diagnostic potential for a serum miRNA neural network for detection of ovarian cancer. Elife 2017, 6, e28932. [Google Scholar] [CrossRef]
- Kandimalla, R.; Wang, W.; Yu, F.; Zhou, N.; Gao, F.; Spillman, M.; Moukova, L.; Slaby, O.; Salhia, B.; Zhou, S.; et al. OCaMIR-A Noninvasive, Diagnostic Signature for Early-Stage Ovarian Cancer: A Multi-cohort Retrospective and Prospective Study. Clin. Cancer Res. 2021, 27, 4277–4286. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Yoshioka, Y.; Hirakawa, A.; Yamamoto, Y.; Ishikawa, M.; Ikeda, S.; Kato, T.; Niimi, K.; Kajiyama, H.; Kikkawa, F.; et al. A combination of circulating miRNAs for the early detection of ovarian cancer. Oncotarget 2017, 8, 89811–89823. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, A.; Matsuzaki, J.; Yamamoto, Y.; Yoneoka, Y.; Takahashi, K.; Shimizu, H.; Uehara, T.; Ishikawa, M.; Ikeda, S.; Sonoda, T.; et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat. Commun. 2018, 9, 4319. [Google Scholar] [CrossRef] [PubMed]
- Garcia, E.; Andrews, C.; Hua, J.; Kim, H.L.; Sukumaran, D.K.; Szyperski, T.; Odunsi, K. Diagnosis of early stage ovarian cancer by 1H NMR metabonomics of serum explored by use of a microflow NMR probe. J. Proteome Res. 2011, 10, 1765–1771. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, Y.; Zhang, X.; Cao, R.; Chen, S.; Huang, Q.; Lu, X.; Wan, X.; Wu, X.; Xu, C.; et al. Application of L-EDA in metabonomics data handling: Global metabolite profiling and potential biomarker discovery of epithelial ovarian cancer prognosis. Metabolomics 2011, 7, 614–622. [Google Scholar] [CrossRef]
- Fan, L.; Zhang, W.; Yin, M.; Zhang, T.; Wu, X.; Zhang, H.; Sun, M.; Li, Z.; Hou, Y.; Zhou, X.; et al. Identification of metabolic biomarkers to diagnose epithelial ovarian cancer using a UPLC/QTOF/MS platform. Acta Oncol. 2012, 51, 473–479. [Google Scholar] [CrossRef]
- Zhang, T.; Wu, X.; Ke, C.; Yin, M.; Li, Z.; Fan, L.; Zhang, W.; Zhang, H.; Zhao, F.; Zhou, X.; et al. Identification of potential biomarkers for ovarian cancer by urinary metabolomic profiling. J. Proteome Res. 2013, 12, 505–512. [Google Scholar] [CrossRef]
- Zhang, Z.; Bast, R.C.; Yu, Y.; Li, J.; Sokoll, L.J.; Rai, A.J.; Rosenzweig, J.M.; Cameron, B.; Wang, Y.Y.; Meng, X.Y.; et al. Three biomarkers identified from serum proteomic analysis for the detection of early stage ovarian cancer. Cancer Res. 2004, 64, 5882–5890. [Google Scholar] [CrossRef]
- Yurkovetsky, Z.; Skates, S.; Lomakin, A.; Nolen, B.; Pulsipher, T.; Modugno, F.; Marks, J.; Godwin, A.; Gorelik, E.; Jacobs, I.; et al. Development of a Multimarker Assay for Early Detection of Ovarian Cancer. J. Clin. Oncol. 2010, 28, 2159. [Google Scholar] [CrossRef]
- Visintin, I.; Feng, Z.; Longton, G.; Ward, D.C.; Alvero, A.B.; Lai, Y.; Tenthorey, J.; Leiser, A.; Flores-Saaib, R.; Yu, H.; et al. Diagnostic markers for early detection of ovarian cancer. Clin. Cancer Res. 2008, 14, 1065–1072. [Google Scholar] [CrossRef]
- Mor, G.; Visintin, I.; Lai, Y.; Zhao, H.; Schwartz, P.; Rutherford, T.; Yue, L.; Bray-Ward, P.; Ward, D.C. Serum protein markers for early detection of ovarian cancer. Proc. Natl. Acad. Sci. USA 2005, 102, 7677–7682. [Google Scholar] [CrossRef] [PubMed]
- Slaughter, L.M. FDA Oversight of Laboratory Developed Tests Essential for Patient Health and Safety. Available online: https://www.ajmc.com/view/fda-oversight-of-laboratory-developed-tests-essential-for-patient-health-and-safety (accessed on 30 May 2022).
- Coates, R.J.; Kolor, K.; Stewart, S.L.; Richardson, L.C. Diagnostic markers for ovarian cancer screening: Not ready for routine clinical use. Clin. Cancer Res. 2008, 14, 7575–7576. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Society of Gynecologic Oncologists Statement on OvaSure. Available online: https://www.sgo.org/wp-content/uploads/2012/09/Statement-on-OvaSure.pdf (accessed on 30 May 2022).
- Gutman, S.I. OvaSureTM Manufacturer Letter | FDA. Available online: https://www.fda.gov/medical-devices/ivd-regulatory-assistance/ovasuretm-manufacturer-letter (accessed on 30 May 2022).
- Guo, J.; Yang, W.L.; Pak, D.; Celestino, J.; Lu, K.H.; Ning, J.; Lokshin, A.E.; Cheng, Z.; Lu, Z.; Bast, R.C. Osteopontin, Macrophage Migration Inhibitory Factor and Anti-Interleukin-8 Autoantibodies Complement CA125 for Detection of Early Stage Ovarian Cancer. Cancers 2019, 11, 596. [Google Scholar] [CrossRef] [PubMed]
- Havrilesky, L.J.; Whitehead, C.M.; Rubatt, J.M.; Cheek, R.L.; Groelke, J.; He, Q.; Malinowski, D.P.; Fischer, T.J.; Berchuck, A. Evaluation of biomarker panels for early stage ovarian cancer detection and monitoring for disease recurrence. Gynecol. Oncol. 2008, 110, 374–382. [Google Scholar] [CrossRef]
- Amonkar, S.D.; Bertenshaw, G.P.; Chen, T.H.; Bergstrom, K.J.; Zhao, J.; Seshaiah, P.; Yip, P.; Mansfield, B.C. Development and Preliminary Evaluation of a Multivariate Index Assay for Ovarian Cancer. PLoS ONE 2009, 4, e4599. [Google Scholar] [CrossRef]
- Enroth, S.; Berggrund, M.; Lycke, M.; Broberg, J.; Lundberg, M.; Assarsson, E.; Olovsson, M.; Stålberg, K.; Sundfeldt, K.; Gyllensten, U. High throughput proteomics identifies a high-accuracy 11 plasma protein biomarker signature for ovarian cancer. Commun. Biol. 2019, 2, 221. [Google Scholar] [CrossRef]
- Edgell, T.; Martin-Roussety, G.; Barker, G.; Autelitano, D.J.; Allen, D.; Grant, P.; Rice, G.E. Phase II biomarker trial of a multimarker diagnostic for ovarian cancer. J. Cancer Res. Clin. Oncol. 2010, 136, 1079. [Google Scholar] [CrossRef]
- Russell, M.R.; D’Amato, A.; Graham, C.; Crosbie, E.J.; Gentry-Maharaj, A.; Ryan, A.; Kalsi, J.K.; Fourkala, E.-O.; Dive, C.; Walker, M.; et al. Novel risk models for early detection and screening of ovarian cancer. Oncotarget 2016, 8, 785–797. [Google Scholar] [CrossRef]
- Russell, M.R.; Graham, C.; D’Amato, A.; Gentry-Maharaj, A.; Ryan, A.; Kalsi, J.K.; Whetton, A.D.; Menon, U.; Jacobs, I.; Graham, R.L.J. Diagnosis of epithelial ovarian cancer using a combined protein biomarker panel. Br. J. Cancer 2019, 121, 483–489. [Google Scholar] [CrossRef]
- Li, A.J.; Madden, A.C.; Cass, I.; Leuchter, R.S.; Lagasse, L.D.; Karlan, B.Y. The prognostic significance of thrombocytosis in epithelial ovarian carcinoma. Gynecol. Oncol. 2004, 92, 211–214. [Google Scholar] [CrossRef]
- Watrowski, R.; Heinze, G.; Jäger, C.; Forster, J.; Zeillinger, R. Usefulness of the preoperative platelet count in the diagnosis of adnexal tumors. Tumour Biol. 2016, 37, 12079–12087. [Google Scholar] [CrossRef] [PubMed]
- Cho, M.S.; Bottsford-Miller, J.; Vasquez, H.G.; Stone, R.; Zand, B.; Kroll, M.H.; Sood, A.K.; Afshar-Kharghan, V. Platelets increase the proliferation of ovarian cancer cells. Blood 2012, 120, 4869–4872. [Google Scholar] [CrossRef] [PubMed]
- Bottsford-Miller, J.; Choi, H.J.; Dalton, H.J.; Stone, R.L.; Cho, M.S.; Haemmerle, M.; Nick, A.M.; Pradeep, S.; Zand, B.; Previs, R.A.; et al. Differential platelet levels affect response to taxane-based therapy in ovarian cancer. Clin. Cancer Res. 2015, 21, 602–610. [Google Scholar] [CrossRef] [PubMed]
- Stone, R.L.; Nick, A.M.; McNeish, I.A.; Balkwill, F.; Han, H.D.; Bottsford-Miller, J.; Rupaimoole, R.; Armaiz-Pena, G.N.; Pecot, C.V.; Coward, J.; et al. Paraneoplastic thrombocytosis in ovarian cancer. N. Engl. J. Med. 2012, 366, 610–618. [Google Scholar] [CrossRef]
- Cho, M.S.; Lee, H.; Gonzalez-Delgado, R.; Li, D.; Sasano, T.; Carlos-Alcalde, W.; Ma, Q.; Liu, J.; Sood, A.K.; Afshar-Kharghan, V. Platelets Increase the Expression of PD-L1 in Ovarian Cancer. Cancers 2022, 14, 2498. [Google Scholar] [CrossRef]
- Lomnytska, M.; Pinto, R.; Becker, S.; Engström, U.; Gustafsson, S.; Björklund, C.; Templin, M.; Bergstrand, J.; Xu, L.; Widengren, J.; et al. Platelet protein biomarker panel for ovarian cancer diagnosis. Biomark. Res. 2018, 6, 2. [Google Scholar] [CrossRef]
- Gyllensten, U.; Hedlund-Lindberg, J.; Svensson, J.; Manninen, J.; Öst, T.; Ramsell, J.; Åslin, M.; Ivansson, E.; Lomnytska, M.; Lycke, M.; et al. Next Generation Plasma Proteomics Identifies High-Precision Biomarker Candidates for Ovarian Cancer. Cancers 2022, 14, 1757. [Google Scholar] [CrossRef]
- Jokerst, J.V.; Raamanathan, A.; Christodoulides, N.; Floriano, P.N.; Pollard, A.A.; Simmons, G.W.; Wong, J.; Gage, C.; Furmaga, W.B.; Redding, S.W.; et al. Nano-bio-chips for high performance multiplexed protein detection: Determinations of cancer biomarkers in serum and saliva using quantum dot bioconjugate labels. Biosens. Bioelectron. 2009, 24, 3622–3629. [Google Scholar] [CrossRef]
- Raamanathan, A.; Simmons, G.W.; Christodoulides, N.; Floriano, P.N.; Furmaga, W.B.; Redding, S.W.; Lu, K.H.; Bast, R.C.; McDevitt, J.T. Programmable Bio-Nano-Chip Systems for Serum CA125 Quantification: Toward Ovarian Cancer Diagnostics at the Point-of-Care. Cancer Prev. Res. 2012, 5, 706–716. [Google Scholar] [CrossRef]
- Shadfan, B.H.; Simmons, A.R.; Simmons, G.W.; Ho, A.; Wong, J.; Lu, K.H.; Bast, R.C.; McDevitt, J.T. A Multiplexable, Microfluidic Platform for the Rapid Quantitation of a Biomarker Panel for Early Ovarian Cancer Detection at the Point-of-Care. Cancer Prev. Res. 2015, 8, 37–48. [Google Scholar] [CrossRef]
- Chen, F.; Liu, Y.; Chen, C.; Gong, H.; Cai, C.; Chen, X. Respective and simultaneous detection tumor markers CA125 and STIP1 using aptamer-based fluorescent and RLS sensors. Sens. Actuators B Chem. 2017, 245, 470–476. [Google Scholar] [CrossRef]
- Xu, X.; Ji, J.; Chen, P.; Wu, J.; Jin, Y.; Zhang, L.; Du, S. Salt-induced gold nanoparticles aggregation lights up fluorescence of DNA-silver nanoclusters to monitor dual cancer markers carcinoembryonic antigen and carbohydrate antigen 125. Anal. Chim. Acta 2020, 1125, 41–49. [Google Scholar] [CrossRef]
- Wang, M.; Hu, M.; Li, Z.; He, L.; Song, Y.; Jia, Q.; Zhang, Z.; Du, M. Construction of Tb-MOF-on-Fe-MOF conjugate as a novel platform for ultrasensitive detection of carbohydrate antigen 125 and living cancer cells. Biosens. Bioelectron. 2019, 142, 111536. [Google Scholar] [CrossRef]
- Jin, H.; Gui, R.; Gong, J.; Huang, W. Aptamer and 5-fluorouracil dual-loading Ag2S quantum dots used as a sensitive label-free probe for near-infrared photoluminescence turn-on detection of CA125 antigen. Biosens. Bioelectron. 2017, 92, 378–384. [Google Scholar] [CrossRef]
- Farzin, L.; Sadjadi, S.; Shamsipur, M.; Sheibani, S.; hasan Mousazadeh, M. Employing AgNPs doped amidoxime-modified polyacrylonitrile (PAN-oxime) nanofibers for target induced strand displacement-based electrochemical aptasensing of CA125 in ovarian cancer patients. Mater. Sci. Eng. C 2019, 97, 679–687. [Google Scholar] [CrossRef]
- Wang, T.H.; Chao, A.; Tsai, C.L.; Chang, C.L.; Chen, S.H.; Lee, Y.S.; Chen, J.K.; Lin, Y.J.; Chang, P.Y.; Wang, C.J.; et al. Stress-induced Phosphoprotein 1 as a Secreted Biomarker for Human Ovarian Cancer Promotes Cancer Cell Proliferation. Mol. Cell. Proteom. 2010, 9, 1873–1884. [Google Scholar] [CrossRef]
- Kim, M.; Chen, C.; Wang, P.; Mulvey, J.J.; Yang, Y.; Wun, C.; Antman-Passig, M.; Luo, H.-B.; Cho, S.; Long-Roche, K.; et al. Detection of ovarian cancer via the spectral fingerprinting of quantum-defect-modified carbon nanotubes in serum by machine learning. Nat. Biomed. Eng. 2022, 6, 267–275. [Google Scholar] [CrossRef]
- Saldova, R.; Struwe, W.B.; Wynne, K.; Elia, G.; Duffy, M.J.; Rudd, P.M. Exploring the Glycosylation of Serum CA125. Int. J. Mol. Sci. 2013, 14, 15636. [Google Scholar] [CrossRef]
- Shang, Y.; Zeng, Y.; Zeng, Y. Integrated Microfluidic Lectin Barcode Platform for High-Performance Focused Glycomic Profiling. Sci. Rep. 2016, 6, 20297. [Google Scholar] [CrossRef]
- Bayoumy, S.; Hyytiä, H.; Leivo, J.; Talha, S.M.; Huhtinen, K.; Poutanen, M.; Hynninen, J.; Perheentupa, A.; Lamminmäki, U.; Gidwani, K.; et al. Glycovariant-based lateral flow immunoassay to detect ovarian cancer–associated serum CA125. Commun. Biol. 2020, 3, 460. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z.; Payne, S.H.; Zhang, B.; McDermott, J.E.; Zhou, J.Y.; Petyuk, V.A.; Chen, L.; Ray, D.; et al. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755–765. [Google Scholar] [CrossRef]
- Bedi, D.G.; Patnana, M.; Ernst, R.D.; Lu, K.H. Sonographic Findings in Early Ovarian Cancer: Preliminary Experience in a Population of High Risk Women Screened with Biannual Ultrasound. In Proceedings of the Radiological Society of North America 2010 Scientific Assembly and Annual Meeting, Chicago, IL, USA, 28 November–3 December 2010. [Google Scholar]
- Barroilhet, L.; Vitonis, A.; Shipp, T.; Muto, M.; Benacerraf, B. Sonographic predictors of ovarian malignancy. J. Clin. Ultrasound 2013, 41, 269–274. [Google Scholar] [CrossRef]
- Khurana, I.; Satia, M.N. Preoperative evaluation of ovarian masses with color Doppler and its correlation with pathological finding. Int. J. Reprod. Contracept. Obstet. Gynecol. 2017, 5, 2084–2092. [Google Scholar] [CrossRef][Green Version]
- Guerriero, S.; Alcazar, J.L.; Ajossa, S.; Galvan, R.; Laparte, C.; Garciá-Manero, M.; Lopez-Garcia, G.; Melis, G.B. Transvaginal Color Doppler Imaging in the Detection of Ovarian Cancer in a Large Study Population. Int. J. Gynecol. Cancer 2010, 20, 781–786. [Google Scholar] [CrossRef]
- Pysz, M.A.; Willmann, J.K. Targeted Contrast-Enhanced Ultrasound: An Emerging Technology in Abdominal and Pelvic Imaging. Gastroenterology 2011, 140, 785–790.e6. [Google Scholar] [CrossRef]
- Szymanski, M.; Socha, M.W.; Kowalkowska, M.E.; Zielińska, I.B.; Eljaszewicz, A.; Szymanski, W. Differentiating between benign and malignant adnexal lesions with contrast-enhanced transvaginal ultrasonography. Int. J. Gynecol. Obstet. 2015, 131, 147–151. [Google Scholar] [CrossRef]
- Mathieu, K.B.; Bedi, D.G.; Thrower, S.L.; Qayyum, A.; Bast, R.C. Screening for ovarian cancer: Imaging challenges and opportunities for improvement. Ultrasound Obstet. Gynecol. 2018, 51, 293–303. [Google Scholar] [CrossRef]
- Zackrisson, S.; van de Ven, S.M.W.Y.; Gambhir, S.S. Light In and Sound Out: Emerging Translational Strategies for Photoacoustic Imaging. Cancer Res. 2014, 74, 979–1004. [Google Scholar] [CrossRef]
- Aguirre, A.; Guo, P.; Gamelin, J.K.; Yan, S.; Sanders, M.M.; Brewer, M.A.; Zhu, Q. Coregistered three-dimensional ultrasound and photoacoustic imaging system for ovarian tissue characterization. J. Biomed. Opt. 2009, 14, 054014. [Google Scholar] [CrossRef]
- Lao, Y.; Xing, D.; Yang, S.; Xiang, L. Noninvasive photoacoustic imaging of the developing vasculature during early tumor growth. Phys. Med. Biol. 2008, 53, 4203. [Google Scholar] [CrossRef]
- Salehi, H.S.; Li, H.; Merkulov, A.; Kumavor, P.D.; Vavadi, H.; Sanders, M.; Kueck, A.; Brewer, M.A.; Zhu, Q. Coregistered photoacoustic and ultrasound imaging and classification of ovarian cancer: Ex vivo and in vivo studies. J. Biomed. Opt. 2016, 21, 046006. [Google Scholar] [CrossRef] [PubMed]
- Nandy, S.; Mostafa, A.; Hagemann, I.S.; Powell, M.A.; Amidi, E.; Robinson, K.; Mutch, D.G.; Siegel, C.; Zhu, Q. Evaluation of ovarian cancer: Initial application of coregistered photoacoustic tomography and US. Radiology 2018, 289, 740–747. [Google Scholar] [CrossRef] [PubMed]
- Grover, V.P.B.; Tognarelli, J.M.; Crossey, M.M.E.; Cox, I.J.; Taylor-Robinson, S.D.; McPhail, M.J.W. Magnetic Resonance Imaging: Principles and Techniques: Lessons for Clinicians. J. Clin. Exp. Hepatol. 2015, 5, 246. [Google Scholar] [CrossRef] [PubMed]
- Sohaib, S.A.A.; Reznek, R.H. MR imaging in ovarian cancer. Cancer Imaging 2007, 7, S119. [Google Scholar] [CrossRef]
- Jung, S.E.; Lee, J.M.; Rha, S.E.; Byun, J.Y.; Jung, J.I.; Hahn, S.T. CT and MR imaging of ovarian tumors with emphasis on differential diagnosis. Radiographics 2002, 22, 1305–1325. [Google Scholar] [CrossRef]
- Wang, R.; Cai, Y.; Lee, I.K.; Hu, R.; Purkayastha, S.; Pan, I.; Yi, T.; Tran, T.M.L.; Lu, S.; Liu, T.; et al. Evaluation of a convolutional neural network for ovarian tumor differentiation based on magnetic resonance imaging. Eur. Radiol. 2021, 31, 4960–4971. [Google Scholar] [CrossRef]
- Rieber, A.; Nüssle, K.; Stöhr, I.; Grab, D.; Fenchel, S.; Kreienberg, R.; Reske, S.N.; Brambs, H.J. Preoperative Diagnosis of Ovarian Tumors with MR ImagingComparison with Transvaginal Sonography, Positron Emission Tomography, and Histologic Findings. AJR Am. J. Roentgenol. 2001, 177, 123–129. [Google Scholar] [CrossRef]
- Grab, D.; Flock, F.; Stöhr, I.; Nüssle, K.; Rieber, A.; Fenchel, S.; Brambs, H.J.; Reske, S.N.; Kreienberg, R. Classification of Asymptomatic Adnexal Masses by Ultrasound, Magnetic Resonance Imaging, and Positron Emission Tomography. Gynecol. Oncol. 2000, 77, 454–459. [Google Scholar] [CrossRef]
- Castellucci, P.; Perrone, A.M.; Picchio, M.; Ghi, T.; Farsad, M.; Nanni, C.; Messa, C.; Meriggiola, M.C.; Pelusi, G.; Al-Nahhas, A.; et al. Diagnostic accuracy of 18F-FDG PET/CT in characterizing ovarian lesions and staging ovarian cancer: Correlation with transvaginal ultrasonography, computed tomography, and histology. Nucl. Med. Commun. 2007, 28, 589–595. [Google Scholar] [CrossRef]
- Nam, E.J.; Yun, M.J.; Oh, Y.T.; Kim, J.W.; Kim, J.H.; Kim, S.; Jung, Y.W.; Kim, S.W.; Kim, Y.T. Diagnosis and staging of primary ovarian cancer: Correlation between PET/CT, Doppler US, and CT or MRI. Gynecol. Oncol. 2010, 116, 389–394. [Google Scholar] [CrossRef]
- Powell, C.B.; Littell, R.D.; Landen, C.N.; Pramanik, S.; Hamilton, I.C.; Suh-Burgmann, E.J. Cytological sampling of fallopian tubes using a hysteroscopic catheter: A multi-center study. Gynecol. Oncol. 2020, 156, 636–640. [Google Scholar] [CrossRef]
- Chen, H.; Klein, R.; Arnold, S.; Wang, Y.; Chambers, S.; Zheng, W. Tubal Cytology of the Fallopian Tube as a Promising Tool for Ovarian Cancer Early Detection. JoVE 2017, 125, e55887. [Google Scholar] [CrossRef]
- Lum, D.; Guido, R.; Rodriguez, E.; Lee, T.; Mansuria, S.; Marshall Austin, R. Brush Cytology of the Fallopian Tube and Implications in Ovarian Cancer Screening. J. Minim. Invasive Gynecol. 2014, 21, 851–856. [Google Scholar] [CrossRef]
- Kerin, J.; Surrey, E.; Daykhovsky, L.; Grundfest, W.S. Development and application of a falloposcope for transvaginal endoscopy of the fallopian tube. J. Laparoendosc. Surg. 1990, 1, 47–56. [Google Scholar] [CrossRef]
- Wong, A.; Walker, S. Falloposcopy-a prerequisite to the proper assessment of tubal infertility. Hong Kong Med. J. 1999, 5, 76–81. [Google Scholar]
- Keenan, M.; Tate, T.H.; Kieu, K.; Black, J.F.; Utzinger, U.; Barton, J.K. Design and characterization of a combined OCT and wide field imaging falloposcope for ovarian cancer detection. Biomed. Opt. Express 2017, 8, 124. [Google Scholar] [CrossRef]
- Optical Imaging Falloposcope for Early Ovarian Cancer Detection: In Vivo Feasibility and Safety. Available online: https://apps.dtic.mil/sti/citations/AD1115246 (accessed on 1 June 2022).
- De Haro, L.P.; Karaulanov, T.; Vreeland, E.C.; Anderson, B.; Hathaway, H.J.; Huber, D.L.; Matlashov, A.N.; Nettles, C.P.; Price, A.D.; Monson, T.C.; et al. Magnetic relaxometry as applied to sensitive cancer detection and localization. Biomed. Tech. 2015, 60, 445–455. [Google Scholar] [CrossRef]
- Mathieu, K.; Lu, Z.; Yang, H.; Pang, L.; Kulp, A.; Hazle, J.; Bast, R.C. Abstract 1864: Feasibility of magnetic relaxometry for early ovarian cancer detection: Preliminary evaluation of sensitivity and specificity in cell culture and in mice. Cancer Res. 2017, 77, 1864. [Google Scholar] [CrossRef]
- Bast, R.C.; Han, C.Y.; Lu, Z.; Lu, K.H. Next steps in the early detection of ovarian cancer. Commun. Med. 2021, 1, 36. [Google Scholar] [CrossRef]
- Zhang, Z.; Chan, D.W. The road from discovery to clinical diagnostics: Lessons learned from the first FDA-cleared in vitro diagnostic multivariate index assay of proteomic biomarkers. Cancer Epidemiol. Biomarkers Prev. 2010, 19, 2995–2999. [Google Scholar] [CrossRef]
- Su, F.; Lang, J.; Kumar, A.; Ng, C.; Hsieh, B.; Suchard, M.A.; Suchard, M.A.; Reddy, S.; Farias-Eisner, R.; Reddy, S.T. Validation of Candidate Serum Ovarian Cancer Biomarkers for Early Detection. Biomark. Insights 2007, 2, 369. [Google Scholar] [CrossRef]
- Forde, G.K.; Hornberger, J.; Michalopoulos, S.; Bristow, R.E. Cost-effectiveness analysis of a multivariate index assay compared to modified American College of Obstetricians and Gynecologists criteria and CA-125 in the triage of women with adnexal masses. Curr. Med. Res. Opin. 2016, 32, 321–329. [Google Scholar] [CrossRef]
- About OVA1plus—Aspira Women’s Health. Available online: https://aspirawh.com/ova1plus-about/ (accessed on 30 May 2022).
- Wang, J.; Gao, J.; Yao, H.; Wu, Z.; Wang, M.; Qi, J. Diagnostic accuracy of serum HE4, CA125 and ROMA in patients with ovarian cancer: A meta-analysis. Tumor Biol. 2014, 35, 6127–6138. [Google Scholar] [CrossRef]
- Van Gorp, T.; Cadron, I.; Despierre, E.; Daemen, A.; Leunen, K.; Amant, F.; Timmerman, D.; De Moor, B.; Vergote, I. HE4 and CA125 as a diagnostic test in ovarian cancer: Prospective validation of the Risk of Ovarian Malignancy Algorithm. Br. J. Cancer 2011, 104, 863–870. [Google Scholar] [CrossRef]
- Jacob, F.; Meier, M.; Caduff, R.; Goldstein, D.; Pochechueva, T.; Hacker, N.; Fink, D.; Heinzelmann-Schwarz, V. No benefit from combining HE4 and CA125 as ovarian tumor markers in a clinical setting. Gynecol. Oncol. 2011, 121, 487–491. [Google Scholar] [CrossRef]
- Jacobs, I.; Oram, D.; Fairbanks, J.; Turner, J.; Frost, C.; Grudzinskas, J.G. A risk of malignancy index incorporating CA 125, ultrasound and menopausal status for the accurate preoperative diagnosis of ovarian cancer. Br. J. Obstet. Gynaecol. 1990, 97, 922–929. [Google Scholar] [CrossRef]
- Tingulstad, S.; Hagen, B.; Skjeldestad, F.E.; Onsrud, M.; Kiserud, T.; Halvorsen, T.; Nustad, K. Evaluation of a risk of malignancy index based on serum CA125, ultrasound findings and menopausal status in the pre-operative diagnosis of pelvic masses. BJOG 1996, 103, 826–831. [Google Scholar] [CrossRef]
- Hayam, F.; Ashraf, M.; Hassan, M. Assessment of the Value of a Modified Risk of Malignancy Index (RMI) in Preoperative Discrimination Between Benign and Malignant Ovarian Masses. Gynecol. Obstet. 2016, 6, 417. [Google Scholar] [CrossRef]
- Aktürk, E.; Karaca, R.E.; Alanbay, I.; Dede, M.; Karaşahin, E.; Yenen, M.C.; Başer, I. Comparison of four malignancy risk indices in the detection of malignant ovarian masses. J. Gynecol. Oncol. 2011, 22, 177–182. [Google Scholar] [CrossRef]
- Al Musalhi, K.; Al Kindi, M.; Al Aisary, F.; Ramadhan, F.; Al Rawahi, T.; Al Hatali, K.; Mula-Abed, W.A. Evaluation of HE4, CA-125, Risk of Ovarian Malignancy Algorithm (ROMA) and Risk of Malignancy Index (RMI) in the Preoperative Assessment of Patients with Adnexal Mass. Oman Med. J. 2016, 31, 336–344. [Google Scholar] [CrossRef]
- Baral, G.; Joshi, R.; Pandit, B. Diagnostic Accuracy of Risk of Malignancy Indices in Ovarian Tumor. J. Nepal Health Res. Counc. 2020, 18, 253–258. [Google Scholar] [CrossRef] [PubMed]
- Nikolova, T.; Zivadinovic, R.; Evtimovska, N.; Klisarovska, V.; Stanojevic, M.; Georgievska, J.; Nikolova, N. Diagnostic performance of human epididymis protein 4 compared to a combination of biophysical and biochemical markers to differentiate ovarian endometriosis from epithelial ovarian cancer in premenopausal women. J. Obstet. Gynaecol. Res. 2017, 43, 1870–1879. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.L.; Breen, K.; Catchings, A.; Ranganathan, M.; Latham, A.; Goldfrank, D.J.; Grisham, R.N.; Long Roche, K.; Frey, M.K.; Chi, D.S.; et al. Risk-Reducing Bilateral Salpingo-Oophorectomy for Ovarian Cancer: A Review and Clinical Guide for Hereditary Predisposition Genes. JCO Oncol. Pract. 2021, 18, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Iranzo, J.; Martincorena, I.; Koonin, E.V. Cancer-mutation network and the number and specificity of driver mutations. Proc. Natl. Acad. Sci. USA 2018, 115, E6010–E6019. [Google Scholar] [CrossRef]
- Salk, J.J.; Loubet-Senear, K.; Maritschnegg, E.; Valentine, C.C.; Williams, L.N.; Higgins, J.E.; Horvat, R.; Vanderstichele, A.; Nachmanson, D.; Baker, K.T.; et al. Ultra-Sensitive TP53 Sequencing for Cancer Detection Reveals Progressive Clonal Selection in Normal Tissue over a Century of Human Lifespan. Cell Rep. 2019, 28, 132–144.e3. [Google Scholar] [CrossRef]
- Krimmel, J.D.; Schmitt, M.W.; Harrell, M.I.; Agnew, K.J.; Kennedy, S.R.; Emond, M.J.; Loeb, L.A.; Swisher, E.M.; Risques, R.A. Ultra-deep sequencing detects ovarian cancer cells in peritoneal fluid and reveals somatic TP53 mutations in noncancerous tissues. Proc. Natl. Acad. Sci. USA 2016, 113, 6005–6010. [Google Scholar] [CrossRef]
- Razavi, P.; Li, B.T.; Brown, D.N.; Jung, B.; Hubbell, E.; Shen, R.; Abida, W.; Juluru, K.; De Bruijn, I.; Hou, C.; et al. High-intensity sequencing reveals the sources of plasma circulating cell-free DNA variants. Nat. Med. 2019, 25, 1928–1937. [Google Scholar] [CrossRef]
- Kinde, I.; Bettegowda, C.; Wang, Y.; Wu, J.; Agrawal, N.; Shih, I.M.; Kurman, R.; Dao, F.; Levine, D.A.; Giuntoli, R.; et al. Evaluation of DNA from the papanicolaou test to detect ovarian and endometrial cancers. Sci. Transl. Med. 2013, 5, 167ra4. [Google Scholar] [CrossRef]
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765. [Google Scholar] [CrossRef]
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238. [Google Scholar] [CrossRef]
- Tug, S.; Helmig, E.; Deichmann, E.R.; Schmeier-Jürchott, A.; Wagner, E.; Zimmerman, T.; Radsak, M.; Giacca, M.; Simon, P. Exercise-induced cell free DNA originates predominantly from haematopoietic cells. Exerc. Immunol. Rev. 2015, 21, 164–173. [Google Scholar] [PubMed]
- Gögenur, M.; Burcharth, J.; Gögenur, I. The role of total cell-free DNA in predicting outcomes among trauma patients in the intensive care unit: A systematic review. Crit. Care 2017, 21, 14. [Google Scholar] [CrossRef] [PubMed]
- Weerakoon, K.G.; McManus, D.P. Cell-Free DNA as a Diagnostic Tool for Human Parasitic Infections. Trends Parasitol. 2016, 32, 378–391. [Google Scholar] [CrossRef] [PubMed]
- Gielis, E.M.; Ledeganck, K.J.; De Winter, B.Y.; Del Favero, J.; Bosmans, J.L.; Claas, F.H.J.; Abramowicz, D.; Eikmans, M. Cell-Free DNA: An Upcoming Biomarker in Transplantation. Am. J. Transplant. 2015, 15, 2541–2551. [Google Scholar] [CrossRef]
- Labidi-Galy, S.I.; Papp, E.; Hallberg, D.; Niknafs, N.; Adleff, V.; Noe, M.; Bhattacharya, R.; Novak, M.; Jones, S.; Phallen, J.; et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat. Commun. 2017, 8, 1903. [Google Scholar] [CrossRef]
- Pisanic, T.R.; Wang, Y.; Sun, H.; Considine, M.; Li, L.; Wang, T.H.; Wang, T.L.; Shih, I.M. Methylomic Landscapes of Ovarian Cancer Precursor Lesions. Clin. Cancer Res. 2020, 26, 6310–6320. [Google Scholar] [CrossRef]
- Maritschnegg, E.; Wang, Y.; Pecha, N.; Horvat, R.; Van Nieuwenhuysen, E.; Vergote, I.; Heitz, F.; Sehouli, J.; Kinde, I.; Diaz, L.A.; et al. Lavage of the uterine cavity for molecular detection of Müllerian duct carcinomas: A proof-of-concept study. J. Clin. Oncol. 2015, 33, 4293–4300. [Google Scholar] [CrossRef]
- Maritschnegg, E.; Heitz, F.; Pecha, N.; Bouda, J.; Trillsch, F.; Grimm, C.; Vanderstichele, A.; Agreiter, C.; Harter, P.; Obermayr, E.; et al. Uterine and Tubal Lavage for Earlier Cancer Detection Using an Innovative Catheter: A Feasibility and Safety Study. Int. J. Gynecol. Cancer 2018, 28, 1692–1698. [Google Scholar] [CrossRef]
- Erickson, B.K.; Kinde, I.; Dobbin, Z.C.; Wang, Y.; Martin, J.Y.; Alvarez, R.D.; Conner, M.G.; Huh, W.K.; Roden, R.B.S.; Kinzler, K.W.; et al. Detection of somatic TP53 mutations in tampons of patients with high-grade serous ovarian cancer. Obstet. Gynecol. 2014, 124, 881–885. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef]
- Hindson, B.J.; Ness, K.D.; Masquelier, D.A.; Belgrader, P.; Heredia, N.J.; Makarewicz, A.J.; Bright, I.J.; Lucero, M.Y.; Hiddessen, A.L.; Legler, T.C.; et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011, 83, 8604–8610. [Google Scholar] [CrossRef] [PubMed]
- Haunschild, C.E.; Tewari, K.S. The current landscape of molecular profiling in the treatment of epithelial ovarian cancer. Gynecol. Oncol. 2021, 160, 333–345. [Google Scholar] [CrossRef] [PubMed]
- Foley, S.B.; Rios, J.J.; Mgbemena, V.E.; Robinson, L.S.; Hampel, H.L.; Toland, A.E.; Durham, L.; Ross, T.S. Use of Whole Genome Sequencing for Diagnosis and Discovery in the Cancer Genetics Clinic. EBioMedicine 2015, 2, 74–81. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhong, P.; Hong, M.; Tan, J.; Yu, X.; Huang, H.; Ouyang, J.; Lin, X.; Chen, P. Applying low coverage whole genome sequencing to detect malignant ovarian mass. J. Transl. Med. 2021, 19, 369. [Google Scholar] [CrossRef]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nature 2019, 571, 489–499. [Google Scholar] [CrossRef]
- Wu, Q.; Lothe, R.A.; Ahlquist, T.; Silins, I.; Tropé, C.G.; Micci, F.; Nesland, J.M.; Suo, Z.; Lind, G.E. DNA methylation profiling of ovarian carcinomas and their in vitro models identifies HOXA9, HOXB5, SCGB3A1, and CRABP1 as novel targets. Mol. Cancer 2007, 6, 45. [Google Scholar] [CrossRef]
- Widschwendter, M.; Apostolidou, S.; Jones, A.A.; Fourkala, E.O.; Arora, R.; Pearce, C.L.; Frasco, M.A.; Ayhan, A.; Zikan, M.; Cibula, D.; et al. HOXA methylation in normal endometrium from premenopausal women is associated with the presence of ovarian cancer: A proof of principle study. Int. J. Cancer 2009, 125, 2214–2218. [Google Scholar] [CrossRef]
- Campan, M.; Moffitt, M.; Houshdaran, S.; Shen, H.; Widschwendter, M.; Daxenbichler, G.; Long, T.; Marth, C.; Laird-Offringa, I.A.; Press, M.F.; et al. Genome-scale screen for DNA methylation-based detection markers for ovarian cancer. PLoS ONE 2011, 6, e28141. [Google Scholar] [CrossRef]
- De Caceres, I.I.; Battagli, C.; Esteller, M.; Herman, J.G.; Dulaimi, E.; Edelson, M.I.; Bergman, C.; Ehya, H.; Eisenberg, B.L.; Cairns, P. Tumor cell-specific BRCA1 and RASSF1A hypermethylation in serum, plasma, and peritoneal fluid from ovarian cancer patients. Cancer Res. 2004, 64, 6476–6481. [Google Scholar] [CrossRef]
- Bondurant, A.E.; Huang, Z.; Whitaker, R.S.; Simel, L.R.; Berchuck, A.; Murphy, S.K. Quantitative detection of RASSF1A DNA promoter methylation in tumors and serum of patients with serous epithelial ovarian cancer. Gynecol. Oncol. 2011, 123, 581–587. [Google Scholar] [CrossRef]
- Liggett, T.E.; Melnikov, A.; Yi, Q.; Replogle, C.; Hu, W.; Rotmensch, J.; Kamat, A.; Sood, A.K.; Levenson, V. Distinctive DNA methylation patterns of cell-free plasma DNA in women with malignant ovarian tumors. Gynecol. Oncol. 2011, 120, 113–120. [Google Scholar] [CrossRef] [PubMed]
- Dammann, R.H.; Kirsch, S.; Schagdarsurengin, U.; Dansranjavin, T.; Gradhand, E.; Schmitt, W.D.; Hauptmann, S. Frequent aberrant methylation of the imprinted IGF2/H19 locus and LINE1 hypomethylation in ovarian carcinoma. Int. J. Oncol. 2010, 36, 171–179. [Google Scholar] [CrossRef] [PubMed]
- Pisanic, T.R.; Asaka, S.; Lin, S.F.; Yen, T.T.; Sun, H.; Bahadirli-Talbott, A.; Wang, T.H.; Burns, K.H.; Wang, T.L.; Shih, I.M. Long Interspersed Nuclear Element 1 Retrotransposons Become Deregulated during the Development of Ovarian Cancer Precursor Lesions. Am. J. Pathol. 2019, 189, 513–520. [Google Scholar] [CrossRef] [PubMed]
- Shih, I.M.; Chen, L.; Wang, C.C.; Gu, J.; Davidson, B.; Cope, L.; Kurman, R.J.; Xuan, J.; Wang, T.L. Distinct DNA methylation profiles in ovarian serous neoplasms and their implications in ovarian carcinogenesis. Am. J. Obstet. Gynecol. 2010, 203, 584.e1. [Google Scholar] [CrossRef]
- Kolbe, D.L.; DeLoia, J.A.; Porter-Gill, P.; Strange, M.; Petrykowska, H.M.; Guirguis, A.; Krivak, T.C.; Brody, L.C.; Elnitski, L. Differential Analysis of Ovarian and Endometrial Cancers Identifies a Methylator Phenotype. PLoS ONE 2012, 7, 32941. [Google Scholar] [CrossRef]
- Zhuang, J.; Jones, A.; Lee, S.H.; Ng, E.; Fiegl, H.; Zikan, M.; Cibula, D.; Sargent, A.; Salvesen, H.B.; Jacobs, I.J.; et al. The Dynamics and Prognostic Potential of DNA Methylation Changes at Stem Cell Gene Loci in Women’s Cancer. PLoS Genet. 2012, 8, 1002517. [Google Scholar] [CrossRef]
- Teschendorff, A.E.; Menon, U.; Gentry-Maharaj, A.; Ramus, S.J.; Gayther, S.A.; Apostolidou, S.; Jones, A.; Lechner, M.; Beck, S.; Jacobs, I.J.; et al. An Epigenetic Signature in Peripheral Blood Predicts Active Ovarian Cancer. PLoS ONE 2009, 4, e8274. [Google Scholar] [CrossRef]
- Li, B.; Pu, K.; Ge, L.; Wu, X. Diagnostic significance assessment of the circulating cell-free DNA in ovarian cancer: An updated meta-analysis. Gene 2019, 714, 143993. [Google Scholar] [CrossRef]
- Bodelon, C.; Keith Killian, J.; Sampson, J.N.; Anderson, W.F.; Matsuno, R.; Brinton, L.A.; Lissowska, J.; Anglesio, M.S.; Bowtell, D.D.L.; Doherty, J.A.; et al. Molecular Classification of Epithelial Ovarian Cancer Based on Methylation Profiling: Evidence for Survival Heterogeneity. Clin. Cancer Res. 2019, 25, 5937–5946. [Google Scholar] [CrossRef]
- Kurman, R.J.; Shih, I.M. The Dualistic Model of Ovarian Carcinogenesis: Revisited, Revised, and Expanded. Am. J. Pathol. 2016, 186, 733–747. [Google Scholar] [CrossRef]
- Nik, N.N.; Vang, R.; Shih, I.M.; Kurman, R.J. Origin and pathogenesis of pelvic (ovarian, tubal, and primary peritoneal) serous carcinoma. Annu. Rev. Pathol. 2014, 9, 27–45. [Google Scholar] [CrossRef] [PubMed]
- Pisanic, T.R.; Cope, L.M.; Lin, S.F.; Yen, T.T.; Athamanolap, P.; Asaka, R.; Nakayama, K.; Fader, A.N.; Wang, T.H.; Shih, I.M.; et al. Methylomic Analysis of Ovarian Cancers Identifies Tumor-Specific Alterations Readily Detectable in Early Precursor Lesions. Clin. Cancer Res. 2018, 24, 6536–6547. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, T.E.; Chindera, K.; McDermott, J.; Breeze, C.E.; Cooke, W.R.; Jones, A.; Reisel, D.; Karegodar, S.T.; Arora, R.; Beck, S.; et al. Epigenetic reprogramming of fallopian tube fimbriae in BRCA mutation carriers defines early ovarian cancer evolution. Nat. Commun. 2016, 7, 11620. [Google Scholar] [CrossRef] [PubMed]
- Ishak, C.A.; de Carvalho, D.D. DNA Methylation Profiling of Premalignant Lesions as a Path to Ovarian Cancer Early Detection. Clin. Cancer Res. 2020, 26, 6083–6085. [Google Scholar] [CrossRef] [PubMed]
- Premarket Approval (PMA) P200006, Food and Drug Administration. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=p200006 (accessed on 23 March 2022).
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V.; Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Smith, D.; Richards, D.; et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759. [Google Scholar] [CrossRef] [PubMed]
- Duffy, M.J.; Diamandis, E.P.; Crown, J. Circulating tumor DNA (ctDNA) as a pan-cancer screening test: Is it finally on the horizon? Clin. Chem. Lab. Med. 2021, 59, 1353–1361. [Google Scholar] [CrossRef] [PubMed]
- Phallen, J.; Sausen, M.; Adleff, V.; Leal, A.; Hruban, C.; White, J.; Anagnostou, V.; Fiksel, J.; Cristiano, S.; Papp, E.; et al. Direct detection of early-stage cancers using circulating tumor DNA. Sci. Transl. Med. 2017, 9, eaan2415. [Google Scholar] [CrossRef]
- Cristiano, S.; Leal, A.; Phallen, J.; Fiksel, J.; Adleff, V.; Bruhm, D.C.; Jensen, S.Ø.; Medina, J.E.; Hruban, C.; White, J.R.; et al. Genome-wide cell-free DNA fragmentation in patients with cancer. Nature 2019, 570, 385–389. [Google Scholar] [CrossRef]
- Fleming, G.F.; Seidman, J.D.; Yemelyanova, A.; Lengyel, E. Epithelial Ovarian Cancer. In Principles and Practice of Gynecologic Oncology; Chi, D.S., Berchuck, A., Dizon, D.S., Yashar, C.M., Eds.; Wolters Kluwer Health: Philadelphia, PA, USA, 2017. [Google Scholar]
- Chen, S.N.; Chang, R.; Lin, L.T.; Chern, C.U.; Tsai, H.W.; Wen, Z.H.; Li, Y.H.; Li, C.J.; Tsui, K.H. MicroRNA in Ovarian Cancer: Biology, Pathogenesis, and Therapeutic Opportunities. Int. J. Environ. Res. Public Health 2019, 16, 1510. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef]
- Hulstaert, E.; Morlion, A.; Levanon, K.; Vandesompele, J.; Mestdagh, P. Candidate RNA biomarkers in biofluids for early diagnosis of ovarian cancer: A systematic review. Gynecol. Oncol. 2021, 160, 633–642. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Ba, Y.; Ma, L.; Cai, X.; Yin, Y.; Wang, K.; Guo, J.; Zhang, Y.; Chen, J.; Guo, X.; et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008, 18, 997–1006. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The MicroRNA Spectrum in 12 Body Fluids. Clin. Chem. 2010, 56, 1733. [Google Scholar] [CrossRef] [PubMed]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Moldovan, L.; Batte, K.E.; Trgovcich, J.; Wisler, J.; Marsh, C.B.; Piper, M. Methodological challenges in utilizing miRNAs as circulating biomarkers. J. Cell. Mol. Med. 2014, 18, 371. [Google Scholar] [CrossRef]
- Cui, M.; Wang, H.; Yao, X.; Zhang, D.; Xie, Y.; Cui, R.; Zhang, X. Circulating MicroRNAs in Cancer: Potential and Challenge. Front. Genet. 2019, 10, 626. [Google Scholar] [CrossRef]
- Tiberio, P.; Callari, M.; Angeloni, V.; Daidone, M.G.; Appierto, V. Challenges in using circulating miRNAs as cancer biomarkers. Biomed Res. Int. 2015, 2015, 731479. [Google Scholar] [CrossRef]
- Cacheux, J.; Bancaud, A.; Leichlé, T.; Cordelier, P. Technological Challenges and Future Issues for the Detection of Circulating MicroRNAs in Patients With Cancer. Front. Chem. 2019, 7, 815. [Google Scholar] [CrossRef]
- Kalra, H.; Drummen, G.P.C.; Mathivanan, S. Focus on Extracellular Vesicles: Introducing the Next Small Big Thing. Int. J. Mol. Sci. 2016, 17, 170. [Google Scholar] [CrossRef]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Théry, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Chang, L.; Ni, J.; Zhu, Y.; Pang, B.; Graham, P.; Zhang, H.; Li, Y. Liquid biopsy in ovarian cancer: Recent advances in circulating extracellular vesicle detection for early diagnosis and monitoring progression. Theranostics 2019, 9, 4130–4140. [Google Scholar] [CrossRef]
- Vaksman, O.; Tropé, C.; Davidson, B.; Reich, R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis 2014, 35, 2113–2120. [Google Scholar] [CrossRef] [PubMed]
- Crow, J.; Atay, S.; Banskota, S.; Artale, B.; Schmitt, S.; Godwin, A.K.; Crow, J.; Atay, S.; Banskota, S.; Artale, B.; et al. Exosomes as mediators of platinum resistance in ovarian cancer. Oncotarget 2017, 8, 11917–11936. [Google Scholar] [CrossRef]
- Li, J.; Sherman-Baust, C.A.; Tsai-Turton, M.; Bristow, R.E.; Roden, R.B.; Morin, P.J. Claudin-containing exosomes in the peripheral circulation of women with ovarian cancer. BMC Cancer 2009, 9, 244. [Google Scholar] [CrossRef]
- Liang, B.; Peng, P.; Chen, S.; Li, L.; Zhang, M.; Cao, D.; Yang, J.; Li, H.; Gui, T.; Li, X.; et al. Characterization and proteomic analysis of ovarian cancer-derived exosomes. J. Proteom. 2013, 80, 171–182. [Google Scholar] [CrossRef]
- Runz, S.; Keller, S.; Rupp, C.; Stoeck, A.; Issa, Y.; Koensgen, D.; Mustea, A.; Sehouli, J.; Kristiansen, G.; Altevogt, P. Malignant ascites-derived exosomes of ovarian carcinoma patients contain CD24 and EpCAM. Gynecol. Oncol. 2007, 107, 563–571. [Google Scholar] [CrossRef]
- Tang, M.K.S.; Yue, P.Y.K.; Ip, P.P.; Huang, R.L.; Lai, H.C.; Cheung, A.N.Y.; Tse, K.Y.; Ngan, H.Y.S.; Wong, A.S.T. Soluble E-cadherin promotes tumor angiogenesis and localizes to exosome surface. Nat. Commun. 2018, 9, 2270. [Google Scholar] [CrossRef]
- Zhao, Z.; Yang, Y.; Zeng, Y.; He, M. A microfluidic ExoSearch chip for multiplexed exosome detection towards blood-based ovarian cancer diagnosis. Lab Chip 2016, 16, 489–496. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.I.; Kang, N.; Leem, S.; Yang, J.; Jo, H.; Lee, M.; Kim, H.S.; Dhanasekaran, D.N.; Kim, Y.K.; Park, T.; et al. Metagenomic Analysis of Serum Microbe-Derived Extracellular Vesicles and Diagnostic Models to Differentiate Ovarian Cancer and Benign Ovarian Tumor. Cancers 2020, 12, 1309. [Google Scholar] [CrossRef] [PubMed]
- Dudas, S.P.; Chatterjee, M.; Tainsky, M.A. Usage of cancer associated autoantibodies in the detection of disease. Cancer Biomark. 2010, 6, 257–270. [Google Scholar] [CrossRef] [PubMed]
- Wilson, A.L.; Moffitt, L.R.; Duffield, N.; Rainczuk, A.; Jobling, T.W.; Plebanski, M.; Stephens, A.N. Autoantibodies against HSF1 and CCDC155 as Biomarkers of Early-Stage, High-Grade Serous Ovarian Cancer. Cancer Epidemiol. Prev. Biomark. 2018, 27, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Blagosklonny, M.V. p53 from complexity to simplicity: Mutant p53 stabilization, gain-of-function, and dominant-negative effect. FASEB J. 2000, 14, 1901–1907. [Google Scholar] [CrossRef]
- Yang, W.L.; Gentry-Maharaj, A.; Simmons, A.; Ryan, A.; Fourkala, E.O.; Lu, Z.; Baggerly, K.A.; Zhao, Y.; Lu, K.H.; Bowtell, D.; et al. Elevation of TP53 Autoantibody Before CA125 in Preclinical Invasive Epithelial Ovarian Cancer. Clin. Cancer Res. 2017, 23, 5912–5922. [Google Scholar] [CrossRef]
- Yoneyama, K.; Kojima, S.; Kodani, Y.; Yamaguchi, N.; Igarashi, A.; Kurose, K.; Kawase, R.; Takeshita, T.; Hattori, S.; Nagata, K. Proteomic Identification of Autoantibodies in Sera from Patients with Ovarian Cancer as Possible Diagnostic Biomarkers. Anticancer Res. 2015, 35, 881–889. [Google Scholar]
- Karabudak, A.A.; Hafner, J.; Shetty, V.; Chen, S.; Secord, A.A.; Morse, M.A.; Philip, R. Autoantibody biomarkers identified by proteomics methods distinguish ovarian cancer from non-ovarian cancer with various CA-125 levels. J. Cancer Res. Clin. Oncol. 2013, 139, 1757–1770. [Google Scholar] [CrossRef]
- Naora, H.; Montz, F.J.; Chai, C.Y.; Roden, R.B.S. Aberrant expression of homeobox gene HOXA7 is associated with müllerian-like differentiation of epithelial ovarian tumors and the generation of a specific autologous antibody response. Proc. Natl. Acad. Sci. USA 2001, 98, 15209–15214. [Google Scholar] [CrossRef]
- Katchman, B.A.; Chowell, D.; Wallstrom, G.; Vitonis, A.F.; LaBaer, J.; Cramer, D.W.; Anderson, K.S. Autoantibody biomarkers for the detection of serous ovarian cancer. Gynecol. Oncol. 2017, 146, 129–136. [Google Scholar] [CrossRef]
- Kim, J.-H.; Herlyn, D.; Wong, K.-K.; Park, D.-C.; Schorge, J.O.; Lu, K.H.; Skates, S.J.; Cramer, D.W.; Berkowitz, R.S.; Mok, S.C. Identification of Epithelial Cell Adhesion Molecule Autoantibody in Patients with Ovarian Cancer. Clin. Cancer Res. 2003, 9, 4782–4791. [Google Scholar] [PubMed]
- Lokshin, A.E.; Winans, M.; Landsittel, D.; Marrangoni, A.M.; Velikokhatnaya, L.; Modugno, F.; Nolen, B.M.; Gorelik, E. Circulating IL-8 and anti-IL-8 autoantibody in patients with ovarian cancer. Gynecol. Oncol. 2006, 102, 244–251. [Google Scholar] [CrossRef] [PubMed]
- Kovács, P.; Csonka, T.; Kovács, T.; Sári, Z.; Ujlaki, G.; Sipos, A.; Karányi, Z.; Szeőcs, D.; Hegedűs, C.; Uray, K.; et al. Lithocholic Acid, a Metabolite of the Microbiome, Increases Oxidative Stress in Breast Cancer. Cancers 2019, 11, 1255. [Google Scholar] [CrossRef] [PubMed]
- Kovács, T.; Mikó, E.; Vida, A.; Sebő, É.; Toth, J.; Csonka, T.; Boratkó, A.; Ujlaki, G.; Lente, G.; Kovács, P.; et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef] [PubMed]
- Mikó, E.; Vida, A.; Kovács, T.; Ujlaki, G.; Trencsényi, G.; Márton, J.; Sári, Z.; Kovács, P.; Boratkó, A.; Hujber, Z.; et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef]
- Ingman, W.V. The Gut Microbiome: A New Player in Breast Cancer Metastasis. Cancer Res. 2019, 79, 3539–3541. [Google Scholar] [CrossRef]
- Kiss, B.; Mikó, E.; Sebő, É.; Toth, J.; Ujlaki, G.; Szabó, J.; Uray, K.; Bai, P.; Árkosy, P. Oncobiosis and Microbial Metabolite Signaling in Pancreatic Adenocarcinoma. Cancers 2020, 12, 1068. [Google Scholar] [CrossRef]
- Zhou, Z.; Zeng, F.; Yuan, J.; Tang, J.; Colditz, G.A.; Tworoger, S.S.; Trabert, B.; Su, X. Pelvic inflammatory disease and the risk of ovarian cancer: A meta-analysis. Cancer Causes Control 2017, 28, 415–428. [Google Scholar] [CrossRef]
- Sipos, A.; Ujlaki, G.; Mikó, E.; Maka, E.; Szabó, J.; Uray, K.; Krasznai, Z.; Bai, P. The role of the microbiome in ovarian cancer: Mechanistic insights into oncobiosis and to bacterial metabolite signaling. Mol. Med. 2021, 27, 33. [Google Scholar] [CrossRef]
- Zhou, B.; Sun, C.; Huang, J.; Xia, M.; Guo, E.; Li, N.; Lu, H.; Shan, W.; Wu, Y.; Li, Y.; et al. The biodiversity Composition of Microbiome in Ovarian Carcinoma Patients. Sci. Rep. 2019, 9, 1691. [Google Scholar] [CrossRef]
- Poore, G.D.; Kopylova, E.; Zhu, Q.; Carpenter, C.; Fraraccio, S.; Wandro, S.; Kosciolek, T.; Janssen, S.; Metcalf, J.; Song, S.J.; et al. Microbiome analyses of blood and tissues suggest cancer diagnostic approach. Nature 2020, 579, 567–574. [Google Scholar] [CrossRef] [PubMed]
- Miao, R.; Badger, T.C.; Groesch, K.; Diaz-Sylvester, P.L.; Wilson, T.; Ghareeb, A.; Martin, J.A.; Cregger, M.; Welge, M.; Bushell, C.; et al. Assessment of peritoneal microbial features and tumor marker levels as potential diagnostic tools for ovarian cancer. PLoS ONE 2020, 15, e0227707. [Google Scholar] [CrossRef] [PubMed]
- Turkoglu, O.; Zeb, A.; Graham, S.; Szyperski, T.; Szender, J.B.; Odunsi, K.; Bahado-Singh, R. Metabolomics of biomarker discovery in ovarian cancer: A systematic review of the current literature. Metabolomics 2016, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Odunsi, K.; Wollman, R.M.; Ambrosone, C.B.; Hutson, A.; McCann, S.E.; Tammela, J.; Geisler, J.P.; Miller, G.; Sellers, T.; Cliby, W.; et al. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int. J. Cancer 2005, 113, 782–788. [Google Scholar] [CrossRef]
- Guan, W.; Zhou, M.; Hampton, C.Y.; Benigno, B.B.; Walker, L.D.E.; Gray, A.; McDonald, J.F.; Fernández, F.M. Ovarian cancer detection from metabolomic liquid chromatography/mass spectrometry data by support vector machines. BMC Bioinform. 2009, 10, 259. [Google Scholar] [CrossRef]
- Zhou, M.; Guan, W.; Walker, L.D.E.; Mezencev, R.; Benigno, B.B.; Gray, A.; Fernández, F.M.; McDonald, J.F. Rapid mass spectrometric metabolic profiling of blood sera detects ovarian cancer with high accuracy. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2262–2271. [Google Scholar] [CrossRef]
- Hilvo, M.; De Santiago, I.; Gopalacharyulu, P.; Schmitt, W.D.; Budczies, J.; Kuhberg, M.; Dietel, M.; Aittokallio, T.; Markowetz, F.; Denkert, C.; et al. Accumulated Metabolites of Hydroxybutyric Acid Serve as Diagnostic and Prognostic Biomarkers of Ovarian High-Grade Serous Carcinomas. Cancer Res. 2016, 76, 796–804. [Google Scholar] [CrossRef]
- Slupsky, C.M.; Steed, H.; Wells, T.H.; Dabbs, K.; Schepansky, A.; Capstick, V.; Faught, W.; Sawyer, M.B. Urine metabolite analysis offers potential early diagnosis of ovarian and breast cancers. Clin. Cancer Res. 2010, 16, 5835–5841. [Google Scholar] [CrossRef]
- Ke, C.; Hou, Y.; Zhang, H.; Fan, L.; Ge, T.; Guo, B.; Zhang, F.; Yang, K.; Wang, J.; Lou, G.; et al. Large-scale profiling of metabolic dysregulation in ovarian cancer. Int. J. Cancer 2015, 136, 516–526. [Google Scholar] [CrossRef]
- Buas, M.F.; Gu, H.; Djukovic, D.; Zhu, J.; Drescher, C.W.; Urban, N.; Raftery, D.; Li, C.I. Identification of novel candidate plasma metabolite biomarkers for distinguishing serous ovarian carcinoma and benign serous ovarian tumors. Gynecol. Oncol. 2016, 140, 138–144. [Google Scholar] [CrossRef]
- Raoof, S.; Lee, R.J.; Jajoo, K.; Mancias, J.D.; Rebbeck, T.R.; Skates, S.J. Multicancer Early Detection Technologies: A Review Informed by Past Cancer Screening Studies. Cancer Epidemiol. Biomarkers Prev. 2022, 31, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.J.; Cunningham, A.P.; Kuchenbaecker, K.B.; Mavaddat, N.; Easton, D.F.; Antoniou, A.C. BOADICEA breast cancer risk prediction model: Updates to cancer incidences, tumour pathology and web interface. Br. J. Cancer 2013, 110, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Antoniou, A.C.; Hardy, R.; Walker, L.; Evans, D.G.; Shenton, A.; Eeles, R.; Shanley, S.; Pichert, G.; Izatt, L.; Rose, S.; et al. Predicting the likelihood of carrying a BRCA1 or BRCA2 mutation: Validation of BOADICEA, BRCAPRO, IBIS, Myriad and the Manchester scoring system using data from UK genetics clinics. J. Med. Genet. 2008, 45, 425–431. [Google Scholar] [CrossRef] [PubMed]
- Lakeman, I.M.M.; Rodríguez-Girondo, M.; Lee, A.; Ruiter, R.; Stricker, B.H.; Wijnant, S.R.A.; Kavousi, M.; Antoniou, A.C.; Schmidt, M.K.; Uitterlinden, A.G.; et al. Validation of the BOADICEA model and a 313-variant polygenic risk score for breast cancer risk prediction in a Dutch prospective cohort. Genet. Med. 2020, 22, 1803–1811. [Google Scholar] [CrossRef]
- National Institute for Health and Care Excellence (NICE). Familial Breast Cancer: Classification, Care and Managing Breast Cancer and Related Risks in People with a Family History of Breast Cancer Clinical Guideline (CG164); NICE: London, UK, 2013. [Google Scholar]
- Blewett, L.A.; Rivera Drew, J.A.; Griffin, R.; King, M.L.; Williams, K.C.W. IPUMS Health Surveys: National Health Interview Survey, Version 6.2; Regents of the University of Minnesota: Twin Cities, MN, USA, 2017. [Google Scholar] [CrossRef]
- Hart, G.R.; Nartowt, B.J.; Muhammad, W.; Liang, Y.; Huang, G.S.; Deng, J. Stratifying Ovarian Cancer Risk Using Personal Health Data. Front. Big Data 2019, 2, 24. [Google Scholar] [CrossRef]
- Stratton, J.F.; Pharoah, P.; Smith, S.K.; Easton, D.; Ponder, B.A.J. A systematic review and meta-analysis of family history and risk of ovarian cancer. BJOG 1998, 105, 493–499. [Google Scholar] [CrossRef]
- Moslehi, R.; Chu, W.; Karlan, B.; Fishman, D.; Risch, H.; Fields, A.; Smotkin, D.; Ben-David, Y.; Rosenblatt, J.; Russo, D.; et al. BRCA1 and BRCA2 Mutation Analysis of 208 Ashkenazi Jewish Women with Ovarian Cancer. Am. J. Hum. Genet. 2000, 66, 1259–1272. [Google Scholar] [CrossRef]
- Premarket Approval (PMA) P190014, Food and Drug Administration. Available online: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P190014 (accessed on 27 April 2022).
- Alsop, K.; Fereday, S.; Meldrum, C.; DeFazio, A.; Emmanuel, C.; George, J.; Dobrovic, A.; Birrer, M.J.; Webb, P.M.; Stewart, C.; et al. BRCA mutation frequency and patterns of treatment response in BRCA mutation-positive women with ovarian cancer: A report from the Australian ovarian cancer study group. J. Clin. Oncol. 2012, 30, 2654–2663. [Google Scholar] [CrossRef]
- Webb, P.M.; Jordan, S.J. Epidemiology of epithelial ovarian cancer. Best Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 3–14. [Google Scholar] [CrossRef]
- Bitler, B.G.; Watson, Z.L.; Wheeler, L.J.; Behbakht, K. PARP inhibitors: Clinical utility and possibilities of overcoming resistance. Gynecol. Oncol. 2017, 147, 695–704. [Google Scholar] [CrossRef]
- Duffy, D.M.; Ko, C.; Jo, M.; Brannstrom, M.; Curry, T.E. Ovulation: Parallels With Inflammatory Processes. Endocr. Rev. 2019, 40, 369–416. [Google Scholar] [CrossRef] [PubMed]
- Gates, M.A.; Rosner, B.A.; Hecht, J.L.; Tworoger, S.S. Risk Factors for Epithelial Ovarian Cancer by Histologic Subtype. Am. J. Epidemiol. 2010, 171, 45–53. [Google Scholar] [CrossRef] [PubMed]
- Risch, H.A.; Marrett, L.D.; Jain, M.; Howe, G.R. Differences in Risk Factors for Epithelial Ovarian Cancer by Histologic TypeResults of a Case-Control Study. Am. J. Epidemiol. 1996, 144, 363–372. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, J.R.; Risch, H.A.; Lubinski, J.; Moller, P.; Ghadirian, P.; Lynch, H.; Karlan, B.; Fishman, D.; Rosen, B.; Neuhausen, S.L.; et al. Reproductive risk factors for ovarian cancer in carriers of BRCA1 or BRCA2 mutations: A case-control study. Lancet Oncol. 2007, 8, 26–34. [Google Scholar] [CrossRef]
- Havrilesky, L.J.; Moorman, P.G.; Lowery, W.J.; Gierisch, J.M.; Coeytaux, R.R.; Urrutia, R.P.; Dinan, M.; McBroom, A.J.; Hasselblad, V.; Sanders, G.D.; et al. Oral contraceptive pills as primary prevention for ovarian cancer: A systematic review and meta-analysis. Obstet. Gynecol. 2013, 122, 139–147. [Google Scholar] [CrossRef]
- Huang, X.; Wang, X.; Shang, J.; Lin, Y.; Yang, Y.; Song, Y.; Yu, S. Association between dietary fiber intake and risk of ovarian cancer: A meta-analysis of observational studies. J. Int. Med. Res. 2018, 46, 3995–4005. [Google Scholar] [CrossRef]
- Zheng, B.; Shen, H.; Han, H.; Han, T.; Qin, Y. Dietary fiber intake and reduced risk of ovarian cancer: A meta-analysis. Nutr. J. 2018, 17, 99. [Google Scholar] [CrossRef]
- Guo, H.; Guo, J.; Xie, W.; Yuan, L.; Sheng, X. The role of vitamin D in ovarian cancer: Epidemiology, molecular mechanism and prevention. J. Ovarian Res. 2018, 11, 71. [Google Scholar] [CrossRef]
- Zhang, M.; Yang, Z.Y.; Binns, C.W.; Lee, A.H. Diet and ovarian cancer risk: A case–control study in China. Br. J. Cancer 2002, 86, 712–717. [Google Scholar] [CrossRef]
- Nagle, C.M.; Dixon, S.C.; Jensen, A.; Kjaer, S.K.; Modugno, F.; DeFazio, A.; Fereday, S.; Hung, J.; Johnatty, S.E.; Fasching, P.A.; et al. Obesity and survival among women with ovarian cancer: Results from the Ovarian Cancer Association Consortium. Br. J. Cancer 2015, 113, 817–826. [Google Scholar] [CrossRef]
- Olsen, C.M.; Green, A.C.; Whiteman, D.C.; Sadeghi, S.; Kolahdooz, F.; Webb, P.M. Obesity and the risk of epithelial ovarian cancer: A systematic review and meta-analysis. Eur. J. Cancer 2007, 43, 690–709. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.Y.; Jeon, I.P.; Kim, J.W.; Song, Y.S.; Yoon, J.M.; Park, S.M. Diabetes mellitus and ovarian cancer risk: A systematic review and meta-analysis of observational studies. Int. J. Gynecol. Cancer 2013, 23, 402–412. [Google Scholar] [CrossRef]
- Akhavan, S.; Ghahghaei-Nezamabadi, A.; Modaresgilani, M.; Mousavi, A.S.; Sepidarkish, M.; Tehranian, A.; Rezayof, E. Impact of diabetes mellitus on epithelial ovarian cancer survival. BMC Cancer 2018, 18, 1246. [Google Scholar] [CrossRef] [PubMed]
- Hansen, J.M.; Nagle, C.M.; Ibiebele, T.I.; Grant, P.T.; Obermair, A.; Friedlander, M.L.; DeFazio, A.; Webb, P.M. A healthy lifestyle and survival among women with ovarian cancer. Int. J. Cancer 2020, 147, 3361–3369. [Google Scholar] [CrossRef] [PubMed]
- Bandera, E.V.; Lee, V.S.; Qin, B.; Rodriguez-Rodriguez, L.; Powell, C.B.; Kushi, L.H. Impact of body mass index on ovarian cancer survival varies by stage. Br. J. Cancer 2017, 117, 282–289. [Google Scholar] [CrossRef]
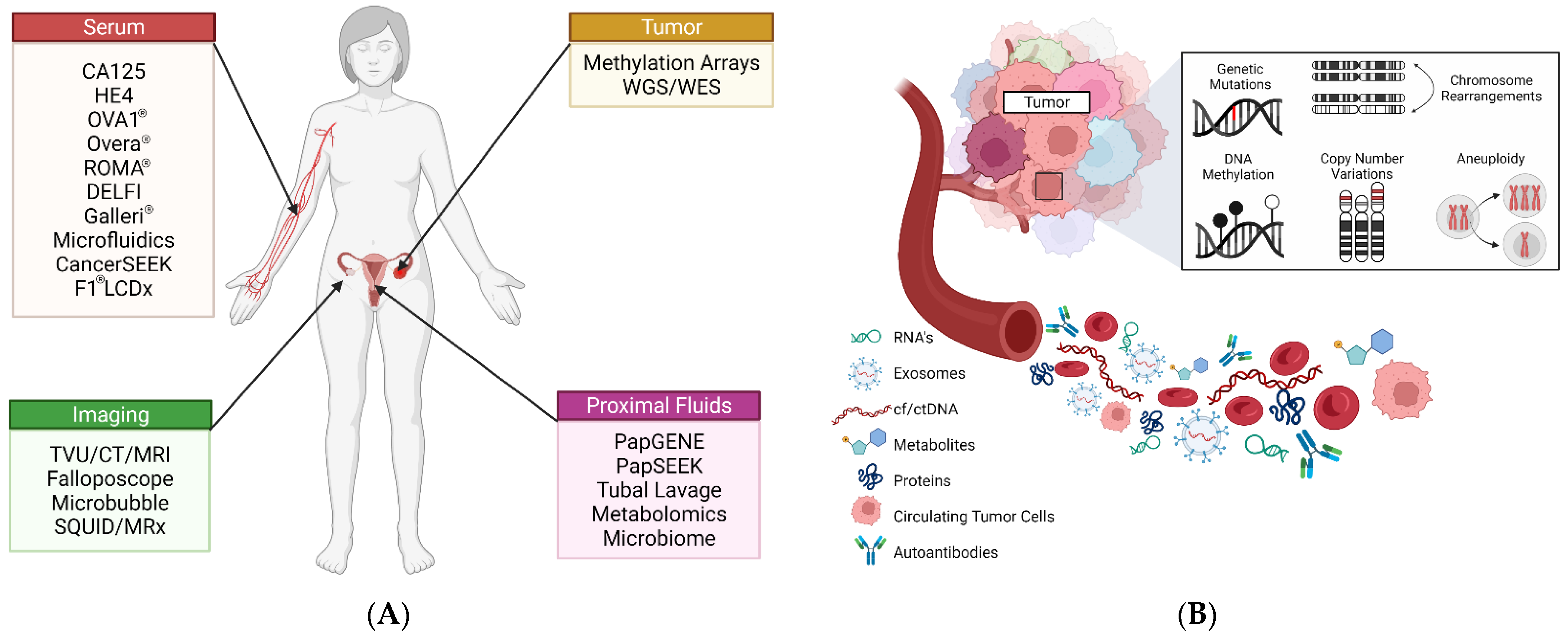
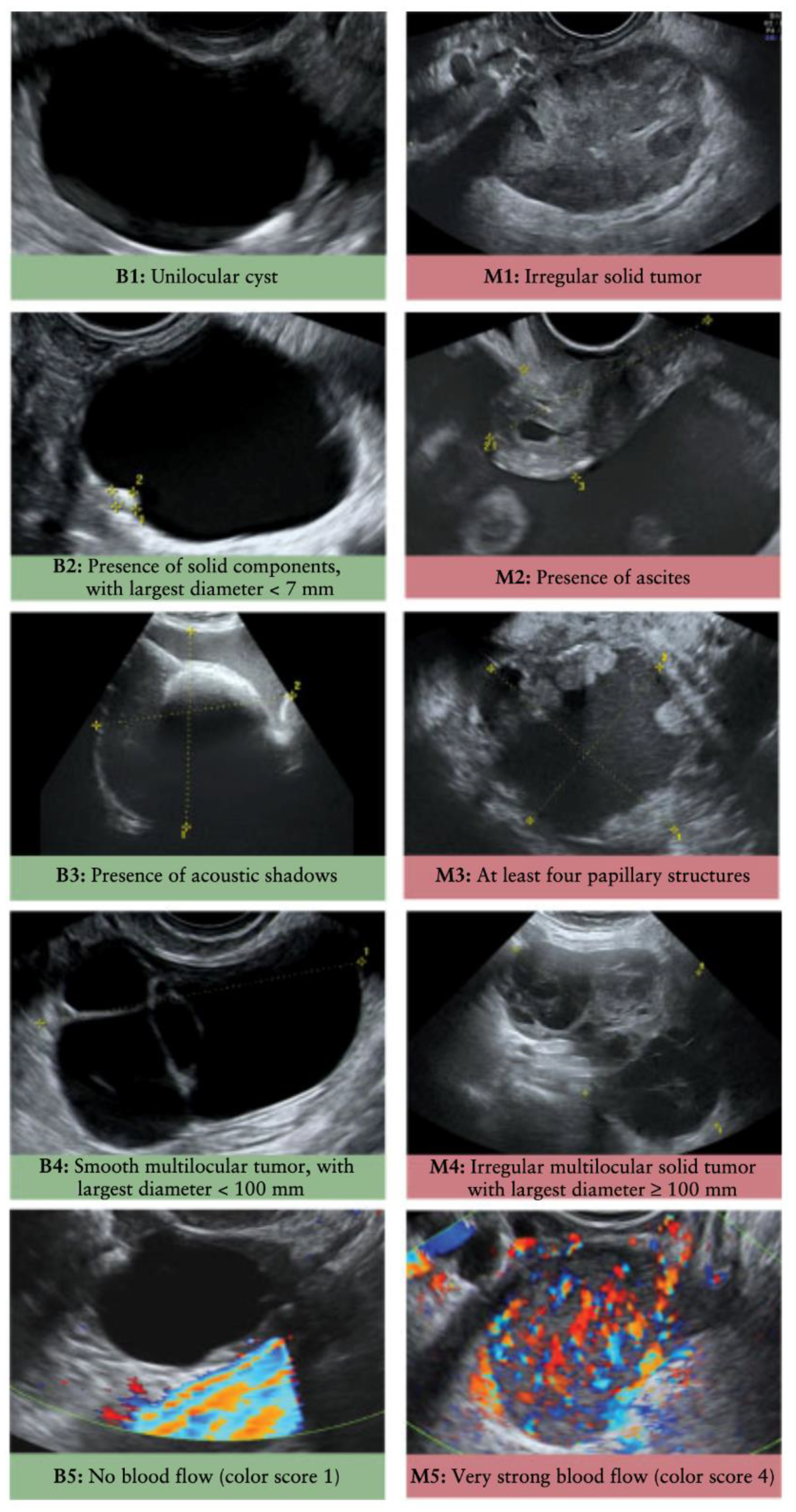
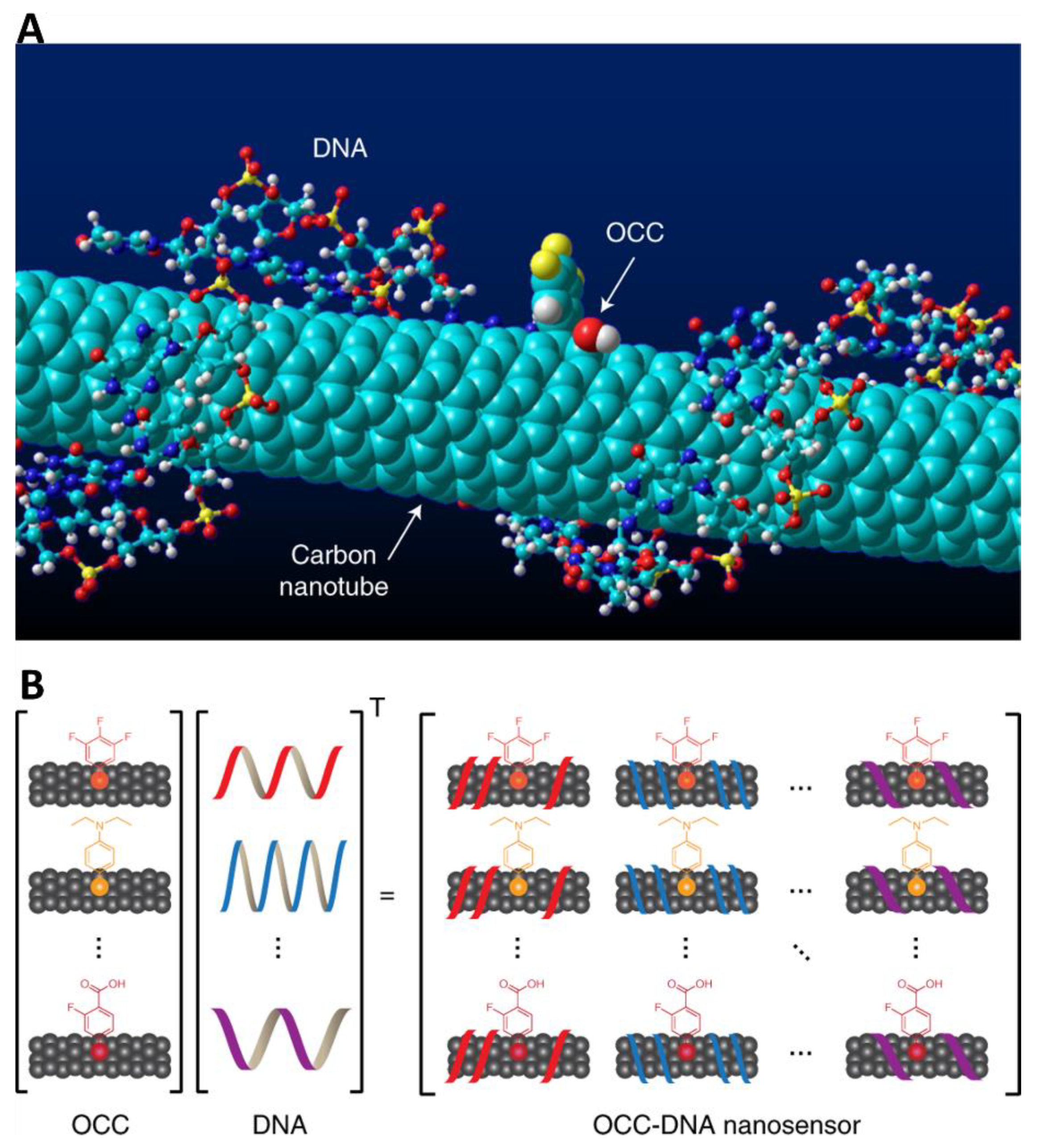
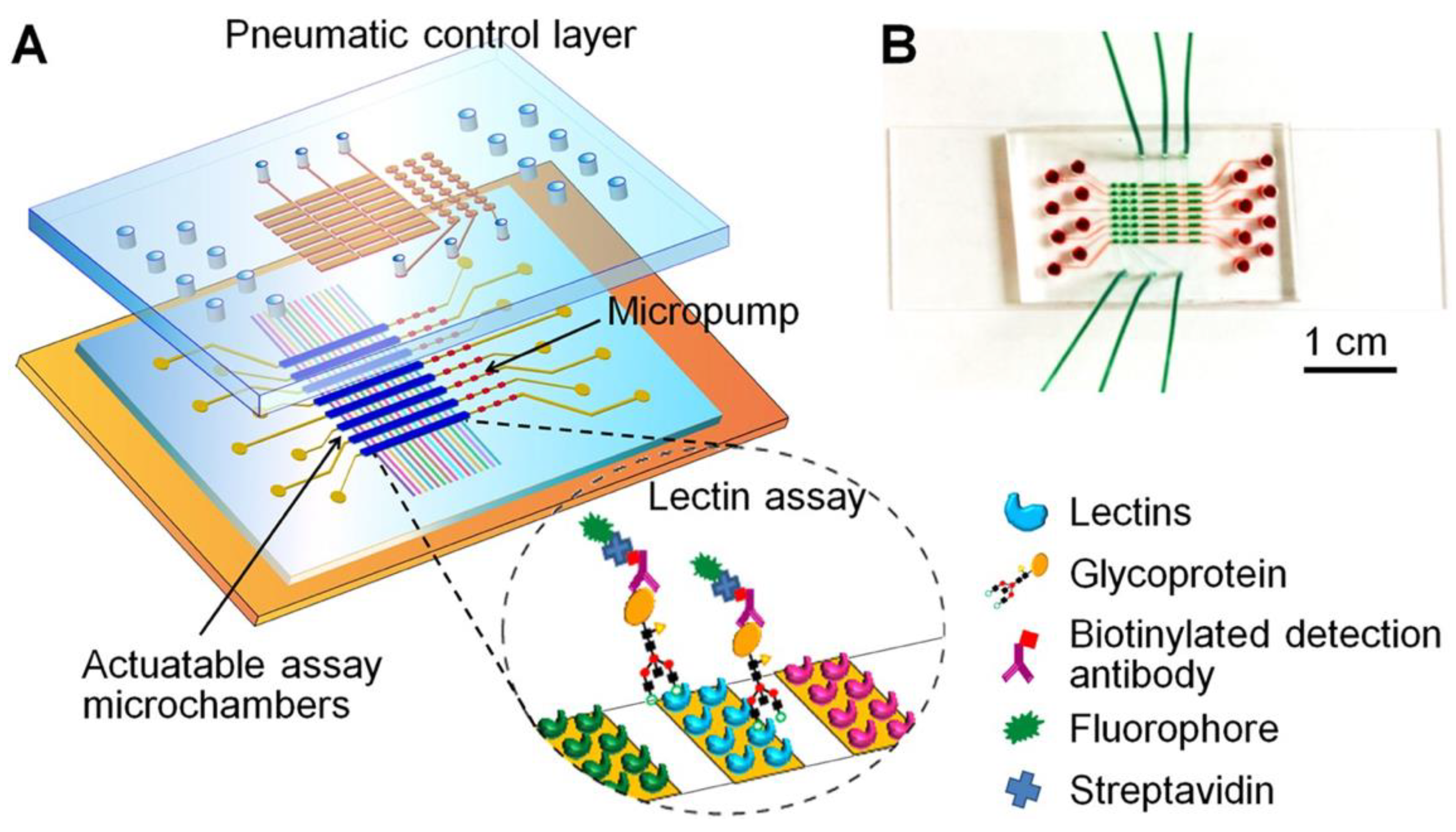

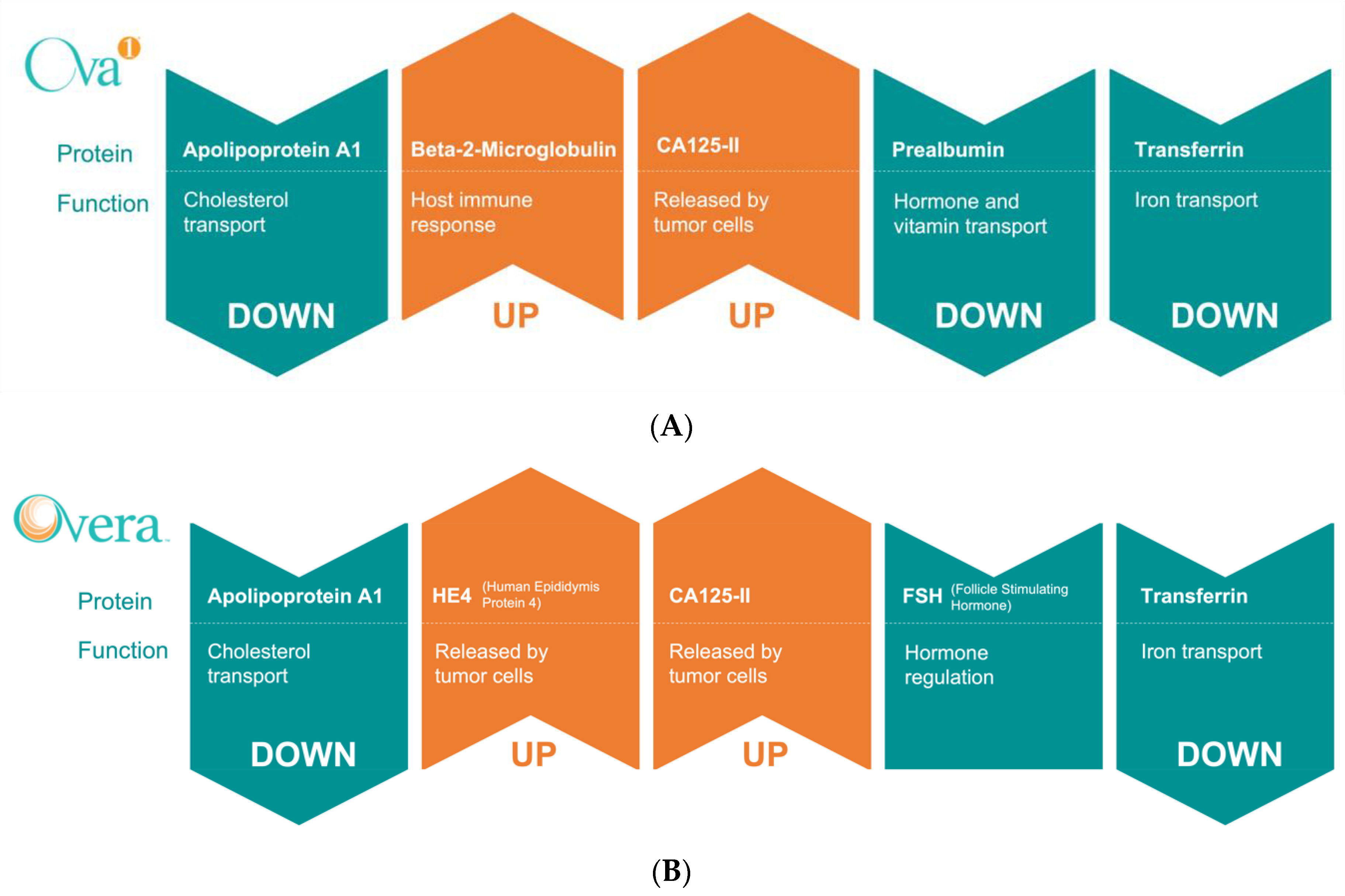
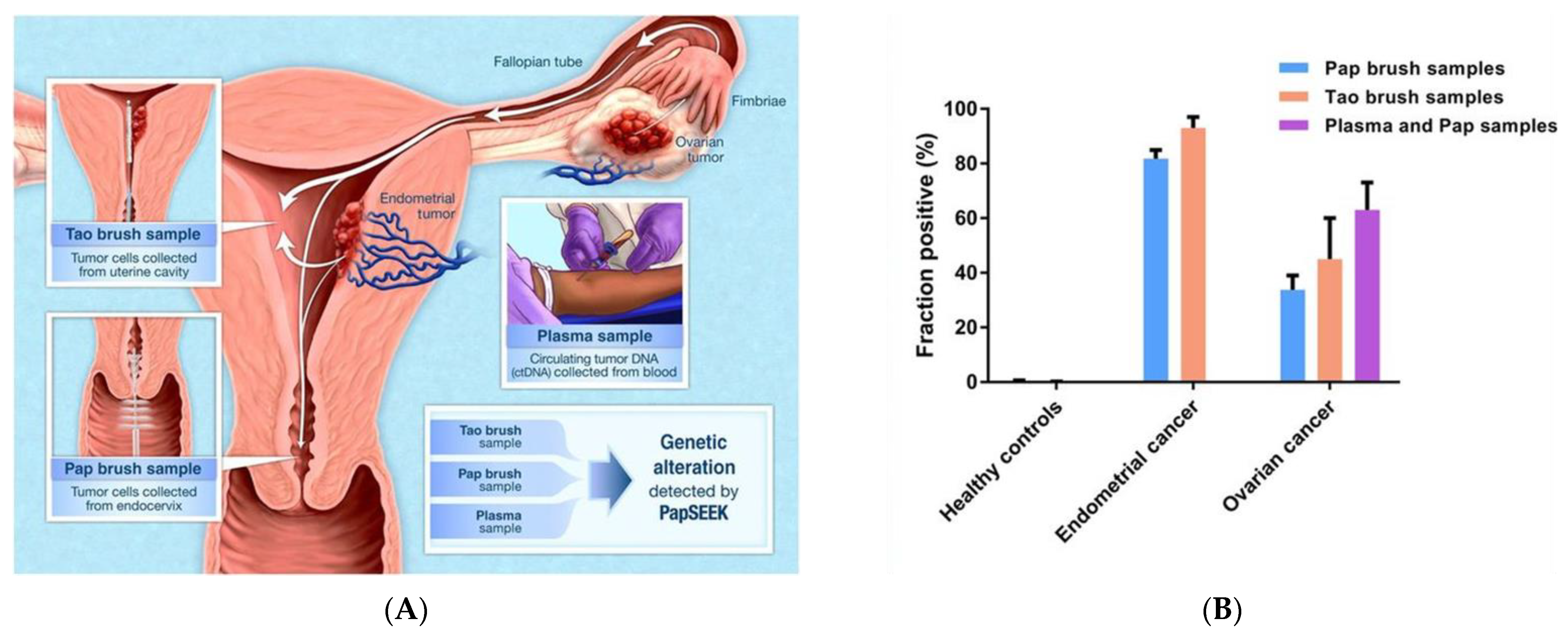
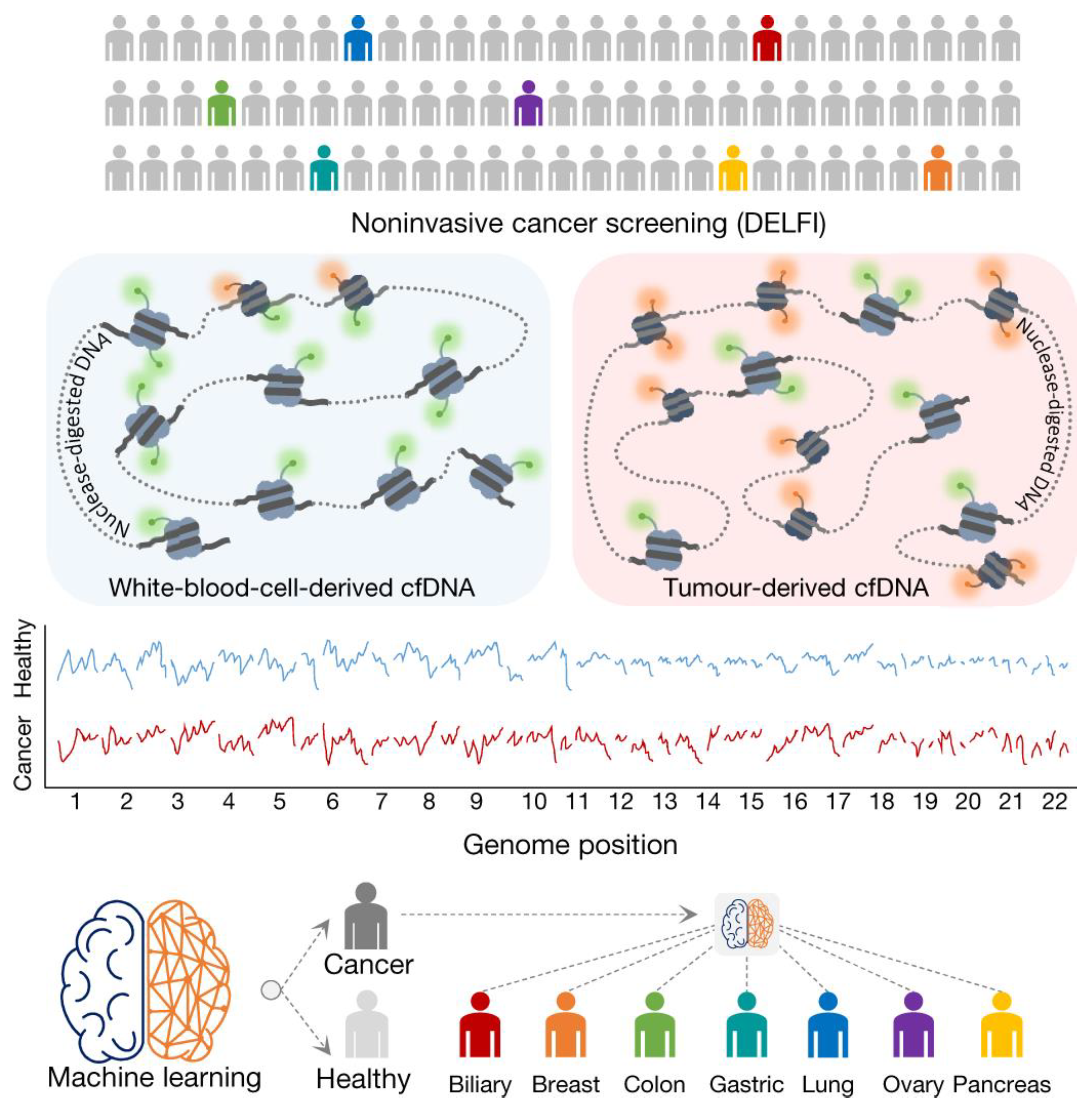
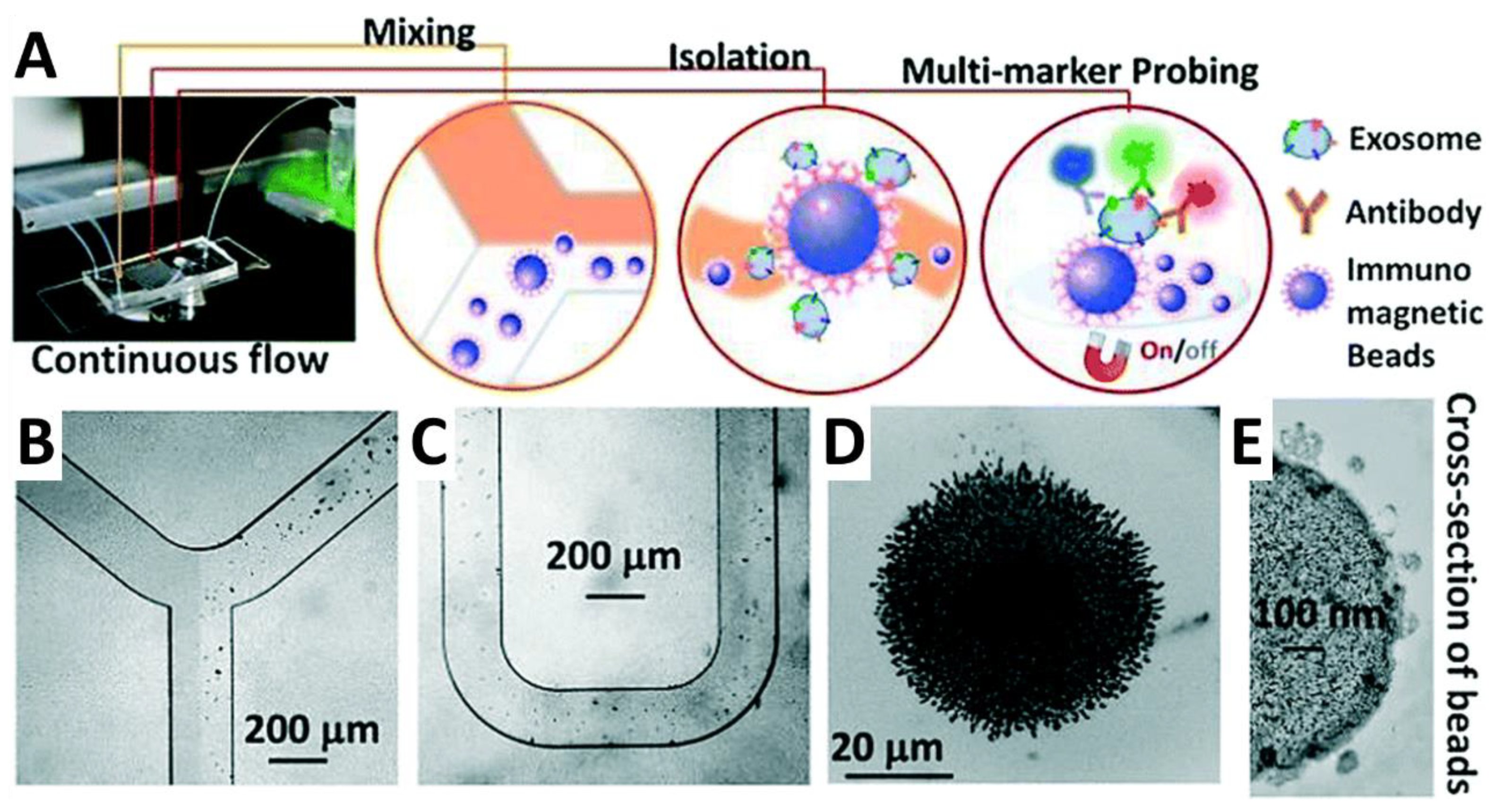
| Test Name | Marker(s) | Modality | Potential Clinical Application(s) | Average Sens./Spec. (%) | Reference(s) |
|---|---|---|---|---|---|
| N/A | CA125 | Protein | Preoperative diagnostic, Prognostic (FDA cleared) | 83.7/86.0 | [66,67,68] |
| ROMA® | CA125, HE4, and Menopause Status | Protein | Preoperative diagnostic, Prognostic (FDA cleared) | 85.3/80.9 | [68,69,70,71,72,73,74,75,76,77,78] |
| CPH-I | CA125, HE4, and Age | Protein | Preoperative diagnostic, Prognostic | 82.2/78.9 | [68,70,71,73,76,77,79] |
| OVA1® | CA125, ApoA-1, TTR, TF, and B2M | Protein | Preoperative diagnostic, Prognostic (FDA cleared) | 87.7/52.6 | [80,81,82,83,84,85] |
| Overa® | CA125, ApoA-1, TF, HE4, and FSH | Protein | Preoperative diagnostic, Prognostic (FDA cleared) | 91.1/67.6 | [86,87] |
| N/A | CA125, CA 15-3, CA72-4, and MCSF | Protein | Diagnostic | 69.5/98.0 | [88,89] |
| PapSEEK | ctDNA mutations | Genetic | Diagnostic, Prognostic | 63.0/99.9 | [90] |
| N/A | OPCML hypermethylation | Epigenetic | Screening, Diagnostic, and Prognostic | 90.1/91.8 | [91,92] |
| N/A | ZNF154 hypermethylation | Epigenetic | Diagnostic | 67.6/93.5 | [93,94,95] |
| CancerSEEK | ctDNA mutations and glycoproteins | Pan-cancer | Screening, Diagnostic, and Prognostic | 96.0/99.0 | [96] |
| N/A | miR-200(a/b/c) + miR-320 + miR-141, among others | MicroRNA | Diagnostic, Prognostic | 85.3/96.0 | [97,98,99,100,101] |
| N/A | VLDL, LDL, lysoPC, valine, alanine, and ceramides | Metabolite | Prognostic | 78.9/93.2 | [102,103,104,105] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liberto, J.M.; Chen, S.-Y.; Shih, I.-M.; Wang, T.-H.; Wang, T.-L.; Pisanic, T.R., II. Current and Emerging Methods for Ovarian Cancer Screening and Diagnostics: A Comprehensive Review. Cancers 2022, 14, 2885. https://doi.org/10.3390/cancers14122885
Liberto JM, Chen S-Y, Shih I-M, Wang T-H, Wang T-L, Pisanic TR II. Current and Emerging Methods for Ovarian Cancer Screening and Diagnostics: A Comprehensive Review. Cancers. 2022; 14(12):2885. https://doi.org/10.3390/cancers14122885
Chicago/Turabian StyleLiberto, Juliane M., Sheng-Yin Chen, Ie-Ming Shih, Tza-Huei Wang, Tian-Li Wang, and Thomas R. Pisanic, II. 2022. "Current and Emerging Methods for Ovarian Cancer Screening and Diagnostics: A Comprehensive Review" Cancers 14, no. 12: 2885. https://doi.org/10.3390/cancers14122885
APA StyleLiberto, J. M., Chen, S.-Y., Shih, I.-M., Wang, T.-H., Wang, T.-L., & Pisanic, T. R., II. (2022). Current and Emerging Methods for Ovarian Cancer Screening and Diagnostics: A Comprehensive Review. Cancers, 14(12), 2885. https://doi.org/10.3390/cancers14122885








