Cellular Damage in the Target and Out-Of-Field Peripheral Organs during VMAT SBRT Prostate Radiotherapy: An In Vitro Phantom-Based Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Radiation Delivery
2.3. Cell Irradiation Setup
2.4. Dose Measurements
2.5. Flow Cytometry Analysis
2.6. Clonogenic Assay
2.7. RNA Isolation and Reverse Transcription
2.8. Microarray Expression Study
2.9. Microarray Data Analysis
2.10. Gene Set Enrichment Analysis (GSEA)
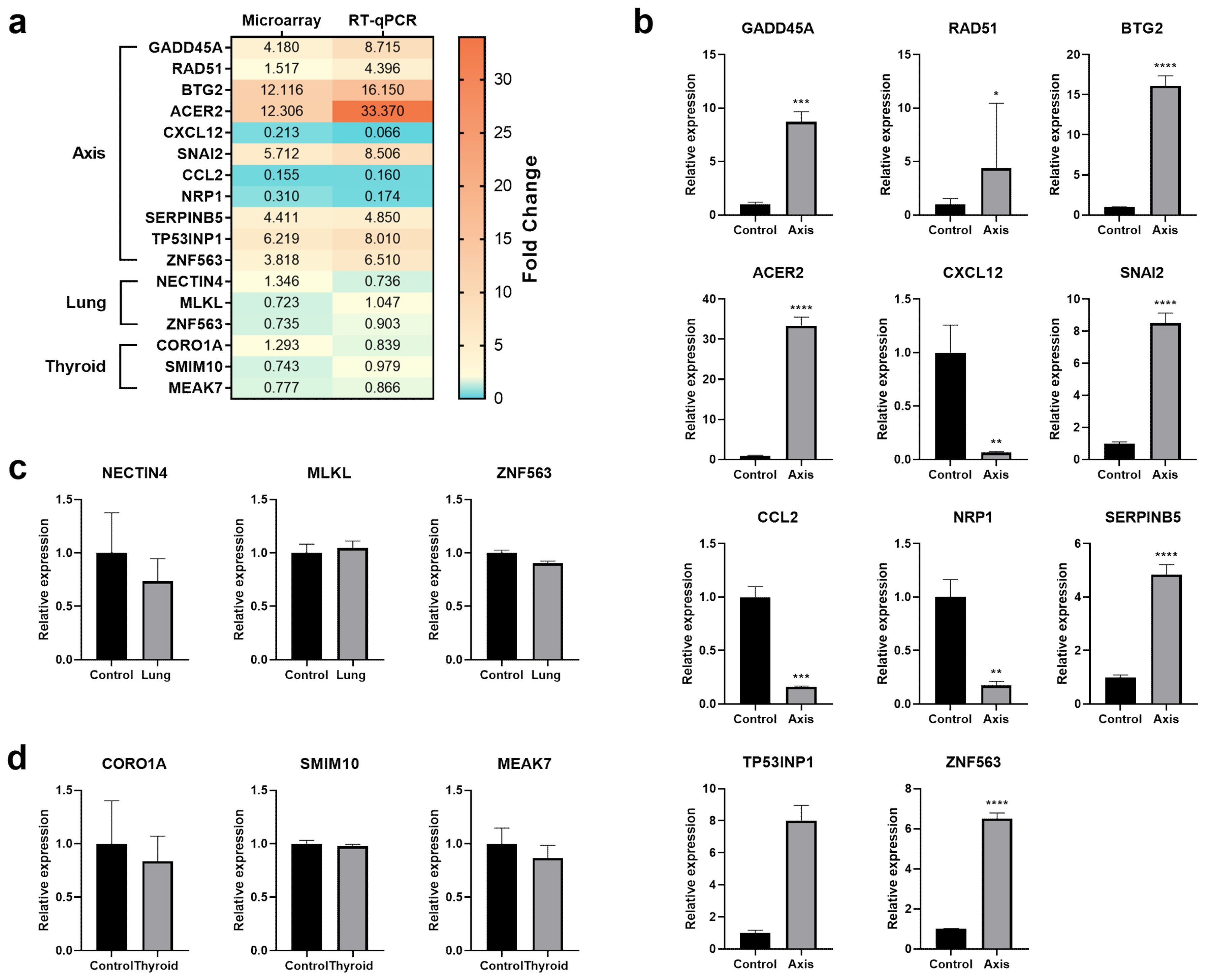
2.11. Real-Time Quantitative PCR
2.12. Statistical Analysis
3. Results
3.1. The Dose Measurement in the Locations of Peripheral Organs during Prostate VMAT SBRT Delivery
3.2. The Effect of Prostate VMAT SBRT Delivery on DNA Damage and Cell Death in Cells Irradiated in Peripheral Organs
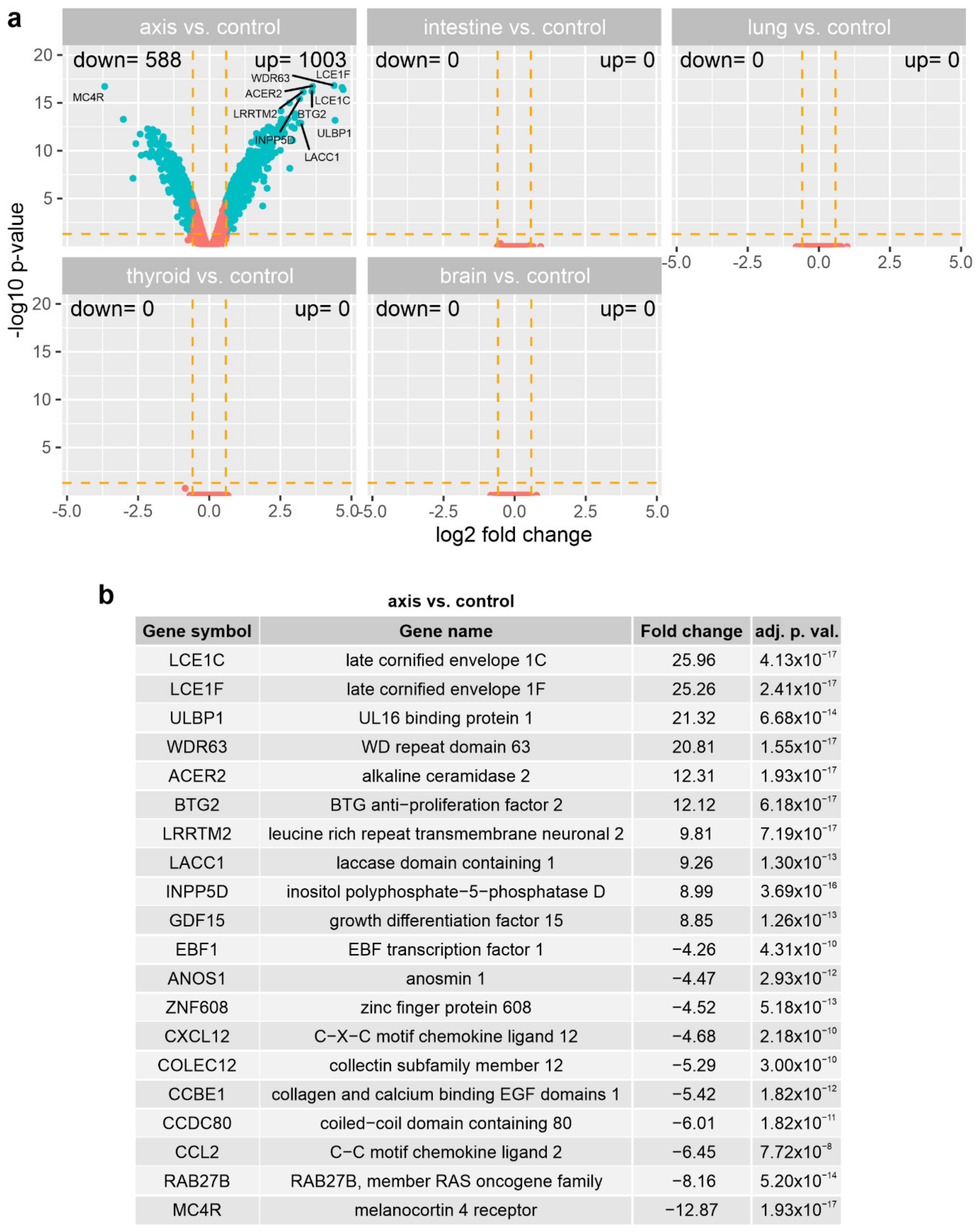
3.3. The Effect of Prostate VMAT SBRT Delivery on the Transcriptomic Profile of Cells Irradiated in Peripheral Organs
3.4. The Effect of Prostate VMAT SBRT Delivery on Enrichment of Biological Processes in Cells Irradiated in Peripheral Organs
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Dasu, A.; Toma-Dasu, I. Prostate alpha/beta revisited—An analysis of clinical results from 14 168 patients. Acta Oncol. 2012, 51, 963–974. [Google Scholar] [CrossRef]
- Kishan, A.U.; Dang, A.; Katz, A.J.; Mantz, C.A.; Collins, S.P.; Aghdam, N.; Chu, F.I.; Kaplan, I.D.; Appelbaum, L.; Fuller, D.B.; et al. Long-term Outcomes of Stereotactic Body Radiotherapy for Low-Risk and Intermediate-Risk Prostate Cancer. JAMA Netw. Open 2019, 2, e188006. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahase, S.S.; D’Angelo, D.; Kang, J.; Hu, J.C.; Barbieri, C.E.; Nagar, H. Trends in the Use of Stereotactic Body Radiotherapy for Treatment of Prostate Cancer in the United States. JAMA Netw. Open 2020, 3, e1920471. [Google Scholar] [CrossRef] [PubMed]
- Dang, A.T.; Levin-Epstein, R.G.; Shabsovich, D.; Cao, M.; King, C.; Chu, F.I.; Mantz, C.A.; Stephans, K.L.; Reddy, C.A.; Loblaw, D.A.; et al. Gantry-Mounted Linear Accelerator-Based Stereotactic Body Radiation Therapy for Low- and Intermediate-Risk Prostate Cancer. Adv. Radiat. Oncol. 2020, 5, 404–411. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.W.; Cho, L.C.; Straka, C.; Christie, A.; Lotan, Y.; Pistenmaa, D.; Kavanagh, B.D.; Nanda, A.; Kueplian, P.; Brindle, J.; et al. Predictors of rectal tolerance observed in a dose-escalated phase 1-2 trial of stereotactic body radiation therapy for prostate cancer. Int. J. Radiat. Oncol. Biol. Phys. 2014, 89, 509–517. [Google Scholar] [CrossRef]
- Kirkby, C.; Field, C.; MacKenzie, M.; Syme, A.; Fallone, B.G. A Monte Carlo study of the variation of electron fluence in water from a 6 MV photon beam outside of the field. Phys. Med. Biol. 2007, 52, 3563–3578. [Google Scholar] [CrossRef]
- Skrobala, A.; Adamczyk, S.; Kruszyna-Mochalska, M.; Skorska, M.; Konefal, A.; Suchorska, W.; Zaleska, K.; Kowalik, A.; Jackowiak, W.; Malicki, J. Low dose out-of-field radiotherapy, part 2: Calculating the mean photon energy values for the out-of-field photon energy spectrum from scattered radiation using Monte Carlo methods. Cancer Radiother. 2017, 21, 352–357. [Google Scholar] [CrossRef]
- Butterworth, K.T.; McGarry, C.K.; Trainor, C.; McMahon, S.J.; O’Sullivan, J.M.; Schettino, G.; Hounsell, A.R.; Prise, K.M. Dose, dose-rate and field size effects on cell survival following exposure to non-uniform radiation fields. Phys. Med. Biol. 2012, 57, 3197–3206. [Google Scholar] [CrossRef]
- Shields, L.; Vega-Carrascal, I.; Singleton, S.; Lyng, F.M.; McClean, B. Cell survival and DNA damage in normal prostate cells irradiated out-of-field. Radiat. Res. 2014, 182, 499–506. [Google Scholar] [CrossRef]
- Murray, L.J.; Thompson, C.M.; Lilley, J.; Cosgrove, V.; Franks, K.; Sebag-Montefiore, D.; Henry, A.M. Radiation-induced second primary cancer risks from modern external beam radiotherapy for early prostate cancer: Impact of stereotactic ablative radiotherapy (SABR), volumetric modulated arc therapy (VMAT) and flattening filter free (FFF) radiotherapy. Phys. Med. Biol. 2015, 60, 1237–1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Agostino, E.; Bogaerts, R.; Defraene, G.; de Freitas Nascimento, L.; Van den Heuvel, F.; Verellen, D.; Duchateau, M.; Schoonjans, W.; Vanhavere, F. Peripheral doses in radiotherapy: A comparison between IMRT, VMAT and Tomotherapy. Radiat. Meas. 2013, 57, 62–67. [Google Scholar] [CrossRef]
- Milano, M.T.; Constine, L.S.; Okunieff, P. Normal tissue tolerance dose metrics for radiation therapy of major organs. Semin. Radiat. Oncol. 2007, 17, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Lee, S.; Sung, J.; Yoon, M. Radiotherapy-induced secondary cancer risk for breast cancer: 3D conformal therapy versus IMRT versus VMAT. J. Radiol. Prot. 2014, 34, 325–331. [Google Scholar] [CrossRef]
- Kruszyna-Mochalska, M.; Skrobala, A.; Romanski, P.; Ryczkowski, A.; Suchorska, W.; Kulcenty, K.; Piotrowski, I.; Borowicz, D.; Matuszak, N.; Malicki, J. Development of a quasi-humanoid phantom to perform dosimetric and radiobiological measurements for out-of-field doses from external beam radiation therapy. J. Appl. Clin. Med. Phys. 2022, 23, e13514. [Google Scholar] [CrossRef]
- Kry, S.F.; Bednarz, B.; Howell, R.M.; Dauer, L.; Followill, D.; Klein, E.; Paganetti, H.; Wang, B.; Wuu, C.S.; George Xu, X. AAPM TG 158: Measurement and calculation of doses outside the treated volume from external-beam radiation therapy. Med. Phys. 2017, 44, e391–e429. [Google Scholar] [CrossRef]
- Niroomand-Rad, A.; Chiu-Tsao, S.T.; Grams, M.P.; Lewis, D.F.; Soares, C.G.; Van Battum, L.J.; Das, I.J.; Trichter, S.; Kissick, M.W.; Massillon-Jl, G.; et al. Report of AAPM Task Group 235 Radiochromic Film Dosimetry: An Update to TG-55. Med. Phys. 2020, 47, 5986–6025. [Google Scholar] [CrossRef]
- Stelcer, E.; Kulcenty, K.; Rucinski, M.; Jopek, K.; Trzeciak, T.; Richter, M.; Wroblewska, J.P.; Suchorska, W.M. Expression of Pluripotency Genes in Chondrocyte-Like Cells Differentiated from Human Induced Pluripotent Stem Cells. Int. J. Mol. Sci. 2018, 19, 550. [Google Scholar] [CrossRef]
- Trejter, M.; Hochol, A.; Tyczewska, M.; Ziolkowska, A.; Jopek, K.; Szyszka, M.; Malendowicz, L.K.; Rucinski, M. Visinin-like peptide 1 in adrenal gland of the rat. Gene Expr. Its Horm. Control. Pept. 2015, 63, 22–29. [Google Scholar] [CrossRef]
- Jopek, K.; Celichowski, P.; Szyszka, M.; Tyczewska, M.; Milecka, P.; Malendowicz, L.K.; Rucinski, M. Transcriptome Profile of Rat Adrenal Evoked by Gonadectomy and Testosterone or Estradiol Replacement. Front. Endocrinol. (Lausanne) 2017, 8, 26. [Google Scholar] [CrossRef] [Green Version]
- Trejter, M.; Jopek, K.; Celichowski, P.; Tyczewska, M.; Malendowicz, L.K.; Rucinski, M. Expression of estrogen, estrogen related and androgen receptors in adrenal cortex of intact adult male and female rats. Folia Histochem. Cytobiol. 2015, 53, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautier, L.; Cope, L.; Bolstad, B.M.; Irizarry, R.A. Affy—Analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 2004, 20, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, B.S.; Irizarry, R.A. A framework for oligonucleotide microarray preprocessing. Bioinformatics 2010, 26, 2363–2367. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Damian, D.; Gorfine, M. Statistical concerns about the GSEA procedure. Nat. Genet. 2004, 36, 663. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdottir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivashkevich, A.; Redon, C.E.; Nakamura, A.J.; Martin, R.F.; Martin, O.A. Use of the gamma-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett. 2012, 327, 123–133. [Google Scholar] [CrossRef] [Green Version]
- Massillon-Jl, G.; Chiu-Tsao, S.; Domingo-Munoz, I.; Chan, M. Energy dependence of the new Gafchromic EBT3 film–dose response curves for 50 kV, 6 and 15 MV x-ray beams. Int. J. Med. Phys. Clin. Eng. Rad. Oncol. 2012, 1, 60–65. [Google Scholar] [CrossRef] [Green Version]
- Kruszyna, M.; Adamczyk, S.; Skrobala, A.; Skorska, M.; Suchorska, W.; Zaleska, K.; Kowalik, A.; Jackowiak, W.; Malicki, J. Low dose out-of-field radiotherapy, part 1: Measurement of scattered doses. Cancer Radiother. 2017, 21, 345–351. [Google Scholar] [CrossRef]
- Dang, T.M.; Peters, M.J.; Hickey, B.; Semciw, A. Efficacy of flattening-filter-free beam in stereotactic body radiation therapy planning and treatment: A systematic review with meta-analysis. J. Med. Imaging Radiat. Oncol. 2017, 61, 379–387. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kry, S.F.; Vassiliev, O.N.; Mohan, R. Out-of-field photon dose following removal of the flattening filter from a medical accelerator. Phys. Med. Biol. 2010, 55, 2155–2166. [Google Scholar] [CrossRef] [PubMed]
- Treutwein, M.; Loeschel, R.; Hipp, M.; Koelbl, O.; Dobler, B. Secondary malignancy risk for patients with localized prostate cancer after intensity-modulated radiotherapy with and without flattening filter. J. Appl. Clin. Med. Phys. 2020, 21, 197–205. [Google Scholar] [CrossRef] [PubMed]
- Jadon, R.; Higgins, E.; Hanna, L.; Evans, M.; Coles, B.; Staffurth, J. A systematic review of dose-volume predictors and constraints for late bowel toxicity following pelvic radiotherapy. Radiat. Oncol. 2019, 14, 57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potten, C.S. A comprehensive study of the radiobiological response of the murine (BDF1) small intestine. Int. J. Radiat. Biol. 1990, 58, 925–973. [Google Scholar] [CrossRef] [PubMed]
- Ulyanenko, S.; Pustovalova, M.; Koryakin, S.; Beketov, E.; Lychagin, A.; Ulyanenko, L.; Kaprin, A.; Grekhova, A.; Ozerova, A.M.; Ozerov, I.V.; et al. Formation of gammaH2AX and pATM Foci in Human Mesenchymal Stem Cells Exposed to Low Dose-Rate Gamma-Radiation. Int. J. Mol. Sci. 2019, 20, 2645. [Google Scholar] [CrossRef] [Green Version]
- Petragnano, F.; Pietrantoni, I.; Di Nisio, V.; Fasciani, I.; Del Fattore, A.; Capalbo, C.; Cheleschi, S.; Tini, P.; Orelli, S.; Codenotti, S.; et al. Modulating the dose-rate differently affects the responsiveness of human epithelial prostate- and mesenchymal rhabdomyosarcoma-cancer cell line to radiation. Int. J. Radiat. Biol. 2020, 96, 823–835. [Google Scholar] [CrossRef]
- Syme, A.; Kirkby, C.; Mirzayans, R.; MacKenzie, M.; Field, C.; Fallone, B.G. Relative biological damage and electron fluence in and out of a 6 MV photon field. Phys. Med. Biol. 2009, 54, 6623–6633. [Google Scholar] [CrossRef]
- Liu, H.H.; Verhaegen, F. An investigation of energy spectrum and lineal energy variations in mega-voltage photon beams used for radiotherapy. Radiat. Prot. Dosim. 2002, 99, 425–427. [Google Scholar] [CrossRef]
- Zaleska, K.; Suchorska, W.M.; Kowalik, A.; Kruszyna, M.; Jackowiak, W.; Skrobala, A.; Skorska, M.; Malicki, J. Low dose out-of-field radiotherapy, part 3: Qualitative and quantitative impact of scattered out-of-field radiation on MDA-MB-231 cell lines. Cancer Radiother. 2017, 21, 358–364. [Google Scholar] [CrossRef]
- Piotrowski, I.; Kulcenty, K.; Suchorska, W.M.; Skrobala, A.; Skorska, M.; Kruszyna-Mochalska, M.; Kowalik, A.; Jackowiak, W.; Malicki, J. Carcinogenesis Induced by Low-dose Radiation. Radiol. Oncol. 2017, 51, 369–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadetzki, S.; Chetrit, A.; Lubina, A.; Stovall, M.; Novikov, I. Risk of thyroid cancer after childhood exposure to ionizing radiation for tinea capitis. J. Clin. Endocrinol. Metab. 2006, 91, 4798–4804. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.C.; Tian, L.L.; Fan, C.X.; Duo, C.H.; Xu, K.M. The Adaptive Responses in Non-Small Cell Lung Cancer A549 Cell Lines Induced by Low-Dose Ionizing Radiation and the Variations of miRNA Expression. Dose Response 2021, 19, 15593258211039931. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.J.; Kang, H.; Hong, E.H.; Kim, J.Y.; Nam, S.Y. Transcriptome analysis of low-dose ionizing radiation-impacted genes in CD4(+) T-cells undergoing activation and regulation of their expression of select cytokines. J. Immunotoxicol. 2018, 15, 137–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mortezaee, K.; Najafi, M.; Farhood, B.; Ahmadi, A.; Shabeeb, D.; Musa, A.E. NF-kappaB targeting for overcoming tumor resistance and normal tissues toxicity. J. Cell Physiol. 2019, 234, 17187–17204. [Google Scholar] [CrossRef]
- Najafi, M.; Motevaseli, E.; Shirazi, A.; Geraily, G.; Rezaeyan, A.; Norouzi, F.; Rezapoor, S.; Abdollahi, H. Mechanisms of inflammatory responses to radiation and normal tissues toxicity: Clinical implications. Int. J. Radiat. Biol. 2018, 94, 335–356. [Google Scholar] [CrossRef]
- Stelcer, E.; Kulcenty, K.; Suchorska, W.M. Chondrocytes differentiated from human induced pluripotent stem cells: Response to ionizing radiation. PLoS ONE 2018, 13, e0205691. [Google Scholar] [CrossRef]
- Klokov, D.; MacPhail, S.M.; Banath, J.P.; Byrne, J.P.; Olive, P.L. Phosphorylated histone H2AX in relation to cell survival in tumor cells and xenografts exposed to single and fractionated doses of X-rays. Radiother. Oncol. 2006, 80, 223–229. [Google Scholar] [CrossRef]
- Alongi, F.; Cozzi, L.; Arcangeli, S.; Iftode, C.; Comito, T.; Villa, E.; Lobefalo, F.; Navarria, P.; Reggiori, G.; Mancosu, P.; et al. Linac based SBRT for prostate cancer in 5 fractions with VMAT and flattening filter free beams: Preliminary report of a phase II study. Radiat. Oncol. 2013, 8, 171. [Google Scholar] [CrossRef] [Green Version]
- Franzese, C.; Badalamenti, M.; Di Brina, L.; D’Agostino, G.; Franceschini, D.; Comito, T.; Clerici, E.; Navarria, P.; Reggiori, G.; Mancosu, P.; et al. Linac-based stereotactic body radiation therapy for low and intermediate-risk prostate cancer : Long-term results and factors predictive for outcome and toxicity. Strahlenther. Onkol. 2020, 196, 608–616. [Google Scholar] [CrossRef]
- Mohamad, O.; Tabuchi, T.; Nitta, Y.; Nomoto, A.; Sato, A.; Kasuya, G.; Makishima, H.; Choy, H.; Yamada, S.; Morishima, T.; et al. Risk of subsequent primary cancers after carbon ion radiotherapy, photon radiotherapy, or surgery for localised prostate cancer: A propensity score-weighted, retrospective, cohort study. Lancet Oncol. 2019, 20, 674–685. [Google Scholar] [CrossRef]
- Aksnessaether, B.Y.; Lund, J.A.; Myklebust, T.A.; Klepp, O.H.; Skovlund, E.; Roth Hoff, S.; Solberg, A. Second cancers in radically treated Norwegian prostate cancer patients. Acta Oncol. 2019, 58, 838–844. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegemann, N.S.; Schlesinger-Raab, A.; Ganswindt, U.; Horl, C.; Combs, S.E.; Holzel, D.; Gschwend, J.E.; Stief, C.; Belka, C.; Engel, J. Risk of second cancer following radiotherapy for prostate cancer: A population-based analysis. Radiat. Oncol. 2017, 12, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikitaki, Z.; Mavragani, I.V.; Laskaratou, D.A.; Gika, V.; Moskvin, V.P.; Theofilatos, K.; Vougas, K.; Stewart, R.D.; Georgakilas, A.G. Systemic mechanisms and effects of ionizing radiation: A new ‘old’ paradigm of how the bystanders and distant can become the players. Semin. Cancer Biol. 2016, 37–38, 77–95. [Google Scholar] [CrossRef] [PubMed]
- McGarry, C.K.; Butterworth, K.T.; Trainor, C.; McMahon, S.J.; O’Sullivan, J.M.; Prise, K.M.; Hounsell, A.R. In-vitro investigation of out-of-field cell survival following the delivery of conformal, intensity-modulated radiation therapy (IMRT) and volumetric modulated arc therapy (VMAT) plans. Phys. Med. Biol. 2012, 57, 6635–6645. [Google Scholar] [CrossRef]
- Zhao, H.; Zhuang, Y.; Li, R.; Liu, Y.; Mei, Z.; He, Z.; Zhou, F.; Zhou, Y. Effects of different doses of X-ray irradiation on cell apoptosis, cell cycle, DNA damage repair and glycolysis in HeLa cells. Oncol. Lett. 2019, 17, 42–54. [Google Scholar] [CrossRef]
- Sudprasert, W.; Navasumrit, P.; Ruchirawat, M. Effects of low-dose gamma radiation on DNA damage, chromosomal aberration and expression of repair genes in human blood cells. Int. J. Hyg. Environ. Health 2006, 209, 503–511. [Google Scholar] [CrossRef]
- Nieri, D.; Berardinelli, F.; Antoccia, A.; Tanzarella, C.; Sgura, A. Comparison between two FISH techniques in the in vitro study of cytogenetic markers for low-dose X-ray exposure in human primary fibroblasts. Front. Genet. 2013, 4, 141. [Google Scholar] [CrossRef] [Green Version]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Piotrowski, I.; Kulcenty, K.; Suchorska, W.; Rucinski, M.; Jopek, K.; Kruszyna-Mochalska, M.; Skrobala, A.; Romanski, P.; Ryczkowski, A.; Borowicz, D.; et al. Cellular Damage in the Target and Out-Of-Field Peripheral Organs during VMAT SBRT Prostate Radiotherapy: An In Vitro Phantom-Based Study. Cancers 2022, 14, 2712. https://doi.org/10.3390/cancers14112712
Piotrowski I, Kulcenty K, Suchorska W, Rucinski M, Jopek K, Kruszyna-Mochalska M, Skrobala A, Romanski P, Ryczkowski A, Borowicz D, et al. Cellular Damage in the Target and Out-Of-Field Peripheral Organs during VMAT SBRT Prostate Radiotherapy: An In Vitro Phantom-Based Study. Cancers. 2022; 14(11):2712. https://doi.org/10.3390/cancers14112712
Chicago/Turabian StylePiotrowski, Igor, Katarzyna Kulcenty, Wiktoria Suchorska, Marcin Rucinski, Karol Jopek, Marta Kruszyna-Mochalska, Agnieszka Skrobala, Piotr Romanski, Adam Ryczkowski, Dorota Borowicz, and et al. 2022. "Cellular Damage in the Target and Out-Of-Field Peripheral Organs during VMAT SBRT Prostate Radiotherapy: An In Vitro Phantom-Based Study" Cancers 14, no. 11: 2712. https://doi.org/10.3390/cancers14112712
APA StylePiotrowski, I., Kulcenty, K., Suchorska, W., Rucinski, M., Jopek, K., Kruszyna-Mochalska, M., Skrobala, A., Romanski, P., Ryczkowski, A., Borowicz, D., Matuszak, N., & Malicki, J. (2022). Cellular Damage in the Target and Out-Of-Field Peripheral Organs during VMAT SBRT Prostate Radiotherapy: An In Vitro Phantom-Based Study. Cancers, 14(11), 2712. https://doi.org/10.3390/cancers14112712







