The Role of Indoleamine 2, 3-Dioxygenase 1 in Regulating Tumor Microenvironment
Abstract
Simple Summary
Abstract
1. Introduction
2. Biology and Essential Role of IDO1
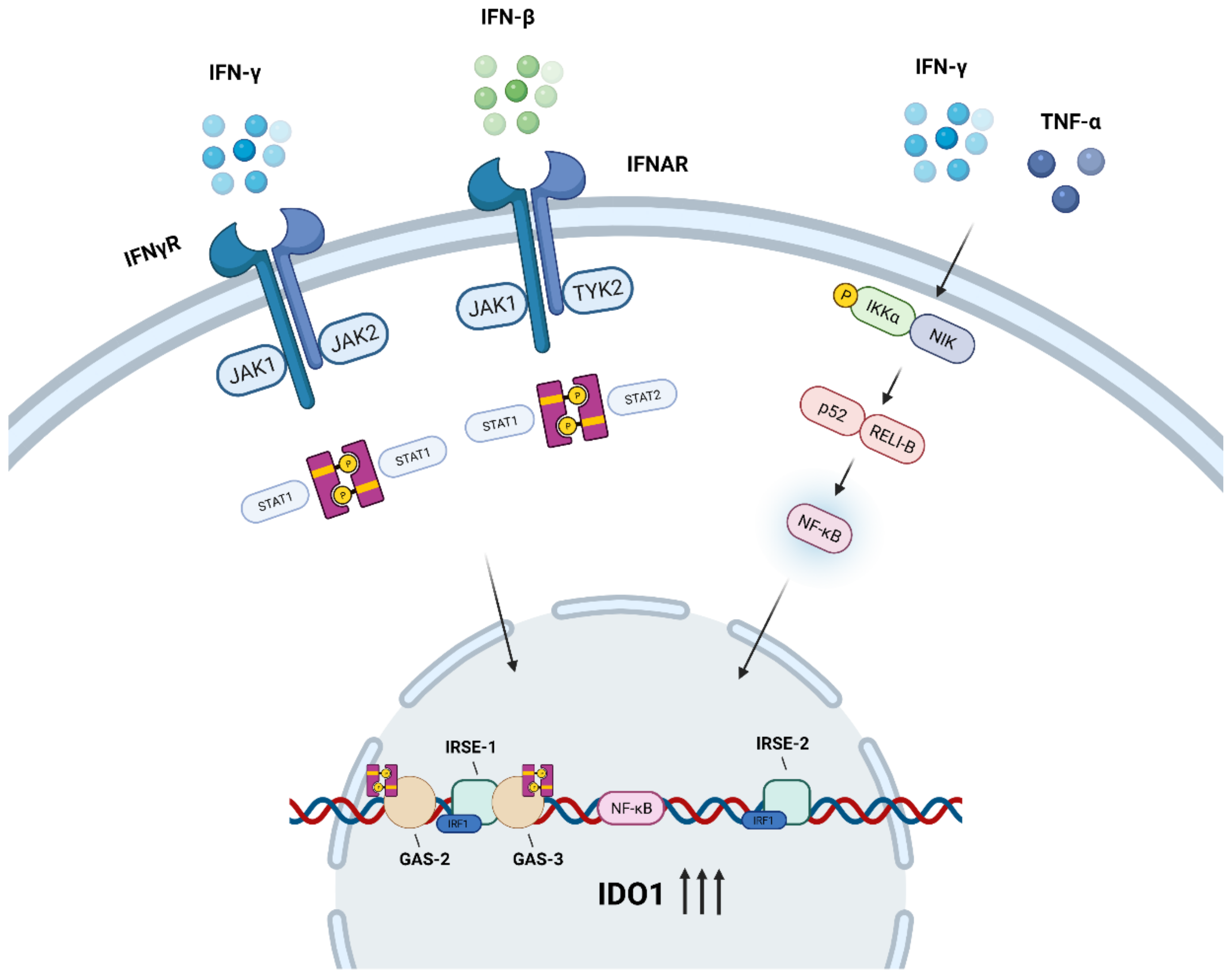
3. Regulation of IDO1
3.1. JAK/STAT Signaling Pathway
3.2. NF-κB Signaling Pathway
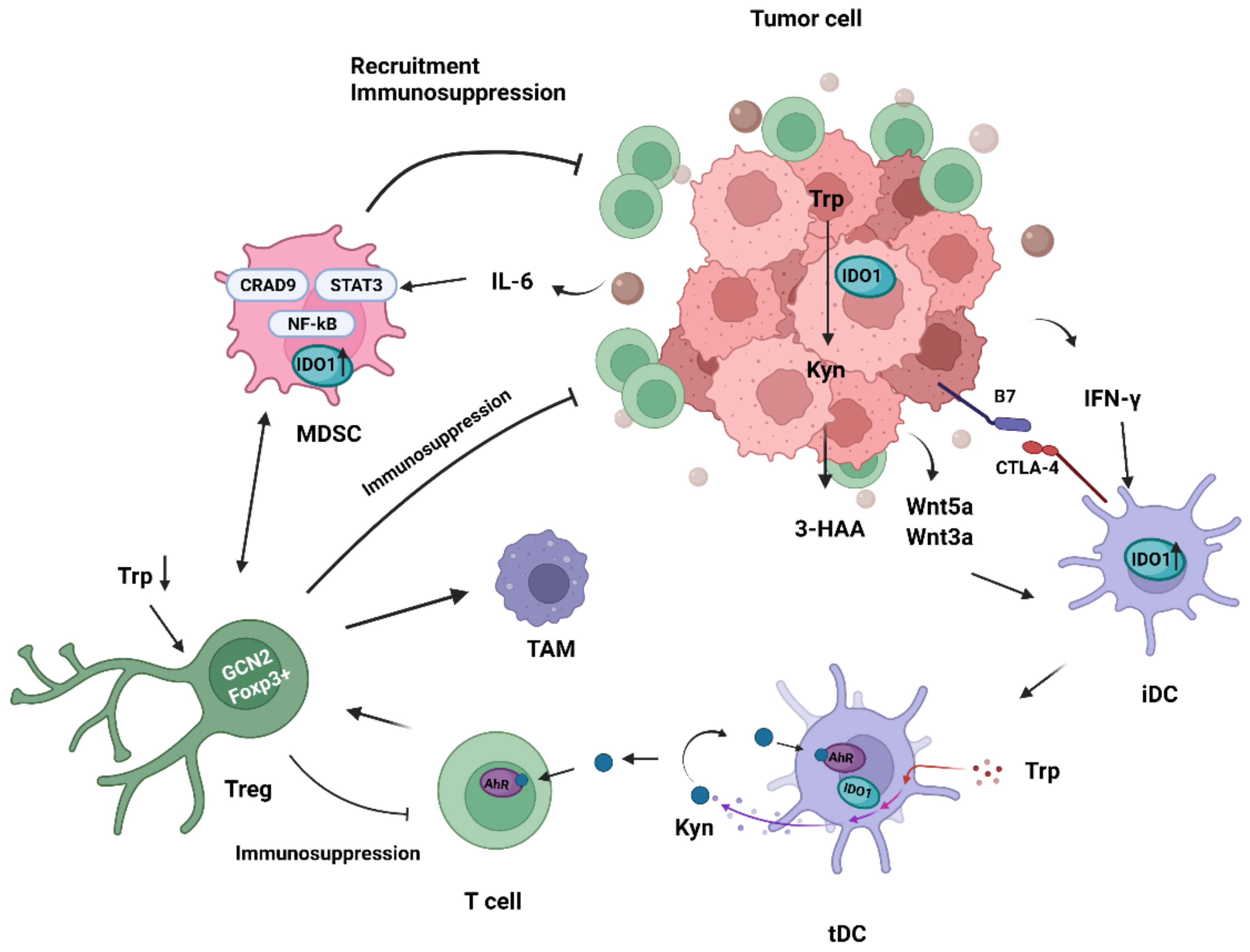
4. Role of IDO1 in TME
4.1. Immunomodulatory Effects of IDO1 on Tumor-Associated Dendritic Cells
4.2. Immunomodulatory Effects of IDO1 on Tregs
4.3. Immunomodulatory Effects of IDO1 on MDSCs
4.4. Immunomodulatory Effects of IDO1 on Natural Killer Cells
4.5. Immunomodulatory Effects of IDO1 on Tumor-Associated Macrophages
4.6. Immunomodulatory Effects of IDO1 on Other TME-Related Cells
5. Current Status of IDO1 and TME in Treatment of Cancer, Future Perspective
6. Conclusions
| Cell Type | Mechanism (Pathway) | Effects | Condition |
|---|---|---|---|
| DCs | IDO1-Kyn-AhR pathway | Endowed with the tolerance phenotype and lose the ability to activate prime CD8 + T cells | Low metastatic lung alveolar carcinoma in vivo [90], mammary carcinoma in vivo [95], and non-tumor in vivo [96] |
| IDO1-Kyn-AhR pathway | Promote formation of Foxp3 + Tregs and induce Foxp3 + Tregs to inhibit normal immune surveillance. | Acute myeloid leukemia in vitro [22], and hepatocellular carcinoma in vitro [23], non-tumor in vitro [24,25], low metastatic lung alveolar carcinoma in vivo [90], acute myeloid leukemia in vitro [102], mesothelioma and lung cancer in vivo [16], mammary carcinoma, lung alveolar carcinoma, and colon carcinoma in vivo [14], and non-tumor in vivo [15] | |
| IDO1-Kyn-AhR pathway and amino acid-responsive GCN2 pathway | Activate pre-existing Tregs suppressive activity | Melanoma in vivo [77] | |
| Tregs | IDO1-Kyn-AhR pathway | Differentiated into CD4 + CD25 + Foxp3 + Tregs | Non-tumor in vitro [1,24,109], melanoma, and colon cancer in vivo [19] |
| IDO1-Kyn-AhR pathway and amino acid-responsive GCN2 pathway | Acquire suppressive ability | Melanoma in vivo [77,110] | |
| IDO1-Kyn-AhR pathway | Inhibit CD8 + T cells proliferation | Melanoma and colon cancer in vivo [19], and Melanoma in vivo [17] | |
| IDO1-Kyn-AhR pathway | Promote expression of immunosuppressive factors | Melanoma and colon cancer in vivo [19] | |
| IDO1-Kyn-AhR pathway | Promote TAMs proliferation | Melanoma and colon cancer in vitro [19] | |
| IDO1-Kyn-AhR pathway | Recruit MDSCs to tumor tissues | Melanoma in vivo [17] | |
| MDSCs | IDO1-Kyn-AhR pathway | Suppress T cells activity and induce suppressive Tregs | Chronic lymphocytic leukemia in vitro [123,124] |
| IDO1-Kyn-AhR pathway | Enhance Tregs suppressive activity | Chronic lymphocytic leukemia in vitro [123,124] | |
| IDO1-Kyn-AhR pathway | Promote formation of Foxp3 + Tregs | Non-tumor in vitro [18] | |
| IDO1-Kyn-AhR pathway | Interfere with B cells proliferation and immune function | Non-tumor in vitro [125] | |
| IDO1-Kyn-AhR pathway | Accelerated tumor outgrowth | Melanoma in vivo [17,119], lung cancer in vivo [120], lewis lung carcinoma in vivo [121], and triple-negative breast cancer in vitro [122] | |
| NK cells | IDO1-Kyn-AhR pathway, JAK-STAT pathway, and IDO1-miR-18a-NKG2D-NKG2DL axis | Cytotoxic killing ability dysfunction | Thyroid cancer in vitro [21], pancreatic cancer in vitro [20], and breast cancer in vitro [138] |
| TAMs | IDO1-Kyn-autophagy pathway | Phagocytic cancer cells with active autophagy | Cervical cancer in vitro [150] |
| IDO1-Kyn-AhR pathway, CD39-CD73-adenosine pathway, and IDO1-Kyn-AhR-CD155 pathway | Impair T cell response | Glioblastoma in vivo [151], non-tumor in vitro and breast cancer in vivo [152] | |
| CAFs | IL-6-STAT3-IDO1 pathway | Educate DCs to acquire an IDO1-express tolerogenic phenotype | Hepatocellular carcinoma in vitro [62] |
| TECs | IDO1-Kyn-AhR pathway | Regulate tumor neovascularization | Clear cell renal cell carcinoma specimen [159], and lung cancer in vivo [160] |
| TRCs | IDO1-Kyn-AhR-p27 pathway | Enter dormancy | Colon cancer, hepatocellular carcinoma, breast cancer, stomach cancer, and liver cancer in vitro, and melanoma both in vitro and in vivo [47,165] |
| B cells | IDO1-Kyn-AhR pathway | Transform into iBreg to regulate T cells and drive the generation of Foxp3 + CD4 + T cells | Non-tumor in vitro [166] |
| Agent | Strategy | NCT Number | Phase | Conditions | Clinical Efficacy |
|---|---|---|---|---|---|
| Indoximod (1-D-MT) | Single agent | NCT03852446 | Early I | Healthy | Unknown |
| NCT03372239 | I | Healthy | Unknown | ||
| NCT00567931 | I | Unspecified adult solid tumor | Unknown | ||
| Sipuleucel-T | NCT01560923 | II | Metastatic prostate cancer | Stabel disease (SD) is 50% | |
| Idarubicin and cytarabine | NCT02835729 | I | Acute myeloid leukemia | Unknown | |
| Temozolomide, cyclophosphamide, etoposide, and radiation | NCT02502708 | I | Glioblastoma multiforme, glioma, gliosarcoma, malignant brain tumor, ependymoma, medulloblastoma, diffuse intrinsic pontine glioma, and primary CNS tumor | Unknown | |
| Nab-Paclitaxel and gemcitabine | NCT02077881 | I/II | Metastatic pancreatic adenocarcinoma and metastatic pancreatic cancer | Unknown | |
| Ipilimumab, nivolumab, and pembrolizumab | NCT02073123 | I/II | Metastatic melanoma and stage III-IV melanoma | Unknown | |
| Docetaxel, indoximod, and paclitaxel | NCT01792050 | II | Metastatic breast cancer | Objective response rate (ORR) is 40% and 37%, respectively (indoximod vs. placebo) [181] | |
| Temozolomide, bevacizumab, and radiation | NCT02052648 | I/II | Glioblastoma multiforme, glioma, gliosarcoma, and malignant brain tumor | Unknown | |
| Docetaxel | NCT01191216 | I | Unspecified adult solid tumor | Unknown | |
| Epacadostat (INCB024360) | Single agent | NCT01195311 | II | Solid tumors and hematologic malignancy | SD lasting ≥16 weeks was observed in 7 of 52 patients [169] |
| NCT01822691 | II | Myelodysplastic syndromes | SD in 12 (80%) patients and progressive disease in 3 (20%) patients [170] | ||
| Pembrolizumab | NCT03322540 | II | Lung cancer | ORR 32.5% | |
| NCT03291054 | II | Gastrointestinal stromal tumors | Unknown | ||
| NCT02364076 | II | Thymic carcinoma, thymus neoplasms, and thymus cancer | SD 52.5% | ||
| NCT03196232 | II | Gastric adenocarcinoma, gastroesophageal junction adenocarcinoma, recurrent esophageal carcinoma, recurrent gastric carcinoma, stage IV esophageal cancer AJCC v7, stage IV gastric cancer AJCC v7, and unresectable esophageal carcinoma | Unknown | ||
| NCT02752074 | III | Melanoma | No significant differences were found between the treatment groups for progression-free survival [178] | ||
| NCT03361865 | III | Urothelial cancer | ORR 31.8% | ||
| NCT03374488 | III | Urothelial cancer | ORR 21.4% | ||
| Pembrolizumab and chemotherapy | NCT02862457 | I | Neoplasms, carcinoma, and non-small-cell lung | Unknown | |
| NCT03322566 | II | Lung cancer | ORR 26.4% | ||
| Pembrolizumab, oxaliplatin, leucovorin, 5-fluorouracil, gemcitabine, nab-paclitaxel, carboplatin, paclitaxel, pemetrexed, cyclophosphamide, and cisplatin | NCT03085914 | I/II | Solid tumor | Partial response (PR): Epa + Pembrolizumab + mFOLFOX6: 55.6%; Epa + Pembrolizumab + 5-FU and Platinum Agent: 45.5% | |
| Durvalumab (MEDI4736) | NCT02318277 | I/II | Solid tumors, head and neck cancer, lung cancer, and urothelial cancer | ORR 12.9% (phase II) | |
| SD-101 and radiation | NCT03322384 | I/II | Advanced solid tumors, lymphoma | Unknown | |
| MK-3475 | NCT02178722 | I/II | Microsatellite-instability high colorectal cancer, endometrial cancer, head and neck cancer, hepatocellular carcinoma, gastric cancer, lung cancer, lymphoma, renal cell carcinoma, ovarian cancer, solid tumors, urothelial cancer, breast cancer, and melanoma | ORR: microsatellite-instability high colorectal cancer: 43.8%; melanoma: 60.5%; non-small cell lung cancer: 30.8%; renal cell carcinoma: 32.4%; squamous cell carcinoma of the head and neck: 33.3%; transitional carcinoma of the genitourinary tract: 30.6% | |
| Nivolumab and chemotherapy | NCT02327078 | I/II | B-cell malignancies, colorectal cancer (CRC), head and neck cancer, lung cancer, lymphoma, melanoma, ovarian cancer, and glioblastoma | Unknown | |
| MELITAC 12.1 Peptide Vaccine | NCT01961115 | II | Stage III–IV melanoma | Unknown | |
| Fludarabine, cyclophosphamide, NK cells, and IL-2 | NCT02118285 | I | Ovarian cancer, fallopian tube carcinoma, and primary peritoneal carcinoma | Unknown | |
| DEC-205, NY-ESO-1 Fusion Protein CDX-1401, and Poly ICLC | NCT02166905 | I/II | Fallopian tube carcinoma, ovarian carcinoma, and primary peritoneal carcinoma | Unknown | |
| BMS-986205 | Single agent | NCT03378310 | I | Healthy | Unknown |
| NCT03312426 | I | Healthy | Unknown | ||
| NCT03374228 | I | Healthy | Unknown | ||
| NCT03362411 | I | Healthy | Unknown | ||
| NCT03247283 | I | Cancer | Unknown | ||
| Nivolumab | NCT03192943 | I | Advanced cancer | Unknown | |
| NCT03792750 | I/II | Advanced cancer | Unknown | ||
| NCT03329846 | III | Melanoma and skin Cancer | Unknown | ||
| Omeprazole | NCT03936374 | I | Healthy | Unknown | |
| Itraconazole and rifampin | NCT03346837 | I | Malignancies multiple | Unknown | |
| navoximod (GDC-0919/NLG919) | Single agent | NCT02048709 | I | Solid tumor | (8) 36% had stable disease and (10) 46% had progressive disease [168] |
| Atezolizumab | NCT02471846 | I | Solid tumor | (6) 9% dose escalation patients achieved PR, (10) 11% expansion patients achieved PR or CR [182] | |
| NLG802 | Single agent | NCT03164603 | I | Solid tumor | Unknown |
| SHR9146 (HTI-1090) | Single agent | NCT03208959 | I | Advanced solid tumor | Unknown |
| Mogamulizumab | NCT02867007 | I | Solid tumor, cancer, and carcinoma | Unknown |
Author Contributions
Funding
Conflicts of Interest
References
- Mezrich, J.D.; Fechner, J.H.; Zhang, X.; Johnson, B.P.; Burlingham, W.J.; Bradfield, C.A. An interaction between kynurenine and the aryl hydrocarbon receptor can generate regulatory T cells. J. Immunol. 2010, 185, 3190–3198. [Google Scholar] [CrossRef] [PubMed]
- Pantouris, G.; Serys, M.; Yuasa, H.J.; Ball, H.J.; Mowat, C.G. Human indoleamine 2,3-dioxygenase-2 has substrate specificity and inhibition characteristics distinct from those of indoleamine 2,3-dioxygenase-1. Amino Acids 2014, 46, 2155–2163. [Google Scholar] [CrossRef] [PubMed]
- Zhai, L.; Bell, A.; Ladomersky, E.; Lauing, K.L.; Bollu, L.; Sosman, J.A.; Zhang, B.; Wu, J.D.; Miller, S.D.; Meeks, J.J.; et al. Immunosuppressive IDO in Cancer: Mechanisms of Action, Animal Models, and Targeting Strategies. Front. Immunol. 2020, 11, 1185. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Xu, W.; Liu, F.; Zhu, H.; Zhang, L.; Ding, Z.; Liang, H.; Song, J. The emerging roles of IDO2 in cancer and its potential as a therapeutic target. Biomed. Pharmacother. 2021, 137, 111295. [Google Scholar] [CrossRef]
- Mondanelli, G.; Mandarano, M.; Belladonna, M.L.; Suvieri, C.; Pelliccia, C.; Bellezza, G.; Sidoni, A.; Carvalho, A.; Grohmann, U.; Volpi, C. Current Challenges for IDO2 as Target in Cancer Immunotherapy. Front. Immunol. 2021, 12, 679953. [Google Scholar] [CrossRef]
- Feng, X.; Liao, D.; Liu, D.; Ping, A.; Li, Z.; Bian, J. Development of Indoleamine 2,3-Dioxygenase 1 Inhibitors for Cancer Therapy and Beyond: A Recent Perspective. J. Med. Chem. 2020, 63, 15115–15139. [Google Scholar] [CrossRef]
- Kiyozumi, Y.; Baba, Y.; Okadome, K.; Yagi, T.; Ishimoto, T.; Iwatsuki, M.; Miyamoto, Y.; Yoshida, N.; Watanabe, M.; Komohara, Y.; et al. IDO1 Expression Is Associated with Immune Tolerance and Poor Prognosis in Patients With Surgically Resected Esophageal Cancer. Ann. Surg. 2019, 269, 1101–1108. [Google Scholar] [CrossRef]
- Zhai, L.; Ladomersky, E.; Lenzen, A.; Nguyen, B.; Patel, R.; Lauing, K.L.; Wu, M.; Wainwright, D.A. IDO1 in cancer: A Gemini of immune checkpoints. Cell Mol. Immunol. 2018, 15, 447–457. [Google Scholar] [CrossRef]
- Zhai, L.; Ladomersky, E.; Lauing, K.L.; Wu, M.; Genet, M.; Gritsina, G.; Gyorffy, B.; Brastianos, P.K.; Binder, D.C.; Sosman, J.A.; et al. Infiltrating T Cells Increase IDO1 Expression in Glioblastoma and Contribute to Decreased Patient Survival. Clin. Cancer Res. 2017, 23, 6650–6660. [Google Scholar] [CrossRef]
- Niu, N.; Shen, W.; Zhong, Y.; Bast, R.C., Jr.; Jazaeri, A.; Sood, A.K.; Liu, J. Expression of B7-H4 and IDO1 is associated with drug re-sistance and poor prognosis in high-grade serous ovarian carcinomas. Hum. Pathol. 2021, 113, 20–27. [Google Scholar] [CrossRef]
- Jarosz-Biej, M.; Smolarczyk, R.; Cichon, T.; Kulach, N. Tumor Microenvironment as A “Game Changer” in Cancer Radiotherapy. Int. J. Mol. Sci. 2019, 20, 3212. [Google Scholar] [CrossRef] [PubMed]
- Albini, A.; Bruno, A.; Noonan, D.M.; Mortara, L. Contribution to Tumor Angiogenesis From Innate Immune Cells Within the Tu-mor Microenvironment: Implications for Immunotherapy. Front. Immunol. 2018, 9, 527. [Google Scholar] [CrossRef] [PubMed]
- Tugues, S.; Ducimetiere, L.; Friebel, E.; Becher, B. Innate lymphoid cells as regulators of the tumor microenvironment. Semin. Immunol. 2019, 41, 101270. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Virtuoso, L.P.; Anderson, C.D.; Egilmez, N.K. Regulatory Rebound in IL-12-Treated Tumors Is Driven by Uncommitted Pe-ripheral Regulatory T Cells. J. Immunol. 2015, 195, 1293–1300. [Google Scholar] [CrossRef]
- Mohammadpour, H.; Pourfathollah, A.A.; Zarif, M.N.; Tahoori, M.T. TNF-alpha modulates the immunosuppressive effects of MSCs on dendritic cells and T cells. Int. Immunopharmacol. 2015, 28, 1009–1017. [Google Scholar] [CrossRef]
- Klampatsa, A.; Leibowitz, M.S.; Sun, J.; Liousia, M.; Arguiri, E.; Albelda, S.M. Analysis and Augmentation of the Immunologic By-stander Effects of CAR T Cell Therapy in a Syngeneic Mouse Cancer Model. Mol. Ther. Oncolytics 2020, 18, 360–371. [Google Scholar] [CrossRef]
- Holmgaard, R.B.; Zamarin, D.; Li, Y.; Gasmi, B.; Munn, D.H.; Allison, J.P.; Merghoub, T.; Wolchok, J.D. Tumor-Expressed IDO Recruits and Activates MDSCs in a Treg-Dependent Manner. Cell Rep. 2015, 13, 412–424. [Google Scholar] [CrossRef]
- Tomic, S.; Joksimovic, B.; Bekic, M.; Vasiljevic, M.; Milanovic, M.; Colic, M.; Vucevic, D. Prostaglanin-E2 Potentiates the Suppressive Functions of Human Mononuclear Myeloid-Derived Suppressor Cells and Increases Their Capacity to Expand IL-10-Producing Regulatory T Cell Subsets. Front. Immunol. 2019, 10, 475. [Google Scholar] [CrossRef]
- Campesato, L.F.; Budhu, S.; Tchaicha, J.; Weng, C.H.; Gigoux, M.; Cohen, I.J.; Redmond, D.; Mangarin, L.; Pourpe, S.; Liu, C.; et al. Block-ade of the AHR restricts a Treg-macrophage suppressive axis induced by L-Kynurenine. Nat. Commun. 2020, 11, 4011. [Google Scholar] [CrossRef]
- Peng, Y.P.; Zhang, J.J.; Liang, W.B.; Tu, M.; Lu, Z.P.; Wei, J.S.; Jiang, K.R.; Gao, W.T.; Wu, J.L.; Xu, Z.K.; et al. Elevation of MMP-9 and IDO induced by pancreatic cancer cells mediates natural killer cell dysfunction. BMC Cancer 2014, 14, 738. [Google Scholar] [CrossRef]
- Park, A.; Yang, Y.; Lee, Y.; Kim, M.S.; Park, Y.-J.; Jung, H.; Kim, T.-D.; Lee, H.G.; Choi, I.; Yoon, S.R. Indoleamine-2, 3-Dioxygenase in Thyroid Cancer Cells Suppresses Natural Killer Cell Function by Inhibiting NKG2D and NKp46 Expression via STAT Signaling Pathways. J. Clin. Med. 2019, 8, 842. [Google Scholar] [CrossRef]
- Curti, A.; Trabanelli, S.; Onofri, C.; Aluigi, M.; Salvestrini, V.; Ocadlikova, D.; Evangelisti, C.; Rutella, S.; De Cristofaro, R.; Ottaviani, E.; et al. Indoleamine 2, 3-dioxygenase-expressing leukemic dendritic cells impair a leukemia-specific immune response by in-ducing potent T regulatory cells. Haematologica 2010, 95, 2022–2030. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Chen, Z.; Yang, Y.; Jiang, Z.; Gu, Y.; Liu, Y.; Lin, C.; Pan, Z.; Yu, Y.; Jiang, M.; et al. Human CD14+ CTLA-4+ regulatory dendrit-ic cells suppress T-cell response by cytotoxic T-lymphocyte antigen-4-dependent IL-10 and indoleamine-2, 3-dioxygenase production in hepatocellular carcinoma. Hepatology 2014, 59, 567–579. [Google Scholar] [CrossRef]
- Chen, W.; Liang, X.; Peterson, A.J.; Munn, D.; Blazar, B.R. The Indoleamine 2,3-Dioxygenase Pathway Is Essential for Human Plasmacytoid Dendritic Cell-Induced Adaptive T Regulatory Cell Generation. J. Immunol. 2008, 181, 5396–5404. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Min, S.Y.; Park, K.S.; Cho, Y.G.; Cho, M.L.; Jung, Y.O.; Park, H.S.; Chang, S.H.; Cho, S.G.; Min, J.K.; et al. Indoleamine 2, 3-dioxygenase-expressing dendritic cells are involved in the generation of CD4+CD25+ regulatory T cells in Peyer’s patches in an orally tolerized, collagen-induced arthritis mouse model. Arthritis Res. Ther. 2008, 10, R11. [Google Scholar] [CrossRef] [PubMed]
- Oh, M.H.; Sun, I.H.; Zhao, L.; Leone, R.D.; Sun, I.M.; Xu, W.; Collins, S.L.; Tam, A.J.; Blosser, R.L.; Patel, C.H.; et al. Targeting glutamine me-tabolism enhances tumor-specific immunity by modulating suppressive myeloid cells. J. Clin. Investig. 2020, 130, 3865–3884. [Google Scholar] [CrossRef] [PubMed]
- Bilir, C.; Sarisozen, C. Indoleamine 2, 3-dioxygenase (IDO): Only an enzyme or a checkpoint controller? J. Oncol. Sci. 2017, 3, 52–56. [Google Scholar] [CrossRef]
- Burkin, D.; Kimbro, K.S.; Barr, B.L.; Jones, C.; Taylor, M.W.; Gupta, S.L. Localization of Human Indoleamine 2,3-Dioxygenase (IDO) Gene to the Pericentromeric Region of Human Chromosome 8. Genomics 1993, 17, 262–263. [Google Scholar] [CrossRef]
- Hornyak, L.; Dobos, N.; Koncz, G.; Karanyi, Z.; Pall, D.; Szabo, Z.; Halmos, G.; Szekvolgyi, L. The Role of Indoleamine-2, 3-Dioxygenase in Cancer Development, Diagnostics, and Therapy. Front. Immunol. 2018, 9, 151. [Google Scholar] [CrossRef]
- Robinson, C.M.; Shirey, K.A.; Carlin, J.M. Synergistic transcriptional activation of indoleamine dioxygenase by IFN-gamma and tumor necrosis factor-alpha. J. Interferon Cytokine Res. 2003, 23, 413–421. [Google Scholar] [CrossRef]
- Munn, D.H.; Mellor, A.L. IDO in the Tumor Microenvironment: Inflammation, Counter-Regulation, and Tolerance. Trends Immunol. 2016, 37, 193–207. [Google Scholar] [CrossRef] [PubMed]
- Karihtala, K.; Leivonen, S.K.; Bruck, O.; Karjalainen-Lindsberg, M.L.; Mustjoki, S.; Pellinen, T.; Leppa, S. Prognostic Impact of Tumor-Associated Macrophages on Survival Is Checkpoint Dependent in Classical Hodgkin Lymphoma. Cancers 2020, 12, 877. [Google Scholar] [CrossRef] [PubMed]
- Chinn, Z.; Stoler, M.H.; Mills, A.M. PD-L1 and IDO expression in cervical and vulvar invasive and intraepithelial squamous neoplasias: Implications for combination immunotherapy. Histopathology 2019, 74, 256–268. [Google Scholar] [CrossRef] [PubMed]
- Dill, E.A.; Dillon, P.M.; Bullock, T.N.; Mills, A.M. IDO expression in breast cancer: An assessment of 281 primary and metastatic cases with comparison to PD-L1. Mod. Pathol. 2018, 31, 1513–1522. [Google Scholar] [CrossRef] [PubMed]
- Moretti, S.; Menicali, E.; Nucci, N.; Voce, P.; Colella, R.; Melillo, R.M.; Liotti, F.; Morelli, S.; Fallarino, F.; Macchiarulo, A.; et al. Signal Transducer and Activator of Transcription 1 Plays a Pivotal Role in RET/PTC3 Oncogene-induced Expression of Indoleamine 2, 3-Dioxygenase 1. J. Biol. Chem. 2017, 292, 1785–1797. [Google Scholar] [CrossRef] [PubMed]
- Luo, Q.; Yan, L.; Xu, P.; Xiong, C.; Yang, Z.; Hu, P.; Hu, H.; Hong, R. Discovery of a polysaccharide from the fruiting bodies of Lepista sordida as potent inhibitors of indoleamine 2, 3-dioxygenase (IDO) in HepG2 cells via blocking of STAT1-mediated JAK-PKC-delta signaling pathways. Carbohydr. Polym. 2018, 197, 540–547. [Google Scholar] [CrossRef]
- Jeong, Y.I.; Kim, S.W.; Jung, I.D.; Lee, J.S.; Chang, J.H.; Lee, C.M.; Chun, S.H.; Yoon, M.S.; Kim, G.T.; Ryu, S.W.; et al. Curcumin suppresses the induction of indoleamine 2, 3-dioxygenase by blocking the Janus-activated kinase-protein kinase Cdelta-STAT1 signaling pathway in interferon-gamma-stimulated murine dendritic cells. J. Biol. Chem. 2009, 284, 3700–3708. [Google Scholar] [CrossRef]
- Pine, R. Convergence of TNFalpha and IFNgamma signalling pathways through synergistic induction of IRF-1/ISGF-2 is me-diated by a composite GAS/kappaB promoter element. Nucleic Acids Res. 1997, 25, 4346–4354. [Google Scholar] [CrossRef]
- Robinson, C.M.; Hale, P.T.; Carlin, J.M. The role of IFN-gamma and TNF-alpha-responsive regulatory elements in the synergistic induction of indoleamine dioxygenase. J. Interferon Cytokine Res. 2005, 25, 20–30. [Google Scholar] [CrossRef]
- Chon, S.Y.; Hassanain, H.H.; Pine, R.; Gupta, S.L. Involvement of two regulatory elements in interferon-gamma-regulated expres-sion of human indoleamine 2, 3-dioxygenase gene. J. Interferon Cytokine Res. 1995, 15, 517–526. [Google Scholar] [CrossRef]
- Konan, K.V.; Taylor, M.W. Importance of the two interferon-stimulated response element (ISRE) sequences in the regulation of the human indoleamine 2, 3-dioxygenase gene. J. Biol. Chem. 1996, 271, 19140–19145. [Google Scholar] [CrossRef]
- Chon, S.Y.; Hassanain, H.H.; Gupta, S.L. Cooperative role of interferon regulatory factor 1 and p91 (STAT1) response elements in interferon-gamma-inducible expression of human indoleamine 2, 3-dioxygenase gene. J. Biol. Chem. 1996, 271, 17247–17252. [Google Scholar] [CrossRef] [PubMed]
- Du, M.X.; Sotero-Esteva, W.D.; Taylor, M.W. Analysis of transcription factors regulating induction of indoleamine 2, 3-dioxygenase by IFN-gamma. J. Interferon Cytokine Res. 2000, 20, 133–142. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E., Jr.; Kerr, I.M.; Stark, G.R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracel-lular signaling proteins. Science 1994, 264, 1415–1421. [Google Scholar] [CrossRef] [PubMed]
- Mounayar, M.; Kefaloyianni, E.; Smith, B.; Solhjou, Z.; Maarouf, O.H.; Azzi, J.; Chabtini, L.; Fiorina, P.; Kraus, M.; Briddell, R.; et al. PI3kalpha and STAT1 Interplay Regulates Human Mesenchymal Stem Cell Immune Polarization. Stem Cells 2015, 33, 1892–1901. [Google Scholar] [CrossRef] [PubMed]
- Tas, S.W.; Vervoordeldonk, M.J.; Hajji, N.; Schuitemaker, J.H.; van der Sluijs, K.F.; May, M.J.; Ghosh, S.; Kapsenberg, M.L.; Tak, P.P.; de Jong, E.C. Noncanonical NF-kappaB signaling in dendritic cells is required for indoleamine 2, 3-dioxygenase (IDO) induction and immune regulation. Blood 2007, 110, 1540–1549. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lv, J.; Liu, J.; Liang, X.; Jin, X.; Xie, J.; Zhang, L.; Chen, D.; Fiskesund, R.; Tang, K.; et al. STAT3/p53 pathway activation disrupts IFN-beta-induced dormancy in tumor-repopulating cells. J. Clin. Investig. 2018, 128, 1057–1073. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lv, J.; Huang, B. Mediating the death of dormant tumor cells. Mol. Cell Oncol. 2018, 5, e1458013. [Google Scholar] [CrossRef]
- Jitschin, R.; Bottcher, M.; Saul, D.; Lukassen, S.; Bruns, H.; Loschinski, R.; Ekici, A.B.; Reis, A.; Mackensen, A.; Mougiakakos, D. Inflammation-induced glycolytic switch controls suppressivity of mesenchymal Stem Cells via STAT1 glycosylation. Leukemia 2019, 33, 1783–1796. [Google Scholar] [CrossRef]
- Chu, W.M.; Ostertag, D.; Li, Z.W.; Chang, L.; Chen, Y.; Hu, Y.; Williams, B.; Perrault, J.; Karin, M. JNK2 and IKKbeta are required for activating the innate response to viral infection. Immunity 1999, 11, 721–731. [Google Scholar] [CrossRef]
- Delhase, M.; Hayakawa, M.; Chen, Y.; Karin, M. Positive and negative regulation of IkappaB kinase activity through IKKbeta subunit phosphorylation. Science 1999, 284, 309–313. [Google Scholar] [CrossRef] [PubMed]
- Senftleben, U.; Cao, Y.; Xiao, G.; Greten, F.R.; Krahn, G.; Bonizzi, G.; Chen, Y.; Hu, Y.; Fong, A.; Sun, S.C.; et al. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 2001, 293, 1495–1499. [Google Scholar] [CrossRef] [PubMed]
- Dejardin, E.; Droin, N.M.; Delhase, M.; Haas, E.; Cao, Y.; Makris, C.; Li, Z.W.; Karin, M.; Ware, C.F.; Green, D.R. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-kappaB pathways. Immunity 2002, 17, 525–535. [Google Scholar] [CrossRef]
- Kinoshita, D.; Hirota, F.; Kaisho, T.; Kasai, M.; Izumi, K.; Bando, Y.; Mouri, Y.; Matsushima, A.; Niki, S.; Han, H.; et al. Essential role of IkappaB kinase alpha in thymic organogenesis required for the establishment of self-tolerance. J. Immunol. 2006, 176, 3995–4002. [Google Scholar] [CrossRef] [PubMed]
- Puccetti, P.; Grohmann, U. IDO and regulatory T cells: A role for reverse signalling and non-canonical NF-kappaB activation. Nat. Rev. Immunol. 2007, 7, 817–823. [Google Scholar] [CrossRef]
- Ogasawara, N.; Oguro, T.; Sakabe, T.; Matsushima, M.; Takikawa, O.; Isobe, K.-I.; Nagase, F. Hemoglobin induces the expression of indoleamine 2,3-dioxygenase in dendritic cells through the activation of PI3K, PKC, and NF-κB and the generation of reactive oxygen species. J. Cell. Biochem. 2009, 108, 716–725. [Google Scholar] [CrossRef]
- Liang, Y.; Yu, Z.; Song, Y.; Wang, T.; Xiao, B. Indoleamine 2, 3-Dioxygenase Activation by Interferon Gamma in Vascular Endothelial Rat Cells Requires Noncanonical NF-kappaB Signaling. Transplant. Proc. 2019, 51, 2141–2145. [Google Scholar] [CrossRef]
- Robinson, C.M.; Hale, P.T.; Carlin, J.M. NF-kappa B activation contributes to indoleamine dioxygenase transcriptional synergy induced by IFN-gamma and tumor necrosis factor-alpha. Cytokine 2006, 35, 53–61. [Google Scholar] [CrossRef]
- Orabona, C.; Grohmann, U.; Belladonna, M.L.; Fallarino, F.; Vacca, C.; Bianchi, R.; Bozza, S.; Volpi, C.; Salomon, B.L.; Fioretti, M.C.; et al. CD28 induces immunostimulatory signals in dendritic cells via CD80 and CD86. Nat. Immunol. 2004, 5, 1134–1142. [Google Scholar] [CrossRef]
- Liu, W.L.; Lin, Y.H.; Xiao, H.; Xing, S.; Chen, H.; Chi, P.D.; Zhang, G. Epstein-Barr virus infection induces indoleamine 2, 3-dioxygenase expression in human monocyte-derived macrophages through p38/mitogen-activated protein kinase and NF-kappaB pathways: Impairment in T cell functions. J. Virol. 2014, 88, 6660–6671. [Google Scholar] [CrossRef]
- Jiang, M.; Chen, J.; Zhang, W.; Zhang, R.; Ye, Y.; Liu, P.; Yu, W.; Wei, F.; Ren, X.; Yu, J. Interleukin-6 Trans-Signaling Pathway Promotes Immunosuppressive Myeloid-Derived Suppressor Cells via Suppression of Suppressor of Cytokine Signaling 3 in Breast Cancer. Front. Immunol. 2017, 8, 1840. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.T.; Deng, Y.N.; Yi, H.M.; Wang, G.Y.; Fu, B.S.; Chen, W.J.; Liu, W.; Tai, Y.; Peng, Y.W.; Zhang, Q. Hepatic carcinoma-associated fibroblasts induce IDO-producing regulatory dendritic cells through IL-6-mediated STAT3 activation. Oncogenesis 2016, 5, e198. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.; Longman, R.S.; Albert, M.L. A two-step induction of indoleamine 2,3 dioxygenase (IDO) activity during dendritic-cell maturation. Blood 2005, 106, 2375–2381. [Google Scholar] [CrossRef] [PubMed]
- Thomas, S.R.; Salahifar, H.; Mashima, R.; Hunt, N.H.; Richardson, D.R.; Stocker, R. Antioxidants inhibit indoleamine 2, 3-dioxygenase in IFN-gamma-activated human macrophages: Posttranslational regulation by pyrrolidine dithiocarbamate. J. Immunol. 2001, 166, 6332–6340. [Google Scholar] [CrossRef]
- Iachininoto, M.G.; Nuzzolo, E.R.; Bonanno, G.; Mariotti, A.; Procoli, A.; Locatelli, F.; De Cristofaro, R.; Rutella, S. Cyclooxygenase-2 (COX-2) inhibition constrains indoleamine 2,3-dioxygenase 1 (IDO1) activity in acute myeloid leukaemia cells. Molecules 2013, 18, 10132–10145. [Google Scholar] [CrossRef]
- Mondanelli, G.; Coletti, A.; Greco, F.A.; Pallotta, M.T.; Orabona, C.; Iacono, A.; Belladonna, M.L.; Albini, E.; Panfili, E.; Fallarino, F.; et al. Positive allosteric modulation of indoleamine 2,3-dioxygenase 1 restrains neuroinflammation. Proc. Natl. Acad. Sci. USA 2020, 117, 3848–3857. [Google Scholar] [CrossRef]
- Friberg, M.; Jennings, R.; Alsarraj, M.; Dessureault, S.; Cantor, A.; Extermann, M.; Mellor, A.L.; Munn, D.H.; Antonia, S.J. Indoleamine 2,3-dioxygenase contributes to tumor cell evasion of T cell-mediated rejection. Int. J. Cancer 2002, 101, 151–155. [Google Scholar] [CrossRef]
- Uyttenhove, C.; Pilotte, L.; Theate, I.; Stroobant, V.; Colau, D.; Parmentier, N.; Boon, T.; Van den Eynde, B.J. Evidence for a tumoral immune resistance mechanism based on tryptophan degradation by indoleamine 2, 3-dioxygenase. Nat. Med. 2003, 9, 1269–1274. [Google Scholar] [CrossRef]
- Mitsuka, K.; Kawataki, T.; Satoh, E.; Asahara, T.; Horikoshi, T.; Kinouchi, H. Expression of indoleamine 2, 3-dioxygenase and cor-relation with pathological malignancy in gliomas. Neurosurgery 2013, 72, 1031–1038, discussion 1038–1039. [Google Scholar] [CrossRef]
- Jiao, R.; Zheng, X.; Sun, Y.; Feng, Z.; Song, S.; Ge, H. IDO1 Expression Increased After Neoadjuvant Therapy Predicts Poor Patho-logic Response and Prognosis in Esophageal Squamous Cell Carcinoma. Front. Oncol. 2020, 10, 1099. [Google Scholar] [CrossRef]
- Lin, D.J.; Ng, J.C.K.; Huang, L.; Robinson, M.; O’Hara, J.; Wilson, J.A.; Mellor, A.L. The immunotherapeutic role of indoleamine 2, 3-dioxygenase in head and neck squamous cell carcinoma: A systematic review. Clin. Otolaryngol. 2021, 46, 919–934. [Google Scholar] [CrossRef]
- Tierney, J.F.; Vogle, A.; Finnerty, B.; Zarnegar, R.; Ghai, R.; Gattuso, P.; Fahey, T.J.; Keutgen, X.M. Indoleamine 2,3-Dioxygenase-1 Expression in Adrenocortical Carcinoma. J. Surg. Res. 2020, 256, 90–95. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Xia, J.; Wang, L.; Wang, X.; Ma, X.; Deng, Q.; Lu, Y.; Kumar, M.; Zhou, Z.; Li, L.; et al. miR-153 suppresses IDO1 expression and enhances CAR T cell immunotherapy. J. Hematol. Oncol. 2018, 11, 58. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Z.; Li, J.; Song, S.; Wang, J.; Cai, W.; Hu, W.; Ji, J.; Zhu, Z.; Zang, L.; Yan, R.; et al. A positive feedback between IDO1 metabolite and COL12A1 via MAPK pathway to promote gastric cancer metastasis. J. Exp. Clin. Cancer Res. 2019, 38, 314. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Wu, N.; Wei, F.; Li, F.; Zhang, Y.; Liu, J.; Ren, X. Prognosis significance of indoleamine 2, 3-dioxygenase, programmed death ligand-1 and tumor-infiltrating immune cells in microenvironment of breast cancer. Int. Immunopharmacol. 2020, 84, 106506. [Google Scholar] [CrossRef]
- Munn, D.H.; Sharma, M.D.; Hou, D.; Baban, B.; Lee, J.R.; Antonia, S.J.; Messina, J.L.; Chandler, P.; Koni, P.A.; Mellor, A.L. Expression of indoleamine 2, 3-dioxygenase by plasmacytoid dendritic cells in tumor-draining lymph nodes. J. Clin. Investig. 2004, 114, 280–290. [Google Scholar] [CrossRef]
- Sharma, M.D.; Baban, B.; Chandler, P.; Hou, D.Y.; Singh, N.; Yagita, H.; Azuma, M.; Blazar, B.R.; Mellor, A.L.; Munn, D.H. Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2, 3-dioxygenase. J. Clin. Investig. 2007, 117, 2570–2582. [Google Scholar] [CrossRef]
- Qian, F.; Villella, J.; Wallace, P.K.; Mhawech-Fauceglia, P.; Tario, J.D., Jr.; Andrews, C.; Matsuzaki, J.; Valmori, D.; Ayyoub, M.; Frederick, P.J.; et al. Efficacy of levo-1-methyl tryptophan and dextro-1-methyl tryptophan in reversing indoleamine-2, 3-dioxygenase-mediated arrest of T-cell proliferation in human epithelial ovarian cancer. Cancer Res. 2009, 69, 5498–5504. [Google Scholar] [CrossRef]
- Chen, B.; Alvarado, D.M.; Iticovici, M.; Kau, N.S.; Park, H.; Parikh, P.J.; Thotala, D.; Ciorba, M.A. Interferon-Induced IDO1 Mediates Radiation Resistance and Is a Therapeutic Target in Colorectal Cancer. Cancer Immunol. Res. 2020, 8, 451–464. [Google Scholar] [CrossRef]
- Lou, Q.; Liu, R.; Yang, X.; Li, W.; Huang, L.; Wei, L.; Tan, H.; Xiang, N.; Chan, K.; Chen, J.; et al. miR-448 targets IDO1 and regulates CD8(+) T cell response in human colon cancer. J. Immunother. Cancer 2019, 7, 210. [Google Scholar] [CrossRef]
- Yentz, S.; Smith, D. Indoleamine 2,3-Dioxygenase (IDO) Inhibition as a Strategy to Augment Cancer Immunotherapy. BioDrugs 2018, 32, 311–317. [Google Scholar] [CrossRef] [PubMed]
- Eleftheriadis, T.; Pissas, G.; Antoniadi, G.; Liakopoulos, V.; Stefanidis, I. Indoleamine 2, 3-dioxygenase depletes tryptophan, activates general control non-derepressible 2 kinase and down-regulates key enzymes involved in fatty acid synthesis in primary human CD4+ T cells. Immunology 2015, 146, 292–300. [Google Scholar] [CrossRef] [PubMed]
- Eleftheriadis, T.; Pissas, G.; Antoniadi, G.; Tsogka, K.; Sounidaki, M.; Liakopoulos, V.; Stefanidis, I. Indoleamine 2, 3-dioxygenase downregulates Tcell receptor complex zetachain and cMyc, and reduces proliferation, lactate dehydrogenase levels and mi-tochondrial glutaminase in human T-cells. Mol. Med. Rep. 2016, 13, 925–932. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mondal, A.; Smith, C.; DuHadaway, J.B.; Sutanto-Ward, E.; Prendergast, G.C.; Bravo-Nuevo, A.; Muller, A.J. IDO1 is an Integral Mediator of Inflammatory Neovascularization. EBioMedicine 2016, 14, 74–82. [Google Scholar] [CrossRef]
- Dey, S.; Mondal, A.; DuHadaway, J.B.; Sutanto-Ward, E.; Laury-Kleintop, L.D.; Thomas, S.; Prendergast, G.C.; Mandik-Nayak, L.; Muller, A.J. IDO1 Signaling through GCN2 in a Subpopulation of Gr-1(+) Cells Shifts the IFNgamma/IL6 Balance to Promote Neovascularization. Cancer Immunol. Res. 2021, 9, 514–528. [Google Scholar] [CrossRef]
- Laoui, D.; Keirsse, J.; Morias, Y.; Van Overmeire, E.; Geeraerts, X.; Elkrim, Y.; Kiss, M.; Bolli, E.; Lahmar, Q.; Sichien, D.; et al. The tumour microenvironment harbours ontogenically distinct dendritic cell populations with opposing effects on tumour immunity. Nat. Commun. 2016, 7, 13720. [Google Scholar] [CrossRef]
- Di Blasio, S.; van Wigcheren, G.F.; Becker, A.; van Duffelen, A.; Gorris, M.; Verrijp, K.; Stefanini, I.; Bakker, G.J.; Bloemendal, M.; Halilovic, A.; et al. The tumour microenvironment shapes dendritic cell plasticity in a human organotypic melanoma culture. Nat. Commun. 2020, 11, 2749. [Google Scholar] [CrossRef]
- Scarlett, U.K.; Rutkowski, M.R.; Rauwerdink, A.M.; Fields, J.; Escovar-Fadul, X.; Baird, J.; Cubillos-Ruiz, J.R.; Jacobs, A.C.; Gonzalez, J.L.; Weaver, J.; et al. Ovarian cancer progression is controlled by phenotypic changes in dendritic cells. J. Exp. Med. 2012, 209, 495–506. [Google Scholar] [CrossRef]
- Hargadon, K.M.; Bishop, J.D.; Brandt, J.P.; Hand, Z.C.; Ararso, Y.T.; Forrest, O.A. Melanoma-derived factors alter the maturation and activation of differentiated tissue-resident dendritic cells. Immunol. Cell Biol. 2016, 94, 24–38. [Google Scholar] [CrossRef]
- Harden, J.L.; Gu, T.; Kilinc, M.O.; Rowswell-Turner, R.B.; Virtuoso, L.P.; Egilmez, N.K. Dichotomous effects of IFN-gamma on den-dritic cell function determine the extent of IL-12-driven antitumor T cell immunity. J. Immunol. 2011, 187, 126–132. [Google Scholar] [CrossRef]
- Fallarino, F.; Vacca, C.; Orabona, C.; Belladonna, M.L.; Bianchi, R.; Marshall, B.; Keskin, D.B.; Mellor, A.L.; Fioretti, M.C.; Grohmann, U.; et al. Functional expression of indoleamine 2, 3-dioxygenase by murine CD8 alpha(+) dendritic cells. Int. Immunol. 2002, 14, 65–68. [Google Scholar] [CrossRef] [PubMed]
- Grohmann, U.; Orabona, C.; Fallarino, F.; Vacca, C.; Calcinaro, F.; Falorni, A.; Candeloro, P.; Belladonna, M.L.; Bianchi, R.; Fioretti, M.C.; et al. CTLA-4-Ig regulates tryptophan catabolism in vivo. Nat. Immunol. 2002, 3, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- Fallarino, F.; Grohmann, U.; Hwang, K.W.; Orabona, C.; Vacca, C.; Bianchi, R.; Belladonna, M.L.; Fioretti, M.C.; Alegre, M.L.; Puccetti, P. Modulation of tryptophan catabolism by regulatory T cells. Nat. Immunol. 2003, 4, 1206–1212. [Google Scholar] [CrossRef] [PubMed]
- Mellor, A.L.; Chandler, P.; Baban, B.; Hansen, A.M.; Marshall, B.; Pihkala, J.; Waldmann, H.; Cobbold, S.; Adams, E.; Munn, D.H. Specific subsets of murine dendritic cells acquire potent T cell regulatory functions following CTLA4-mediated induction of in-doleamine 2, 3 dioxygenase. Int. Immunol. 2004, 16, 1391–1401. [Google Scholar] [CrossRef]
- Li, Q.; Harden, J.L.; Anderson, C.D.; Egilmez, N.K. Tolerogenic Phenotype of IFN-gamma-Induced IDO+ Dendritic Cells Is Main-tained via an Autocrine IDO-Kynurenine/AhR-IDO Loop. J. Immunol. 2016, 197, 962–970. [Google Scholar] [CrossRef]
- Pallotta, M.T.; Orabona, C.; Volpi, C.; Vacca, C.; Belladonna, M.L.; Bianchi, R.; Servillo, G.; Brunacci, C.; Calvitti, M.; Bicciato, S.; et al. Indoleamine 2,3-dioxygenase is a signaling protein in long-term tolerance by dendritic cells. Nat. Immunol. 2011, 12, 870–878. [Google Scholar] [CrossRef]
- Vogel, C.F.; Goth, S.R.; Dong, B.; Pessah, I.N.; Matsumura, F. Aryl hydrocarbon receptor signaling mediates expression of indoleam-ine 2, 3-dioxygenase. Biochem. Biophys. Res. Commun. 2008, 375, 331–335. [Google Scholar] [CrossRef]
- Gargaro, M.; Vacca, C.; Massari, S.; Scalisi, G.; Manni, G.; Mondanelli, G.; Mazza, E.M.C.; Bicciato, S.; Pallotta, M.T.; Orabona, C.; et al. Engagement of Nuclear Coactivator 7 by 3-Hydroxyanthranilic Acid Enhances Activation of Aryl Hydrocarbon Receptor in Immunoregulatory Dendritic Cells. Front. Immunol. 2019, 10, 1973. [Google Scholar] [CrossRef]
- Manicassamy, S.; Reizis, B.; Ravindran, R.; Nakaya, H.; Salazar-Gonzalez, R.M.; Wang, Y.C.; Pulendran, B. Activation of beta-catenin in dendritic cells regulates immunity versus tolerance in the intestine. Science 2010, 329, 849–853. [Google Scholar] [CrossRef]
- Oderup, C.; LaJevic, M.; Butcher, E.C. Canonical and noncanonical Wnt proteins program dendritic cell responses for tolerance. J. Immunol. 2013, 190, 6126–6134. [Google Scholar] [CrossRef]
- Holtzhausen, A.; Zhao, F.; Evans, K.S.; Tsutsui, M.; Orabona, C.; Tyler, D.S.; Hanks, B.A. Melanoma-Derived Wnt5a Promotes Local Dendritic-Cell Expression of IDO and Immunotolerance: Opportunities for Pharmacologic Enhancement of Immunotherapy. Cancer Immunol. Res. 2015, 3, 1082–1095. [Google Scholar] [CrossRef] [PubMed]
- Zhao, F.; Xiao, C.; Evans, K.S.; Theivanthiran, T.; DeVito, N.; Holtzhausen, A.; Liu, J.; Liu, X.; Boczkowski, D.; Nair, S.; et al. Paracrine Wnt5a-beta-Catenin Signaling Triggers a Metabolic Program that Drives Dendritic Cell Tolerization. Immunity 2018, 48, 147–160.e147. [Google Scholar] [CrossRef] [PubMed]
- Valencia, J.; Hernandez-Lopez, C.; Martinez, V.G.; Hidalgo, L.; Zapata, A.G.; Vicente, A.; Varas, A.; Sacedon, R. Wnt5a skews den-dritic cell differentiation to an unconventional phenotype with tolerogenic features. J. Immunol. 2011, 187, 4129–4139. [Google Scholar] [CrossRef] [PubMed]
- Cortez, J.T.; Montauti, E.; Shifrut, E.; Gatchalian, J.; Zhang, Y.; Shaked, O.; Xu, Y.; Roth, T.L.; Simeonov, D.R.; Zhang, Y.; et al. CRISPR screen in regulatory T cells reveals modulators of Foxp3. Nature 2020, 582, 416–420. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Shima, T.; Saeki, A.; Hidaka, T.; Nakashima, A.; Takikawa, O.; Saito, S. Expression of indoleamine 2, 3-dioxygenase and the recruitment of Foxp3-expressing regulatory T cells in the development and progression of uterine cervical cancer. Cancer Sci. 2007, 98, 874–881. [Google Scholar] [CrossRef]
- Brody, J.R.; Costantino, C.L.; Berger, A.C.; Sato, T.; Lisanti, M.P.; Yeo, C.J.; Emmons, R.V.; Witkiewicz, A.K. Expression of indoleamine 2, 3-dioxygenase in metastatic malignant melanoma recruits regulatory T cells to avoid immune detection and affects survival. Cell Cycle 2009, 8, 1930–1934. [Google Scholar] [CrossRef]
- Asghar, K.; Loya, A.; Rana, I.A.; Abu Bakar, M.; Farooq, A.; Tahseen, M.; Ishaq, M.; Masood, I.; Rashid, M.U. Forkhead box P3 and indoleamine 2,3-dioxygenase co-expression in Pakistani triple negative breast cancer patients. World J. Clin. Oncol. 2020, 11, 1018–1028. [Google Scholar] [CrossRef]
- Mansour, I.; Zayed, R.A.; Said, F.; Latif, L.A. Indoleamine 2,3-dioxygenase and regulatory T cells in acute myeloid leukemia. Hematology 2016, 21, 447–453. [Google Scholar] [CrossRef]
- Takei, H.; Yasuoka, H.; Yoshimoto, K.; Takeuchi, T. Aryl hydrocarbon receptor signals attenuate lung fibrosis in the bleomy-cin-induced mouse model for pulmonary fibrosis through increase of regulatory T cells. Arthritis Res. Ther. 2020, 22, 20. [Google Scholar] [CrossRef]
- Sharma, M.D.; Shinde, R.; McGaha, T.L.; Huang, L.; Holmgaard, R.B.; Wolchok, J.D.; Mautino, M.R.; Celis, E.; Sharpe, A.H.; Francisco, L.M.; et al. The PTEN pathway in Tregs is a critical driver of the suppressive tumor microenvironment. Sci. Adv. 2015, 1, e1500845. [Google Scholar] [CrossRef]
- Crellin, N.K.; Garcia, R.V.; Levings, M.K. Altered activation of AKT is required for the suppressive function of human CD4+CD25+ T regulatory cells. Blood 2007, 109, 2014–2022. [Google Scholar] [CrossRef] [PubMed]
- Vidotto, T.; Saggioro, F.P.; Jamaspishvili, T.; Chesca, D.L.; de Picanco Albuquerque, C.G.; Reis, R.B.; Graham, C.H.; Berman, D.M.; Siemens, D.R.; Squire, J.A.; et al. PTEN-deficient prostate cancer is associated with an immunosuppressive tumor microenvironment mediated by increased expression of IDO1 and infiltrating FoxP3+ T regulatory cells. Prostate 2019, 79, 969–979. [Google Scholar] [CrossRef]
- Porta, C.; Consonni, F.M.; Morlacchi, S.; Sangaletti, S.; Bleve, A.; Totaro, M.G.; Larghi, P.; Rimoldi, M.; Tripodo, C.; Strauss, L.; et al. Tumor-Derived Prostaglandin E2 Promotes p50 NF-kappaB-Dependent Differentiation of Monocytic MDSCs. Cancer Res. 2020, 80, 2874–2888. [Google Scholar] [CrossRef]
- Mao, Y.; Poschke, I.; Wennerberg, E.; de Pico Coana, Y.; Egyhazi Brage, S.; Schultz, I.; Hansson, J.; Masucci, G.; Lundqvist, A.; Kiessling, R. Melanoma-educated CD14+ cells acquire a myeloid-derived suppressor cell phenotype through COX-2-dependent mecha-nisms. Cancer Res. 2013, 73, 3877–3887. [Google Scholar] [CrossRef] [PubMed]
- Donkor, M.K.; Lahue, E.; Hoke, T.A.; Shafer, L.R.; Coskun, U.; Solheim, J.C.; Gulen, D.; Bishay, J.; Talmadge, J.E. Mammary tumor heter-ogeneity in the expansion of myeloid-derived suppressor cells. Int. Immunopharmacol. 2009, 9, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Wang, Y.; Yan, F.; Zhang, P.; Li, H.; Zhao, H.; Yan, C.; Yan, F.; Ren, X. Noncanonical NF-κB Activation Mediates STAT3-Stimulated IDO Upregulation in Myeloid-Derived Suppressor Cells in Breast Cancer. J. Immunol. 2014, 193, 2574–2586. [Google Scholar] [CrossRef]
- Qu, J.; Liu, L.; Xu, Q.; Ren, J.; Xu, Z.; Dou, H.; Shen, S.; Hou, Y.; Mou, Y.; Wang, T. CARD9 prevents lung cancer development by sup-pressing the expansion of myeloid-derived suppressor cells and IDO production. Int. J. Cancer 2019, 145, 2225–2237. [Google Scholar] [CrossRef]
- Wu, X.; Li, F.; Deng, Y.; Fan, X. Analysis of the control mechanism of lung cancer of caspase recruitment domain-containing protein 9 and myeloid-derived suppressor cell in Lewis lung cancer mice model. Saudi J. Biol. Sci. 2019, 26, 2037–2042. [Google Scholar] [CrossRef]
- Holmgaard, R.B.; Zamarin, D.; Lesokhin, A.; Merghoub, T.; Wolchok, J.D. Targeting myeloid-derived suppressor cells with colony stimulating factor-1 receptor blockade can reverse immune resistance to immunotherapy in indoleamine 2, 3-dioxygenase-expressing tumors. EBioMedicine 2016, 6, 50–58. [Google Scholar] [CrossRef]
- Li, A.; Barsoumian, H.B.; Schoenhals, J.E.; Cushman, T.R.; Caetano, M.S.; Wang, X.; Valdecanas, D.R.; Niknam, S.; Younes, A.I.; Li, G.; et al. Indoleamine 2,3-dioxygenase 1 inhibition targets anti-PD1-resistant lung tumors by blocking myeloid-derived suppressor cells. Cancer Lett. 2018, 431, 54–63. [Google Scholar] [CrossRef]
- Li, A.; Barsoumian, H.B.; Schoenhals, J.E.; Caetano, M.S.; Wang, X.; Menon, H.; Valdecanas, D.R.; Niknam, S.; Younes, A.I.; Cortez, M.A.; et al. IDO1 Inhibition Overcomes Radiation-Induced “Rebound Immune Suppression” by Reducing Numbers of IDO1-Expressing Myeloid-Derived Suppressor Cells in the Tumor Microenvironment. Int. J. Radiat. Oncol. Biol. Phys. 2019, 104, 903–912. [Google Scholar] [CrossRef] [PubMed]
- Salvador-Coloma, C.; Santaballa, A.; Sanmartin, E.; Calvo, D.; Garcia, A.; Hervas, D.; Cordon, L.; Quintas, G.; Ripoll, F.; Panadero, J.; et al. Immunosuppressive profiles in liquid biopsy at diagnosis predict response to neoadjuvant chemotherapy in triple-negative breast cancer. Eur. J. Cancer 2020, 139, 119–134. [Google Scholar] [CrossRef]
- Jitschin, R.; Braun, M.; Buttner, M.; Dettmer-Wilde, K.; Bricks, J.; Berger, J.; Eckart, M.J.; Krause, S.W.; Oefner, P.J.; Le Blanc, K.; et al. CLL-cells induce IDOhi CD14+HLA-DRlo myeloid-derived suppressor cells that inhibit T-cell responses and promote TRegs. Blood 2014, 124, 750–760. [Google Scholar] [CrossRef] [PubMed]
- Ozturk, S.; Kalter, V.; Roessner, P.M.; Sunbul, M.; Seiffert, M. IDO1-Targeted Therapy Does Not Control Disease Development in the Emicro-TCL1 Mouse Model of Chronic Lymphocytic Leukemia. Cancers 2021, 13, 1899. [Google Scholar] [CrossRef] [PubMed]
- Jaufmann, J.; Lelis, F.J.N.; Teschner, A.C.; Fromm, K.; Rieber, N.; Hartl, D.; Beer-Hammer, S. Human monocytic myeloid-derived sup-pressor cells impair B-cell phenotype and function in vitro. Eur. J. Immunol. 2020, 50, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Parkhurst, M.R.; Riley, J.P.; Dudley, M.E.; Rosenberg, S.A. Adoptive transfer of autologous natural killer cells leads to high levels of circulating natural killer cells but does not mediate tumor regression. Clin. Cancer Res. 2011, 17, 6287–6297. [Google Scholar] [CrossRef]
- Yang, Y.; Lim, O.; Kim, T.M.; Ahn, Y.O.; Choi, H.; Chung, H.; Min, B.; Her, J.H.; Cho, S.Y.; Keam, B.; et al. Phase I Study of Random Healthy Donor-Derived Allogeneic Natural Killer Cell Therapy in Patients with Malignant Lymphoma or Advanced Solid Tumors. Cancer Immunol. Res. 2016, 4, 215–224. [Google Scholar] [CrossRef]
- Liu, E.; Marin, D.; Banerjee, P.; Macapinlac, H.A.; Thompson, P.; Basar, R.; Nassif Kerbauy, L.; Overman, B.; Thall, P.; Kaplan, M.; et al. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. New Engl. J. Med. 2020, 382, 545–553. [Google Scholar] [CrossRef]
- Nayyar, G.; Chu, Y.; Cairo, M.S. Overcoming Resistance to Natural Killer Cell Based Immunotherapies for Solid Tumors. Front. Oncol. 2019, 9, 51. [Google Scholar] [CrossRef]
- Habif, G.; Crinier, A.; Andre, P.; Vivier, E.; Narni-Mancinelli, E. Targeting natural killer cells in solid tumors. Cell Mol. Immunol. 2019, 16, 415–422. [Google Scholar] [CrossRef]
- Ghiringhelli, F.; Menard, C.; Terme, M.; Flament, C.; Taieb, J.; Chaput, N.; Puig, P.E.; Novault, S.; Escudier, B.; Vivier, E.; et al. CD4+CD25+ regulatory T cells inhibit natural killer cell functions in a transforming growth factor-beta-dependent manner. J. Exp. Med. 2005, 202, 1075–1085. [Google Scholar] [CrossRef]
- Pietra, G.; Manzini, C.; Rivara, S.; Vitale, M.; Cantoni, C.; Petretto, A.; Balsamo, M.; Conte, R.; Benelli, R.; Minghelli, S.; et al. Melanoma cells inhibit natural killer cell function by modulating the expression of activating receptors and cytolytic activity. Cancer Res. 2012, 72, 1407–1415. [Google Scholar] [CrossRef] [PubMed]
- Stojanovic, A.; Correia, M.P.; Cerwenka, A. Shaping of NK cell responses by the tumor microenvironment. Cancer Microenviron. 2013, 6, 135–146. [Google Scholar] [CrossRef] [PubMed]
- Lanier, L.L. NK cell receptors. Annu. Rev. Immunol. 1998, 16, 359–393. [Google Scholar] [CrossRef] [PubMed]
- Kalina, U.; Kauschat, D.; Koyama, N.; Nuernberger, H.; Ballas, K.; Koschmieder, S.; Bug, G.; Hofmann, W.K.; Hoelzer, D.; Ottmann, O.G. IL-18 activates STAT3 in the natural killer cell line 92, augments cytotoxic activity, and mediates IFN-gamma production by the stress kinase p38 and by the extracellular regulated kinases p44erk-1 and p42erk-21. J. Immunol. 2000, 165, 1307–1313. [Google Scholar] [CrossRef]
- Wang, K.S.; Frank, D.A.; Ritz, J. Interleukin-2 enhances the response of natural killer cells to interleukin-12 through up-regulation of the interleukin-12 receptor and STAT4. Blood 2000, 95, 3183–3190. [Google Scholar] [CrossRef]
- Gotthardt, D.; Sexl, V. STATs in NK-Cells: The Good, the Bad, and the Ugly. Front. Immunol. 2016, 7, 694. [Google Scholar] [CrossRef]
- Zhang, J.; Han, X.; Hu, X.; Jin, F.; Gao, Z.; Yin, L.; Qin, J.; Yin, F.; Li, C.; Wang, Y. IDO1 impairs NK cell cytotoxicity by decreasing NKG2D/NKG2DLs via promoting miR-18a. Mol. Immunol. 2018, 103, 144–155. [Google Scholar] [CrossRef]
- Nausch, N.; Galani, I.E.; Schlecker, E.; Cerwenka, A. Mononuclear myeloid-derived “suppressor” cells express RAE-1 and activate natural killer cells. Blood 2008, 112, 4080–4089. [Google Scholar] [CrossRef]
- French, A.R.; Yokoyama, W.M. Natural killer cells and viral infections. Curr. Opin. Immunol. 2003, 15, 45–51. [Google Scholar] [CrossRef]
- Kim, K.J.; Wen, X.Y.; Yang, H.K.; Kim, W.H.; Kang, G.H. Prognostic Implication of M2 Macrophages Are Determined by the Propor-tional Balance of Tumor Associated Macrophages and Tumor Infiltrating Lymphocytes in Microsatellite-Unstable Gastric Carcinoma. PLoS ONE 2015, 10, e0144192. [Google Scholar] [CrossRef] [PubMed]
- Petty, A.J.; Li, A.; Wang, X.; Dai, R.; Heyman, B.; Hsu, D.; Huang, X.; Yang, Y. Hedgehog signaling promotes tumor-associated mac-rophage polarization to suppress intratumoral CD8+ T cell recruitment. J. Clin. Investig. 2019, 129, 5151–5162. [Google Scholar] [CrossRef] [PubMed]
- Tjiu, J.W.; Chen, J.S.; Shun, C.T.; Lin, S.J.; Liao, Y.H.; Chu, C.Y.; Tsai, T.F.; Chiu, H.C.; Dai, Y.S.; Inoue, H.; et al. Tumor-associated macro-phage-induced invasion and angiogenesis of human basal cell carcinoma cells by cyclooxygenase-2 induction. J. Investig. Dermatol. 2009, 129, 1016–1025. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Qiu, X.; Li, J.; Zheng, S.; Li, L.; Zhao, H. TGF-beta secreted by tumor-associated macrophages promotes proliferation and invasion of colorectal cancer via miR-34a-VEGF axis. Cell Cycle 2018, 17, 2766–2778. [Google Scholar] [CrossRef]
- Murdoch, C.; Muthana, M.; Coffelt, S.B.; Lewis, C.E. The role of myeloid cells in the promotion of tumour angiogenesis. Nat. Rev. Cancer 2008, 8, 618–631. [Google Scholar] [CrossRef]
- Tian, Y.; Guo, Y.; Zhu, P.; Zhang, D.; Liu, S.; Tang, M.; Wang, Y.; Jin, Z.; Li, D.; Yan, D.; et al. TRIM59 loss in M2 macrophages promotes melanoma migration and invasion by upregulating MMP-9 and Madcam1. Aging 2019, 11, 8623–8641. [Google Scholar] [CrossRef]
- Liu, L.; Ye, Y.; Zhu, X. MMP-9 secreted by tumor associated macrophages promoted gastric cancer metastasis through a PI3K/AKT/Snail pathway. Biomed. Pharmacother. 2019, 117, 109096. [Google Scholar] [CrossRef]
- Kessenbrock, K.; Plaks, V.; Werb, Z. Matrix metalloproteinases: Regulators of the tumor microenvironment. Cell 2010, 141, 52–67. [Google Scholar] [CrossRef]
- Petty, A.J.; Yang, Y. Tumor-associated macrophages: Implications in cancer immunotherapy. Immunotherapy 2017, 9, 289–302. [Google Scholar] [CrossRef]
- Yang, S.L.; Tan, H.X.; Niu, T.T.; Liu, Y.K.; Gu, C.J.; Li, D.J.; Li, M.Q.; Wang, H.Y. The IFN-gamma-IDO1-kynureine pathway-induced au-tophagy in cervical cancer cell promotes phagocytosis of macrophage. Int. J. Biol. Sci. 2021, 17, 339–352. [Google Scholar] [CrossRef]
- Takenaka, M.C.; Gabriely, G.; Rothhammer, V.; Mascanfroni, I.D.; Wheeler, M.A.; Chao, C.-C.; Gutiérrez-Vázquez, C.; Kenison, J.; Tjon, E.C.; Barroso, A.; et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nature Neuro-Sci. 2019, 22, 729–740. [Google Scholar] [CrossRef] [PubMed]
- McKay, Z.P.; Brown, M.C.; Gromeier, M. Aryl Hydrocarbon Receptor Signaling Controls CD155 Expression on Macrophages and Mediates Tumor Immunosuppression. J. Immunol. 2021, 206, 1385–1394. [Google Scholar] [CrossRef] [PubMed]
- Chandramohan, V.; Bryant, J.D.; Piao, H.; Keir, S.T.; Lipp, E.S.; Lefaivre, M.; Perkinson, K.; Bigner, D.D.; Gromeier, M.; McLendon, R.E. Validation of an Immunohistochemistry Assay for Detection of CD155, the Poliovirus Receptor, in Malignant Gliomas. Arch. Pathol. Lab. Med. 2017, 141, 1697–1704. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, J.S.; Madore, J.; Li, X.Y.; Smyth, M.J. Tumor intrinsic and extrinsic immune functions of CD155. Semin. Cancer Biol. 2020, 65, 189–196. [Google Scholar] [CrossRef]
- Chen, X.; Song, E. Turning foes to friends: Targeting cancer-associated fibroblasts. Nat. Rev. Drug Discov. 2019, 18, 99–115. [Google Scholar] [CrossRef]
- Omary, M.B.; Lugea, A.; Lowe, A.W.; Pandol, S.J. The pancreatic stellate cell: A star on the rise in pancreatic diseases. J. Clin. Investig. 2007, 117, 50–59. [Google Scholar] [CrossRef]
- Cui, G.; Li, C.; Xu, G.; Sun, Z.; Zhu, L.; Li, Z.; Zheng, W.; Li, J.; Yuan, A. Tumor-Associated Fibroblasts and Microvessels Contribute to the Expression of Immunosuppressive Factor Indoleamine 2, 3-Dioxygenase in Human Esophageal Cancers. Pathol. Oncol. Res. 2018, 24, 269–275. [Google Scholar] [CrossRef]
- Wei, L.; Zhu, S.; Li, M.; Li, F.; Wei, F.; Liu, J.; Ren, X. High Indoleamine 2,3-Dioxygenase Is Correlated With Microvessel Density and Worse Prognosis in Breast Cancer. Front. Immunol. 2018, 9, 724. [Google Scholar] [CrossRef]
- Seeber, A.; Klinglmair, G.; Fritz, J.; Steinkohl, F.; Zimmer, K.C.; Aigner, F.; Horninger, W.; Gastl, G.; Zelger, B.; Brunner, A.; et al. High IDO-1 expression in tumor endothelial cells is associated with response to immunotherapy in metastatic renal cell carcinoma. Cancer Sci. 2018, 109, 1583–1591. [Google Scholar] [CrossRef]
- Pan, J.; Yuan, K.; Peng, S.; Huang, Y.; Zhang, Y.; Hu, Y.; Feng, Y.; Shi, Y.; Liu, Y.; Wang, H.; et al. Gene silencing of indoleamine 2,3-dioxygenase hinders tumor growth through angiogenesis inhibition. Int. J. Oncol. 2017, 50, 2136–2144. [Google Scholar] [CrossRef]
- Strauss, D.C.; Thomas, J.M. Transmission of donor melanoma by organ transplantation. Lancet Oncol. 2010, 11, 790–796. [Google Scholar] [CrossRef]
- Milton, C.A.; Barbara, J.; Cooper, J.; Rao, M.; Russell, C.; Russ, G. The transmission of donor-derived malignant melanoma to a renal allograft recipient. Clin. Transplant. 2006, 20, 547–550. [Google Scholar] [CrossRef] [PubMed]
- MacKie, R.M.; Reid, R.; Junor, B. Fatal melanoma transferred in a donated kidney 16 years after melanoma surgery. New Engl. J. Med. 2003, 348, 567–568. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.L.; Zhang, M.; Tang, Y.L.; Liang, X.H. Cancer cell dormancy: Mechanisms and implications of cancer recurrence and metastasis. Oncol. Targets Ther. 2017, 10, 5219–5228. [Google Scholar] [CrossRef]
- Liu, Y.; Liang, X.; Yin, X.; Lv, J.; Tang, K.; Ma, J.; Ji, T.; Zhang, H.; Dong, W.; Jin, X.; et al. Blockade of IDO-kynurenine-AhR metabolic circuitry abrogates IFN-gamma-induced immunologic dormancy of tumor-repopulating cells. Nat. Commun. 2017, 8, 15207. [Google Scholar] [CrossRef]
- Nouel, A.; Pochard, P.; Simon, Q.; Segalen, I.; Le Meur, Y.; Pers, J.O.; Hillion, S. B-Cells induce regulatory T cells through TGF-beta/IDO production in A CTLA-4 dependent manner. J. Autoimmun. 2015, 59, 53–60. [Google Scholar] [CrossRef]
- Cheong, J.E.; Sun, L. Targeting the IDO1/TDO2-KYN-AhR Pathway for Cancer Immunotherapy-Challenges and Opportunities. Trends Pharmacol. Sci. 2018, 39, 307–325. [Google Scholar] [CrossRef]
- Nayak-Kapoor, A.; Hao, Z.; Sadek, R.; Dobbins, R.; Marshall, L.; Vahanian, N.N.; Jay Ramsey, W.; Kennedy, E.; Mautino, M.R.; Link, C.J.; et al. Phase Ia study of the indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor navoximod (GDC-0919) in patients with recur-rent advanced solid tumors. J. Immunother. Cancer 2018, 6, 61. [Google Scholar] [CrossRef]
- Beatty, G.L.; O’Dwyer, P.J.; Clark, J.; Shi, J.G.; Bowman, K.J.; Scherle, P.A.; Newton, R.C.; Schaub, R.; Maleski, J.; Leopold, L.; et al. First-in-Human Phase I Study of the Oral Inhibitor of Indoleamine 2,3-Dioxygenase-1 Epacadostat (INCB024360) in Patients with Advanced Solid Malignancies. Clin. Cancer Res. 2017, 23, 3269–3276. [Google Scholar] [CrossRef]
- Komrokji, R.S.; Wei, S.; Mailloux, A.W.; Zhang, L.; Padron, E.; Sallman, D.; Lancet, J.E.; Tinsley, S.; Nardelli, L.A.; Pinilla-Ibarz, J.; et al. A Phase II Study to Determine the Safety and Efficacy of the Oral Inhibitor of Indoleamine 2, 3-Dioxygenase (IDO) Enzyme INCB024360 in Patients with Myelodysplastic Syndromes. Clin. Lymphoma Myeloma Leuk. 2019, 19, 157–161. [Google Scholar] [CrossRef]
- Zhang, M.L.; Kem, M.; Mooradian, M.J.; Eliane, J.P.; Huynh, T.G.; Iafrate, A.J.; Gainor, J.F.; Mino-Kenudson, M. Differential expression of PD-L1 and IDO1 in association with the immune microenvironment in resected lung adenocarcinomas. Mod. Pathol. 2019, 32, 511–523. [Google Scholar] [CrossRef] [PubMed]
- Miyasato, Y.; Takashima, Y.; Takeya, H.; Yano, H.; Hayano, A.; Nakagawa, T.; Makino, K.; Takeya, M.; Yamanaka, R.; Komohara, Y. The expression of PD-1 ligands and IDO1 by macrophage/microglia in primary central nervous system lymphoma. J. Clin. Exp. Hematop. 2018, 58, 95–101. [Google Scholar] [CrossRef] [PubMed]
- Ludovini, V.; Bianconi, F.; Siggillino, A.; Vannucci, J.; Baglivo, S.; Berti, V.; Tofanetti, F.R.; Reda, M.S.; Bellezza, G.; Mandarano, M.; et al. High PD-L1/IDO-2 and PD-L2/IDO-1 Co-Expression Levels Are Associated with Worse Overall Survival in Resected Non-Small Cell Lung Cancer Patients. Genes 2021, 12, 273. [Google Scholar] [CrossRef]
- Hacking, S.; Chavarria, H.; Jin, C.; Perry, A.; Nasim, M. Landscape of Immune Checkpoint Inhibition in Carcinosarcoma (MMMT): Analysis of IDO-1, PD-L1 and PD-1. Pathol. Res. Pract. 2020, 216, 152847. [Google Scholar] [CrossRef] [PubMed]
- Gomes, B.; Driessens, G.; Bartlett, D.; Cai, D.; Cauwenberghs, S.; Crosignani, S.; Dalvie, D.; Denies, S.; Dillon, C.P.; Fantin, V.R.; et al. Characterization of the Selective Indoleamine 2, 3-Dioxygenase-1 (IDO1) Catalytic Inhibitor EOS200271/PF-06840003 Sup-ports IDO1 as a Critical Resistance Mechanism to PD-(L)1 Blockade Therapy. Mol. Cancer Ther. 2018, 17, 2530–2542. [Google Scholar] [CrossRef] [PubMed]
- Brown, Z.J.; Yu, S.J.; Heinrich, B.; Ma, C.; Fu, Q.; Sandhu, M.; Agdashian, D.; Zhang, Q.; Korangy, F.; Greten, T.F. Indoleamine 2,3-dioxygenase provides adaptive resistance to immune checkpoint inhibitors in hepatocellular carcinoma. Cancer Immunol. Immunother. 2018, 67, 1305–1315. [Google Scholar] [CrossRef]
- Platten, M.; Nollen, E.A.A.; Rohrig, U.F.; Fallarino, F.; Opitz, C.A. Tryptophan metabolism as a common therapeutic target in cancer, neurodegeneration and beyond. Nat. Rev. Drug Discov. 2019, 18, 379–401. [Google Scholar] [CrossRef]
- Long, G.V.; Dummer, R.; Hamid, O.; Gajewski, T.F.; Caglevic, C.; Dalle, S.; Arance, A.; Carlino, M.S.; Grob, J.J.; Kim, T.M.; et al. Epacadostat plus pembrolizumab versus placebo plus pembrolizumab in patients with unresectable or metastatic melanoma (ECHO-301/KEYNOTE-252): A phase 3, randomised, double-blind study. Lancet Oncol. 2019, 20, 1083–1097. [Google Scholar] [CrossRef]
- Opitz, C.A.; Somarribas Patterson, L.F.; Mohapatra, S.R.; Dewi, D.L.; Sadik, A.; Platten, M.; Trump, S. The therapeutic potential of targeting tryptophan catabolism in cancer. Br. J. Cancer 2020, 122, 30–44. [Google Scholar] [CrossRef]
- Sadik, A.; Somarribas Patterson, L.F.; Ozturk, S.; Mohapatra, S.R.; Panitz, V.; Secker, P.F.; Pfander, P.; Loth, S.; Salem, H.; Prentzell, M.T.; et al. IL4I1 Is a Metabolic Immune Checkpoint that Activates the AHR and Promotes Tumor Progression. Cell 2020, 182, 1252–1270.e1234. [Google Scholar] [CrossRef]
- Mariotti, V.; Han, H.; Ismail-Khan, R.; Tang, S.C.; Dillon, P.; Montero, A.J.; Poklepovic, A.; Melin, S.; Ibrahim, N.K.; Kennedy, E.; et al. Effect of Taxane Chemotherapy With or Without Indoximod in Metastatic Breast Cancer: A Randomized Clinical Trial. JAMA Oncol. 2021, 7, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; LoRusso, P.; Burris, H.; Gordon, M.; Bang, Y.-J.; Hellmann, M.D.; Cervantes, A.; de Olza, M.O.; Marabelle, A.; Hodi, F.S.; et al. Phase I Study of the Indoleamine 2,3-Dioxygenase 1 (IDO1) Inhibitor Navoximod (GDC-0919) Administered with PD-L1 Inhibitor (Atezolizumab) in Advanced Solid Tumors. Clin. Cancer Res. 2019, 25, 3220–3228. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X.; Zhang, F.; Wang, X.; Liu, K. The Role of Indoleamine 2, 3-Dioxygenase 1 in Regulating Tumor Microenvironment. Cancers 2022, 14, 2756. https://doi.org/10.3390/cancers14112756
Huang X, Zhang F, Wang X, Liu K. The Role of Indoleamine 2, 3-Dioxygenase 1 in Regulating Tumor Microenvironment. Cancers. 2022; 14(11):2756. https://doi.org/10.3390/cancers14112756
Chicago/Turabian StyleHuang, Xinting, Feng Zhang, Xiaobo Wang, and Ke Liu. 2022. "The Role of Indoleamine 2, 3-Dioxygenase 1 in Regulating Tumor Microenvironment" Cancers 14, no. 11: 2756. https://doi.org/10.3390/cancers14112756
APA StyleHuang, X., Zhang, F., Wang, X., & Liu, K. (2022). The Role of Indoleamine 2, 3-Dioxygenase 1 in Regulating Tumor Microenvironment. Cancers, 14(11), 2756. https://doi.org/10.3390/cancers14112756






