Genome-Wide Association Study Suggests the Variant rs7551288*A within the DHCR24 Gene Is Associated with Poor Overall Survival in Melanoma Patients
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Recruitment and Measures
2.2. Genotyping
2.3. Statistics
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schadendorf, D.; Fisher, D.E.; Garbe, C.; Gershenwald, J.E.; Grob, J.J.; Halpern, A.; Herlyn, M.; Marchetti, M.A.; McArthur, G.; Ribas, A.; et al. Melanoma. Nat. Rev. Dis. Primers 2015, 1, 15003. [Google Scholar] [CrossRef]
- Bastian, B.C. The molecular pathology of melanoma: An integrated taxonomy of melanocytic neoplasia. Annu. Rev. Pathol. 2014, 9, 239–271. [Google Scholar] [CrossRef] [Green Version]
- Burnet, N.G.; Jefferies, S.J.; Benson, R.J.; Hunt, D.P.; Treasure, F.P. Years of life lost (YLL) from cancer is an important measure of population burden—And should be considered when allocating research funds. Br. J. Cancer 2005, 92, 241–245. [Google Scholar] [CrossRef]
- Karimkhani, C.; Green, A.C.; Nijsten, T.; Weinstock, M.A.; Dellavalle, R.P.; Naghavi, M.; Fitzmaurice, C. The global burden of melanoma: Results from the Global Burden of Disease Study 2015. Br. J. Dermatol. 2017, 177, 134–140. [Google Scholar] [CrossRef] [Green Version]
- Bauer, J.; Garbe, C. Acquired melanocytic nevi as risk factor for melanoma development. A comprehensive review of epidemiological data. Pigment. Cell Res. 2003, 16, 297–306. [Google Scholar] [CrossRef]
- Davies, J.R.; Chang, Y.M.; Bishop, D.T.; Armstrong, B.K.; Bataille, V.; Bergman, W.; Berwick, M.; Bracci, P.M.; Elwood, J.M.; Ernstoff, M.S.; et al. Development and validation of a melanoma risk score based on pooled data from 16 case-control studies. Cancer Epidemiol. Biomark. Prev. 2015, 24, 817–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amos, C.I.; Wang, L.E.; Lee, J.E.; Gershenwald, J.E.; Chen, W.V.; Fang, S.; Kosoy, R.; Zhang, M.; Qureshi, A.A.; Vattathil, S.; et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet. 2011, 20, 5012–5023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, J.H.; Iles, M.M.; Harland, M.; Taylor, J.C.; Aitken, J.F.; Andresen, P.A.; Akslen, L.A.; Armstrong, B.K.; Avril, M.F.; Azizi, E.; et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat. Genet. 2011, 43, 1108–1113. [Google Scholar] [CrossRef] [PubMed]
- Bishop, D.T.; Demenais, F.; Iles, M.M.; Harland, M.; Taylor, J.C.; Corda, E.; Randerson-Moor, J.; Aitken, J.F.; Avril, M.F.; Azizi, E.; et al. Genome-wide association study identifies three loci associated with melanoma risk. Nat. Genet. 2009, 41, 920–925. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.M.; Macgregor, S.; Montgomery, G.W.; Craig, D.W.; Zhao, Z.Z.; Iyadurai, K.; Henders, A.K.; Homer, N.; Campbell, M.J.; Stark, M.; et al. Common sequence variants on 20q11.22 confer melanoma susceptibility. Nat. Genet. 2008, 40, 838–840. [Google Scholar] [CrossRef] [Green Version]
- Gudbjartsson, D.F.; Sulem, P.; Stacey, S.N.; Goldstein, A.M.; Rafnar, T.; Sigurgeirsson, B.; Benediktsdottir, K.R.; Thorisdottir, K.; Ragnarsson, R.; Sveinsdottir, S.G.; et al. ASIP and TYR pigmentation variants associate with cutaneous melanoma and basal cell carcinoma. Nat. Genet. 2008, 40, 886–891. [Google Scholar] [CrossRef] [PubMed]
- Law, M.H.; Bishop, D.T.; Lee, J.E.; Brossard, M.; Martin, N.G.; Moses, E.K.; Song, F.; Barrett, J.H.; Kumar, R.; Easton, D.F.; et al. Genome-wide meta-analysis identifies five new susceptibility loci for cutaneous malignant melanoma. Nat. Genet. 2015, 47, 987–995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Macgregor, S.; Montgomery, G.W.; Liu, J.Z.; Zhao, Z.Z.; Henders, A.K.; Stark, M.; Schmid, H.; Holland, E.A.; Duffy, D.L.; Zhang, M.; et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat. Genet. 2011, 43, 1114–1118. [Google Scholar] [CrossRef] [PubMed]
- Nan, H.; Xu, M.; Zhang, J.; Zhang, M.; Kraft, P.; Qureshi, A.A.; Chen, C.; Guo, Q.; Hu, F.B.; Rimm, E.B.; et al. Genome-wide association study identifies nidogen 1 (NID1) as a susceptibility locus to cutaneous nevi and melanoma risk. Hum. Mol. Genet. 2011, 20, 2673–2679. [Google Scholar] [CrossRef]
- Ransohoff, K.J.; Wu, W.; Cho, H.G.; Chahal, H.C.; Lin, Y.; Dai, H.J.; Amos, C.I.; Lee, J.E.; Tang, J.Y.; Hinds, D.A.; et al. Two-stage genome-wide association study identifies a novel susceptibility locus associated with melanoma. Oncotarget 2017, 8, 17586–17592. [Google Scholar] [CrossRef]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.; Ruas, M.; Gregory, F.; Moulin, S.; Delia, D.; Manoukian, S.; Rowe, J.; Brookes, S.; Peters, G. A CDKN2A mutation in familial melanoma that abrogates binding of p16INK4a to CDK4 but not CDK6. Cancer Res. 2007, 67, 9134–9141. [Google Scholar] [CrossRef] [Green Version]
- Ainger, S.A.; Jagirdar, K.; Lee, K.J.; Soyer, H.P.; Sturm, R.A. Skin Pigmentation Genetics for the Clinic. Dermatology 2017, 233, 1–15. [Google Scholar] [CrossRef]
- Globocan. Available online: http://globocan.iarc.fr (accessed on 26 April 2022).
- Leiter, U.; Meier, F.; Schittek, B.; Garbe, C. The natural course of cutaneous melanoma. J. Surg. Oncol. 2004, 86, 172–178. [Google Scholar] [CrossRef]
- Gershenwald, J.E.; Scolyer, R.A.; Hess, K.R.; Sondak, V.K.; Long, G.V.; Ross, M.I.; Lazar, A.J.; Faries, M.B.; Kirkwood, J.M.; McArthur, G.A.; et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA Cancer J. Clin. 2017, 67, 472–492. [Google Scholar] [CrossRef] [Green Version]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daley, G.M.; Duffy, D.L.; Pflugfelder, A.; Jagirdar, K.; Lee, K.J.; Yong, X.L.; Eigentler, T.K.; Weide, B.; Smithers, B.M.; Martin, N.G.; et al. GSTP1 does not modify MC1R effects on melanoma risk. Exp. Dermatol. 2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Battle, A.; Brown, C.D.; Engelhardt, B.E.; Montgomery, S.B. Genetic effects on gene expression across human tissues. Nature 2017, 550, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, T.; Gasch, A.; Nishizawa, N.; Chua, N.H. The DIMINUTO gene of Arabidopsis is involved in regulating cell elongation. Genes Dev. 1995, 9, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Greeve, I.; Hermans-Borgmeyer, I.; Brellinger, C.; Kasper, D.; Gomez-Isla, T.; Behl, C.; Levkau, B.; Nitsch, R.M. The human DIMINUTO/DWARF1 homolog seladin-1 confers resistance to Alzheimer’s disease-associated neurodegeneration and oxidative stress. J. Neurosci. 2000, 20, 7345–7352. [Google Scholar] [CrossRef]
- Waterham, H.R.; Koster, J.; Romeijn, G.J.; Hennekam, R.C.; Vreken, P.; Andersson, H.C.; FitzPatrick, D.R.; Kelley, R.I.; Wanders, R.J. Mutations in the 3beta-hydroxysterol Delta24-reductase gene cause desmosterolosis, an autosomal recessive disorder of cholesterol biosynthesis. Am. J. Hum. Genet. 2001, 69, 685–694. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.; Miloslavskaya, I.; Demontis, S.; Maestro, R.; Galaktionov, K. Regulation of cellular response to oncogenic and oxidative stress by Seladin-1. Nature 2004, 432, 640–645. [Google Scholar] [CrossRef] [Green Version]
- Tsukiyama-Kohara, K. Role of oxidative stress in hepatocarcinogenesis induced by hepatitis C virus. Int. J. Mol. Sci. 2012, 13, 15271–15278. [Google Scholar] [CrossRef] [Green Version]
- Lee, G.T.; Ha, Y.S.; Jung, Y.S.; Moon, S.K.; Kang, H.W.; Lee, O.J.; Joung, J.Y.; Choi, Y.H.; Yun, S.J.; Kim, W.J.; et al. DHCR24 is an independent predictor of progression in patients with non-muscle-invasive urothelial carcinoma, and its functional role is involved in the aggressive properties of urothelial carcinoma cells. Ann. Surg. Oncol. 2014, 21 (Suppl. S4), 538–545. [Google Scholar] [CrossRef]
- Dai, M.; Zhu, X.L.; Liu, F.; Xu, Q.Y.; Ge, Q.L.; Jiang, S.H.; Yang, X.M.; Li, J.; Wang, Y.H.; Wu, Q.K.; et al. Cholesterol Synthetase DHCR24 Induced by Insulin Aggravates Cancer Invasion and Progesterone Resistance in Endometrial Carcinoma. Sci. Rep. 2017, 7, 41404. [Google Scholar] [CrossRef]
- Luciani, P.; Ferruzzi, P.; Arnaldi, G.; Crescioli, C.; Benvenuti, S.; Nesi, G.; Valeri, A.; Greeve, I.; Serio, M.; Mannelli, M.; et al. Expression of the novel adrenocorticotropin-responsive gene selective Alzheimer’s disease indicator-1 in the normal adrenal cortex and in adrenocortical adenomas and carcinomas. J. Clin. Endocrinol. Metab. 2004, 89, 1332–1339. [Google Scholar] [CrossRef] [PubMed]
- Frances, D.; Sharma, N.; Pofahl, R.; Maneck, M.; Behrendt, K.; Reuter, K.; Krieg, T.; Klein, C.A.; Haase, I.; Niemann, C. A role for Rac1 activity in malignant progression of sebaceous skin tumors. Oncogene 2015, 34, 5505–5512. [Google Scholar] [CrossRef] [PubMed]
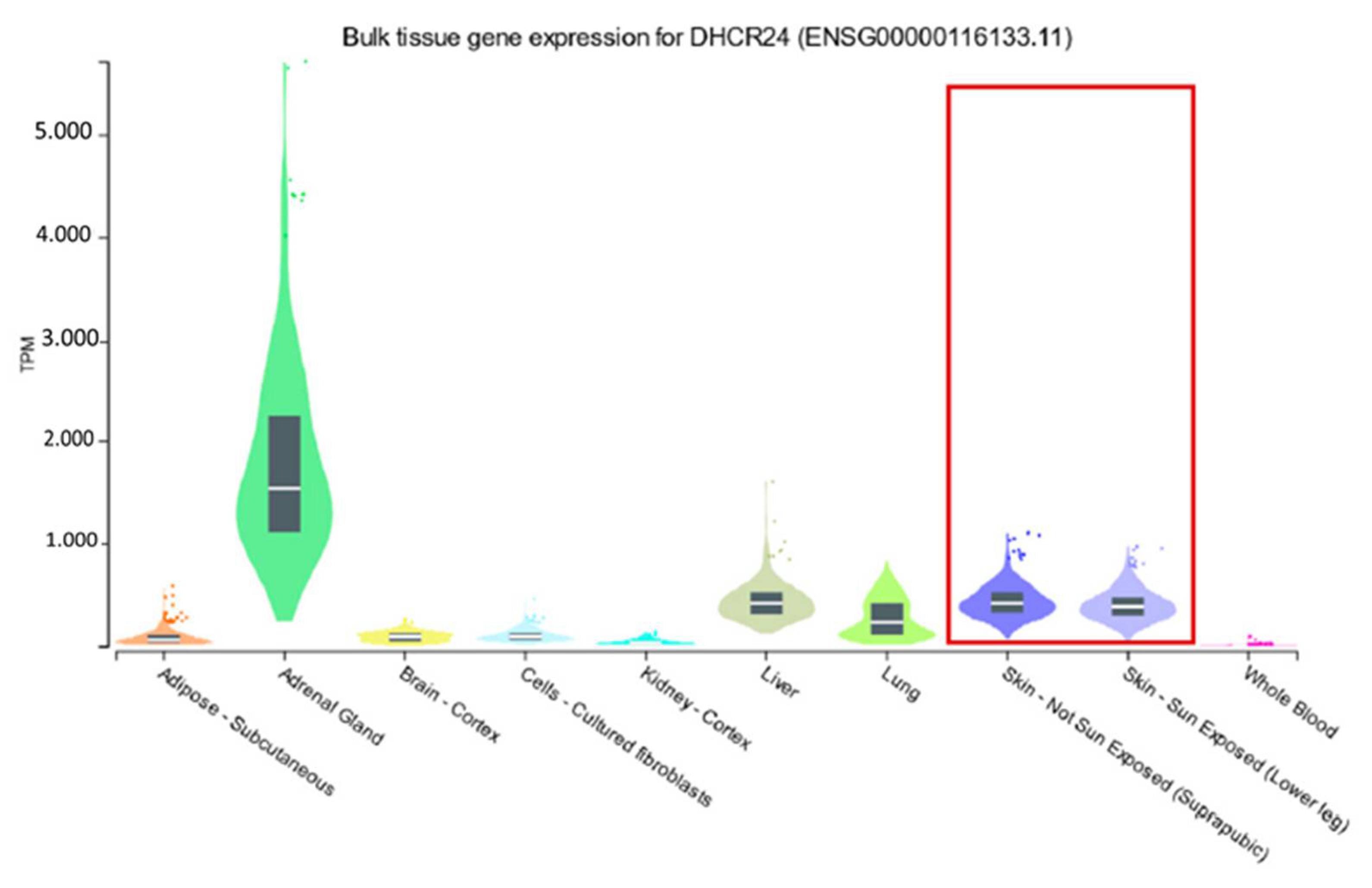
- Di Stasi, D.; Vallacchi, V.; Campi, V.; Ranzani, T.; Daniotti, M.; Chiodini, E.; Fiorentini, S.; Greeve, I.; Prinetti, A.; Rivoltini, L.; et al. DHCR24 gene expression is upregulated in melanoma metastases and associated to resistance to oxidative stress-induced apoptosis. Int. J. Cancer 2005, 115, 224–230. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, V.N.; Hei, T.K. Regulation of apoptosis in human melanoma and neuroblastoma cells by statins, sodium arsenite and TRAIL: A role of combined treatment versus monotherapy. Apoptosis 2011, 16, 1268–1284. [Google Scholar] [CrossRef]
- Hajar, R. Statins: Past and present. Heart Views 2011, 12, 121–127. [Google Scholar] [CrossRef]
- Dellavalle, R.P.; Nicholas, M.K.; Schilling, L.M. Melanoma chemoprevention: A role for statins or fibrates? Am. J. Ther. 2003, 10, 203–210. [Google Scholar] [CrossRef]
- Freeman, S.R.; Drake, A.L.; Heilig, L.F.; Graber, M.; McNealy, K.; Schilling, L.M.; Dellavalle, R.P. Statins, fibrates, and melanoma risk: A systematic review and meta-analysis. J. Natl. Cancer Inst. 2006, 98, 1538–1546. [Google Scholar] [CrossRef]
- Jacobs, E.J.; Newton, C.C.; Thun, M.J.; Gapstur, S.M. Long-term use of cholesterol-lowering drugs and cancer incidence in a large United States cohort. Cancer Res. 2011, 71, 1763–1771. [Google Scholar] [CrossRef] [Green Version]
- von Schuckmann, L.A.; Smith, D.; Hughes, M.C.B.; Malt, M.; van der Pols, J.C.; Khosrotehrani, K.; Smithers, B.M.; Green, A.C. Associations of Statins and Diabetes with Diagnosis of Ulcerated Cutaneous Melanoma. J. Investig. Dermatol. 2017, 137, 2599–2605. [Google Scholar] [CrossRef] [Green Version]
- Livingstone, E.; Hollestein, L.M.; van Herk-Sukel, M.P.; van de Poll-Franse, L.; Joosse, A.; Schilling, B.; Nijsten, T.; Schadendorf, D.; de Vries, E. Statin use and its effect on all-cause mortality of melanoma patients: A population-based Dutch cohort study. Cancer Med. 2014, 3, 1284–1293. [Google Scholar] [CrossRef]
- Stamatakos, S.; Beretta, G.L.; Vergani, E.; Dugo, M.; Corno, C.; Corna, E.; Tinelli, S.; Frigerio, S.; Ciusani, E.; Rodolfo, M.; et al. Deregulated FASN Expression in BRAF Inhibitor-Resistant Melanoma Cells Unveils New Targets for Drug Combinations. Cancers 2021, 13, 2284. [Google Scholar] [CrossRef] [PubMed]
- Muller, C.; Hemmers, S.; Bartl, N.; Plodek, A.; Korner, A.; Mirakaj, V.; Giera, M.; Bracher, F. New chemotype of selective and potent inhibitors of human delta 24-dehydrocholesterol reductase. Eur. J. Med. Chem. 2017, 140, 305–320. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | All Patients | rs7551288 Genotype | p * | |||
|---|---|---|---|---|---|---|
| A/A | A/G | G/G | ||||
| n = 556 (%) | n = 96 (%) | n = 284 (%) | n = 176 (%) | |||
| Gender | Male | 308 (55.4) | 54 (56.3) | 156 (54.9) | 98 (55.7) | 0.971 |
| Female | 248 (44.6) | 42 (43.7) | 128 (45.1) | 78 (44.3) | ||
| Age | 0–30 | 32 (5.7) | 3 (3.1) | 21 (7.4) | 8 (4.5) | 0.502 |
| 31–50 | 154 (27.7) | 28 (29.2) | 81 (28.5) | 45 (25.6) | ||
| 51–70 | 249 (44.8) | 46 (47.9) | 125 (44.0) | 78 (44.3) | ||
| >70 | 121 (21.8) | 19 (19.8) | 57 (20.1) | 45 (25.6) | ||
| Hair colour | red | 33 (5.9) | 9 (9.4) | 18 (6.3) | 6 (3.4) | 0.568 |
| blonde | 239 (43.0) | 38 (39.6) | 125 44.0() | 76 (43.2) | ||
| brown | 242 (43.5 | 41 (42.7) | 119 (41.9) | 82 (46.6) | ||
| black | 32 (5.8) | 6 (6.3) | 16 (5.6) | 10 (5.7) | ||
| na | 10 (1.8) | 2 (2.1) | 6 (2.1) | 2 (1.1) | ||
| Eye colour | blue | 218 (39.2) | 37 (38.5) | 114 (40.1) | 67 (38.1) | 0.863 |
| grey | 110 (19.8) | 22 (22.9) | 55 (19.4) | 33 (18.8) | ||
| green | 91 (16.4) | 18 (18.8) | 42 (14.8) | 31 (17.6) | ||
| brown | 133 (23.9) | 19 (19.8) | 72 (25.4) | 42 (23.9) | ||
| na | 4 (0.7) | - | 1 (0.4) | 3 (1.7) | ||
| UV tanning response | no tan | 97 (17.4) | 20 (20.8) | 52 (18.3) | 25 (14.2) | 0.400 |
| light tan | 271 (48.7) | 49 (51.0) | 141 (49.6) | 81 (46.0) | ||
| strong tan | 130 (23.4) | 20 (20.8) | 59 (20.8) | 51 (29.0) | ||
| always tan | 52 (9.4) | 7 (7.3) | 27 (9.5) | 18 (10.2) | ||
| na | 6 (1.1) | - | 5 (1.8) | 1 (0.6) | ||
| Total naevus count | 0–10 | 134 (24.1) | 18 (18.8) | 77 (27.1) | 39 (22.2) | 0.326 |
| 11–30 | 198 (35.6) | 43 (44.8) | 92 (32.4) | 63 (35.8) | ||
| 31–50 | 105 (18.9) | 18 (18.8) | 49 (17.3) | 38 (21.6) | ||
| 51–100 | 79 (14.2) | 13 (13.5) | 45 (15.8) | 21 (11.9) | ||
| >100 | 38 (6.8) | 4 (4.2) | 20 (7.0) | 14 (8.0) | ||
| na | 2 (0.4) | - | 1 (0.4) | 1 (0.6) | ||
| Obesity | BMI ≤ 30 | 426 (76.6) | 77 (80.2) | 220 (77.5) | 129 (73.3) | 0.606 |
| BMI > 30 | 100 (18.0) | 14 (14.6) | 53 (18.7) | 33 (18.8) | ||
| na | 30 (5.4) | 5 (5.2) | 11 (3.9) | 14 (8.0) | ||
| Second cancer | yes | 37 (6.7) | 7 (7.3) | 14 (4.9) | 16 (9.1) | 0.212 |
| no | 519 (93.3) | 89 (92.7) | 270 (95.1) | 160 (90.9) | ||
| Family history of Melanoma | yes | 29 (5.2) | 7 (7.3) | 9 (3.2) | 13 (7.4) | 0.094 |
| no | 508 (91.4) | 88 (91.7) | 263 (92.6) | 157 (89.2) | ||
| na | 19 (3.4) | 1 (1.0) | 12 (4.2) | 6 (3.4) | ||
| Characteristics | All Patients | rs7551288 Genotype | p * | |||
|---|---|---|---|---|---|---|
| A/A | A/G | G/G | ||||
| n = 556 (%) | n = 96 (%) | n = 284 (%) | n = 176 (%) | |||
| Tumour thickness | <1.0 mm | 127 (22.8) | 20 (20.8) | 69 (24.3) | 38 (21.6) | 0.034 |
| 1.01–2.0 mm | 213 (38.3) | 31 (32.3) | 100 (35.2) | 82 (46.6) | ||
| 2.01–4 mm | 126 (22.7) | 22 (22.9) | 73 (25.7) | 31 (17.6) | ||
| >4 mm | 53 (9.5) | 15 (15.6) | 26 (9.2) | 12 (6.8) | ||
| na | 37 (6.7) | 8 (8.3) | 16 (5.6) | 13 (7.4) | ||
| Histology | SSM | 307 (55.2) | 57 (59.4) | 149 (52.5) | 101 (57.4) | 0.287 |
| NM | 90 (16.2) | 12 (12.5) | 53 (18.7) | 25 (14.2) | ||
| LMM | 26 (4.7) | 5 (5.2) | 18 (6.3) | 3 (1.7) | ||
| ALM | 28 (5.0) | 6 (6.3) | 11 (3.9) | 11 (6.3) | ||
| others | 51 (9.2) | 9 (9.4) | 26 (9.2) | 16 (9.1) | ||
| na | 54 (9.7) | 7 (7.3) | 27 (9.5) | 20 (11.4) | ||
| Ulceration | no | 322 (57.9) | 58 (60.4) | 165 (58.1) | 99 (56.3) | 0.914 |
| yes | 122 (21.9) | 24 (25.0) | 62 (21.8) | 36 (20.5) | ||
| na | 112 (20.1) | 14 (14.6) | 57 (20.1) | 41 (23.3) | ||
| Stage at Diagnoses | Stage I | 289 (52.0) | 39 (40.6) | 147 (51.8) | 103 (58.5) | 0.041 |
| Stage II | 169 (30.4) | 31 (32.3) | 93 (32.7) | 45 (25.6) | ||
| Stage III | 81 (14.6) | 22 (22.9) | 36 (12.7) | 23 (13.1) | ||
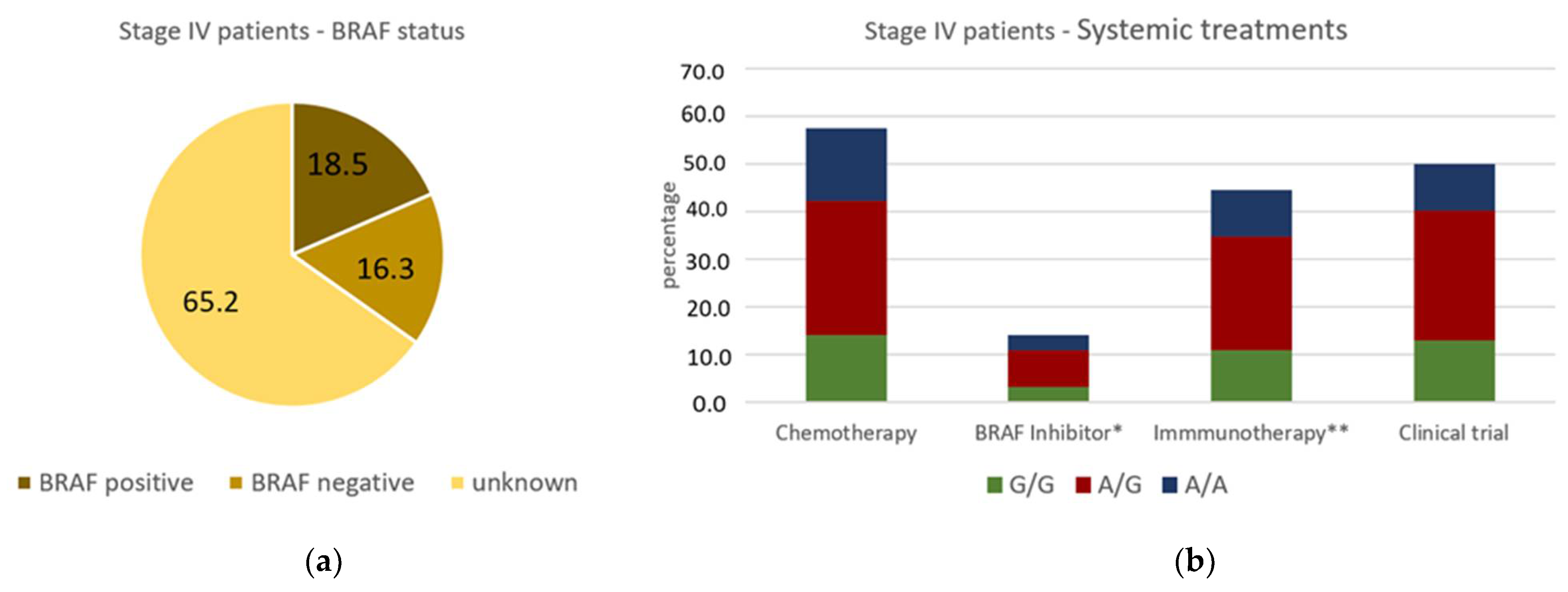
| Stage IV | 6 (1.1) | 0 (0) | 3 (1.1) | 3 (1.7) | ||
| na | 11 (2.0) | 4 (4.2) | 5 (1.8) | 2 (1.1) | ||
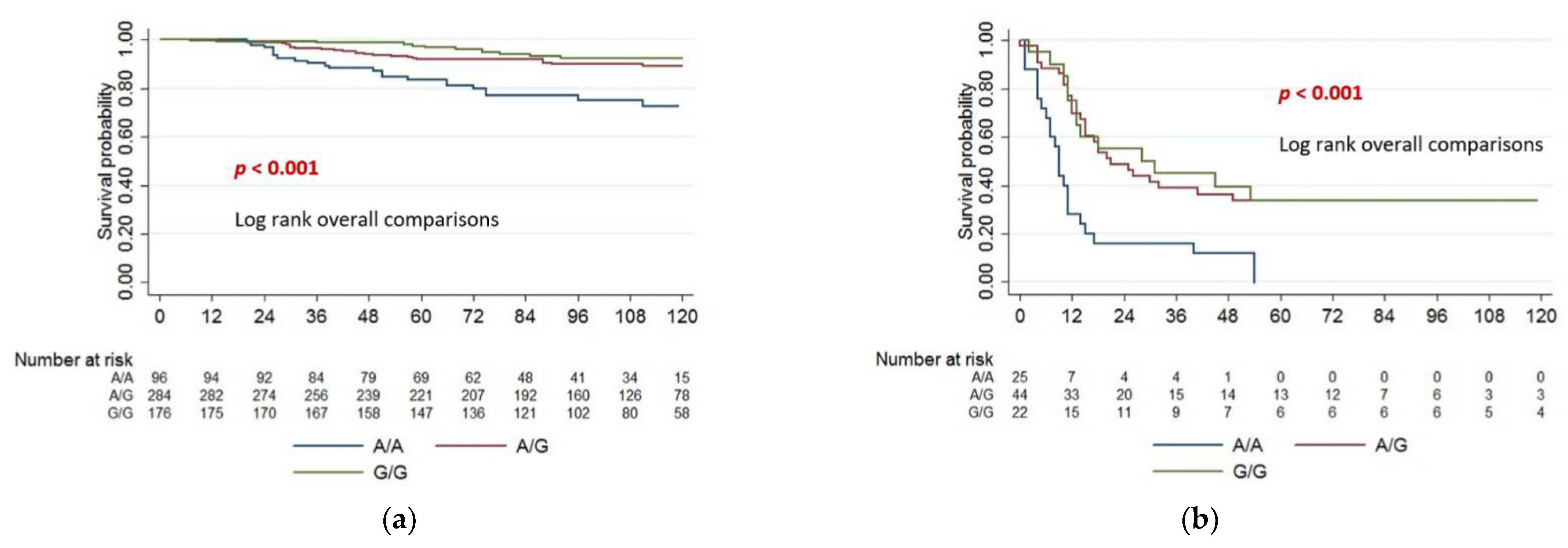
| Melanoma Death | yes | 64 (11.5) | 23 (24.0) | 28 (9.9) | 13 (7.4) | <0.001 |
| no | 492 (88.5) | 73 (76.0) | 256 (90.1) | 163 (92.6) | ||
| Overall Survival | |||||
|---|---|---|---|---|---|
| Definition | Univariate HR (95% CI) | p-Value | Multivariate HR (95% CI) | p-Value | |
| Tumour thickness | ≤2.0 mm | 1 | 1 | ||
| >2.0 mm | 1.84 (1.08–3.12) | 0.024 | 1.5 (0.84–2.74) | 0.164 | |
| Ulceration | no | 1 | 1 | ||
| yes | 2.49 (1.44–4.31) | 0.001 | 2.07 (1.15–3.73) | 0.016 | |
| Stage at Diagnoses | Stage I + II | 1 | 1 | ||
| Stage III + IV | 3.62 (2.16–6.08) | <0.001 | 2.37 (1.25–4.49) | 0.008 | |
| rs7551288 Genotype | G/G | 1 | 1 | ||
| G/A | 1.46 (0.76–2.83) | 0.26 | 2.13 (0.93–4.86) | 0.07 | |
| A/A | 3.95 (1.99–7.83) | <0.001 | 5.31 (2.30–12.25) | <0.001 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pflugfelder, A.; Yong, X.L.H.; Jagirdar, K.; Eigentler, T.K.; Soyer, H.P.; Sturm, R.A.; Flatz, L.; Duffy, D.L. Genome-Wide Association Study Suggests the Variant rs7551288*A within the DHCR24 Gene Is Associated with Poor Overall Survival in Melanoma Patients. Cancers 2022, 14, 2410. https://doi.org/10.3390/cancers14102410
Pflugfelder A, Yong XLH, Jagirdar K, Eigentler TK, Soyer HP, Sturm RA, Flatz L, Duffy DL. Genome-Wide Association Study Suggests the Variant rs7551288*A within the DHCR24 Gene Is Associated with Poor Overall Survival in Melanoma Patients. Cancers. 2022; 14(10):2410. https://doi.org/10.3390/cancers14102410
Chicago/Turabian StylePflugfelder, Annette, Xuan Ling Hilary Yong, Kasturee Jagirdar, Thomas K. Eigentler, H. Peter Soyer, Richard A. Sturm, Lukas Flatz, and David L. Duffy. 2022. "Genome-Wide Association Study Suggests the Variant rs7551288*A within the DHCR24 Gene Is Associated with Poor Overall Survival in Melanoma Patients" Cancers 14, no. 10: 2410. https://doi.org/10.3390/cancers14102410
APA StylePflugfelder, A., Yong, X. L. H., Jagirdar, K., Eigentler, T. K., Soyer, H. P., Sturm, R. A., Flatz, L., & Duffy, D. L. (2022). Genome-Wide Association Study Suggests the Variant rs7551288*A within the DHCR24 Gene Is Associated with Poor Overall Survival in Melanoma Patients. Cancers, 14(10), 2410. https://doi.org/10.3390/cancers14102410





