Auger Emitter Conjugated PARP Inhibitor for Therapy in Triple Negative Breast Cancers: A Comparative In-Vitro Study
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
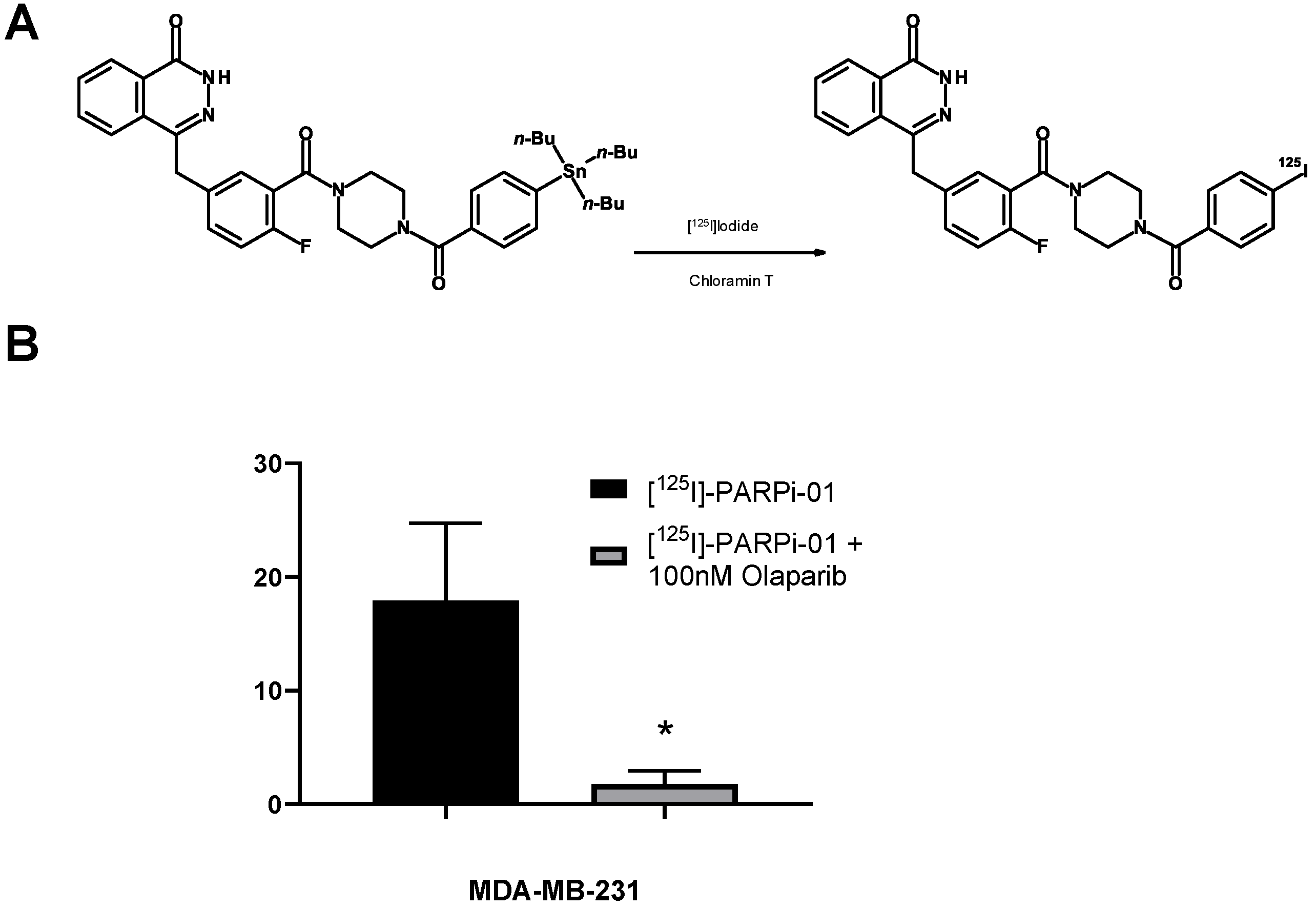
2.1. Synthesis and Characterization of PARP-01 Precursor
2.2. Radiosynthesis of [125I]-PARPi-01
2.3. Cell Culture
2.4. Treatment Strategies
2.5. Subcellular Fractionation
2.6. SDS/Western Blot
2.7. Cell Uptake Assay
2.8. Flow Cytometry
2.9. Microscopy
2.10. Colony Formation Assay
2.11. Statistical Analysis
3. Results
3.1. [125I]-PARPi-01 Is Specific to PARP1
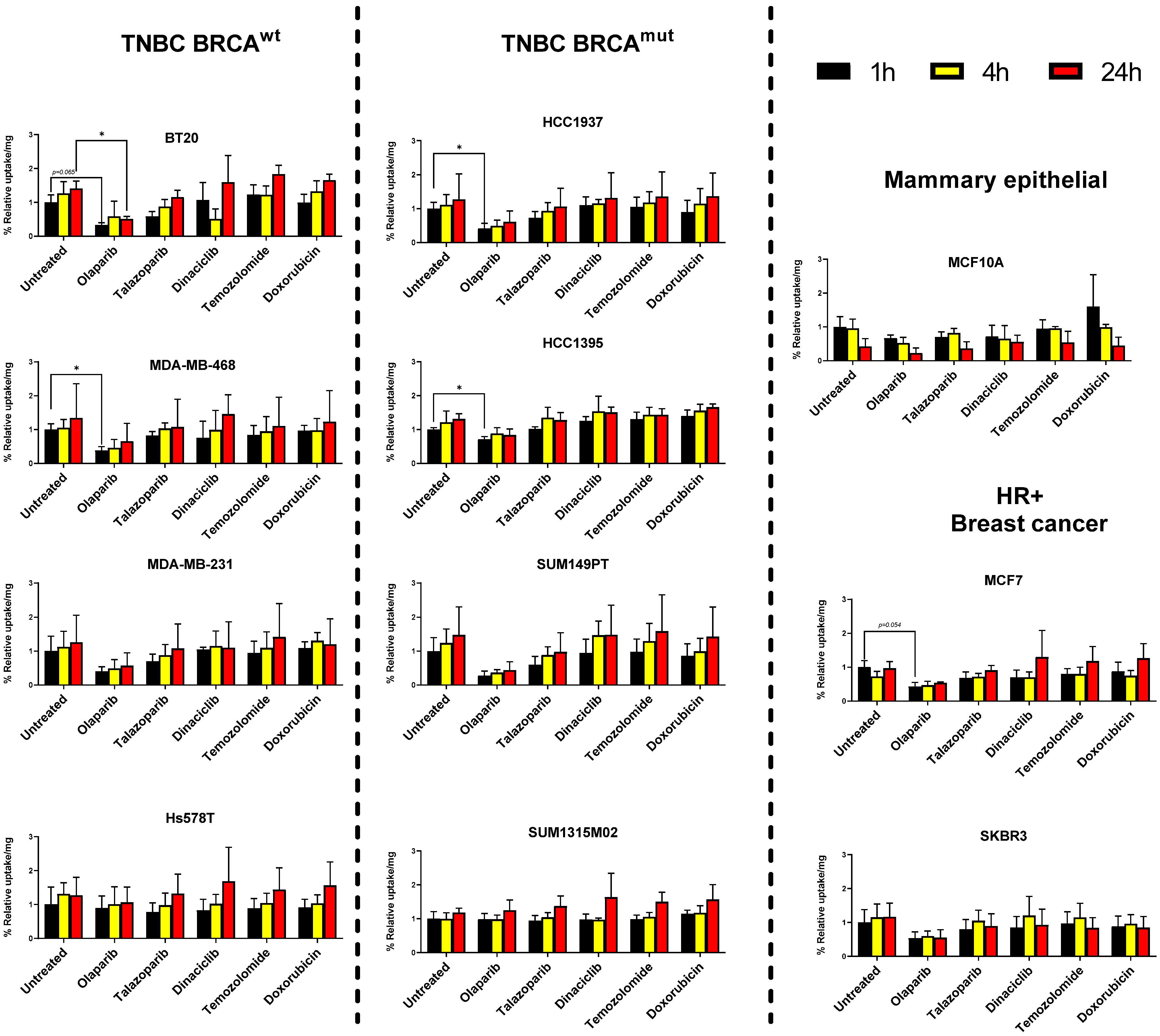
3.2. Breast Cancer Cell Lines but Not Mammary Epithelial Cell Line Show Retention of [125I]-PARPi-01
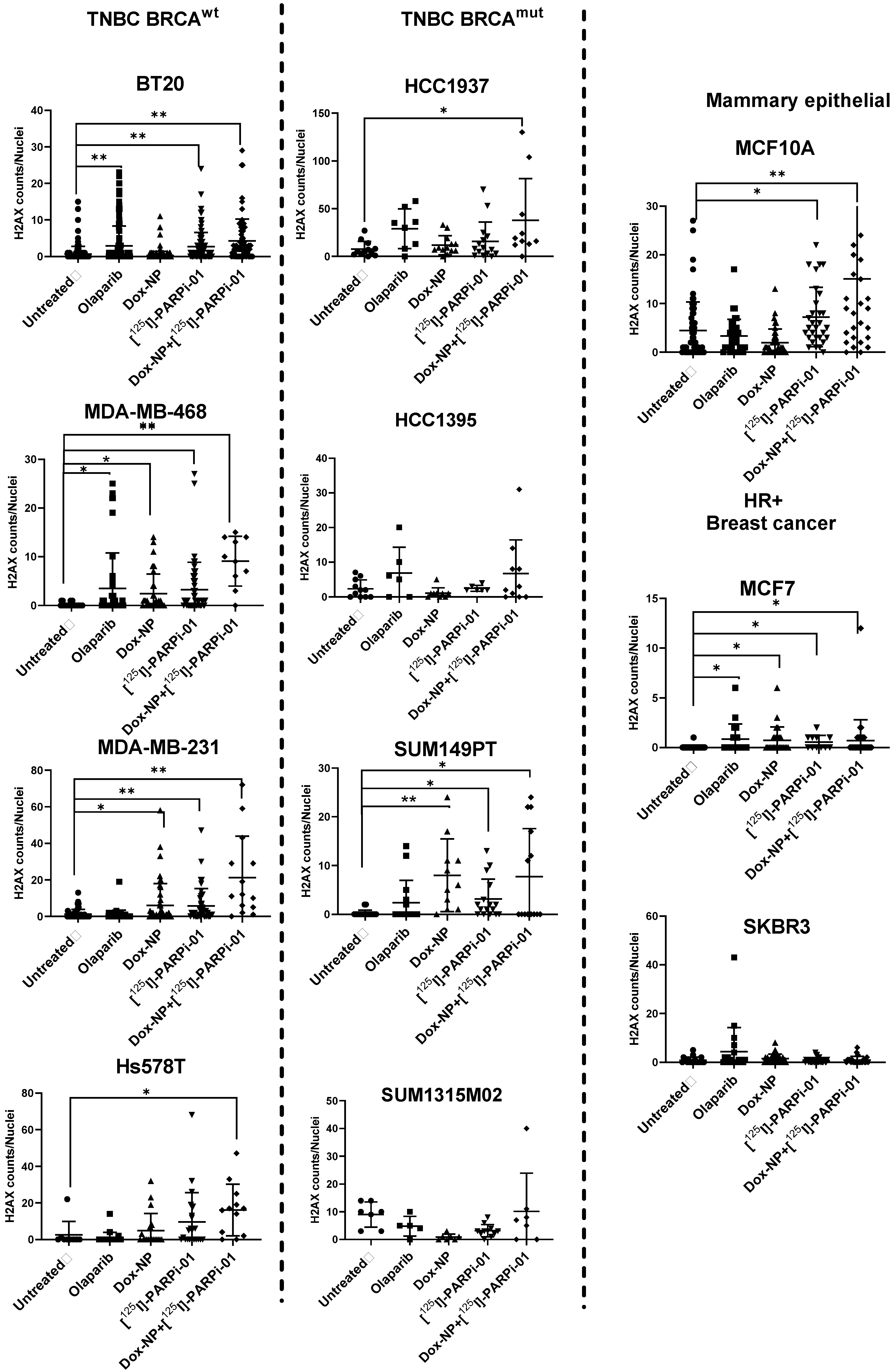
3.3. Dox-NP Combination with [125I]-PARPi-01 Causes High DNA Damage (γH2AX Foci) in Resistant Breast Cancer Cell Lines
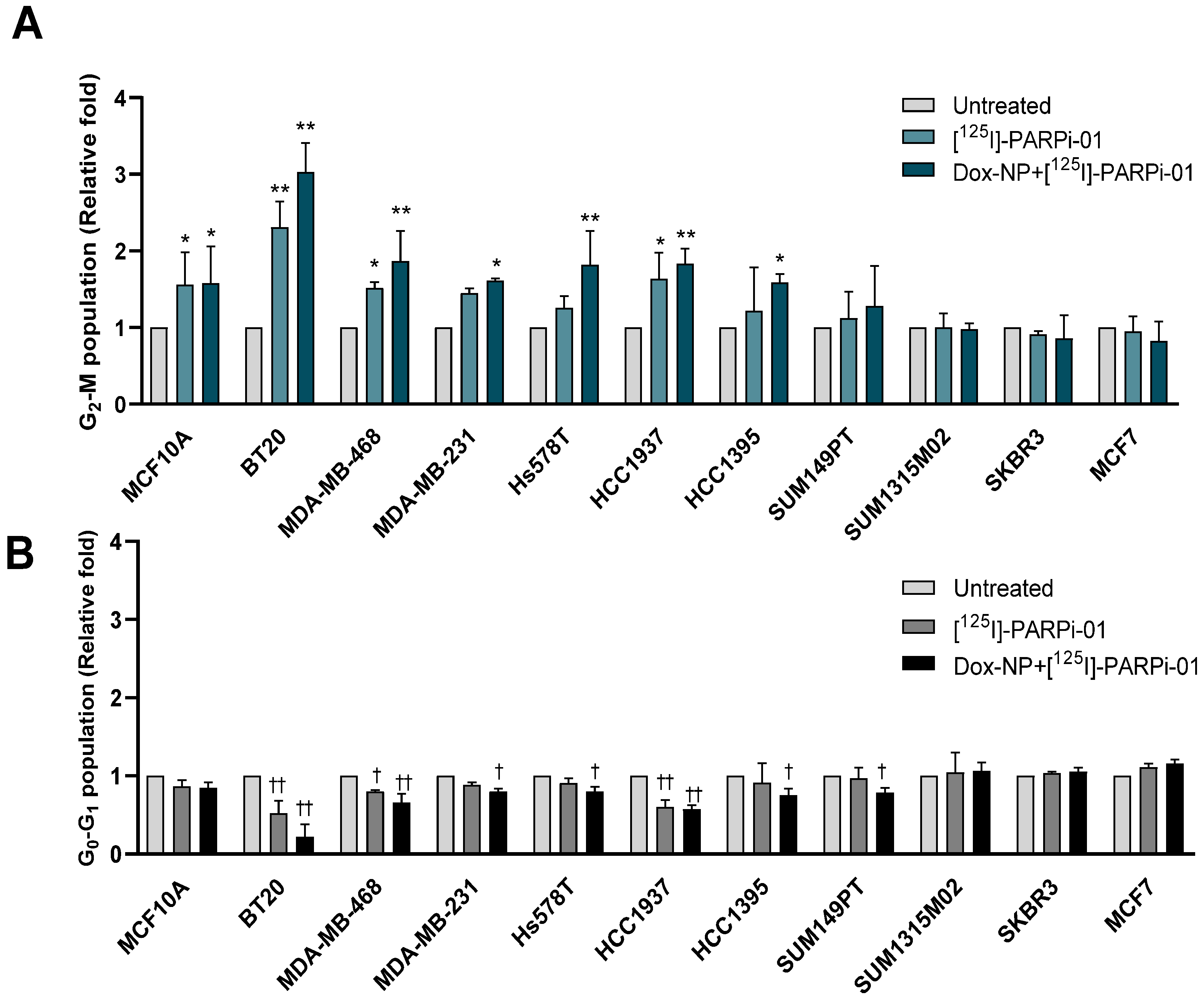
3.4. Pre-Treatment with Dox-NP Significantly Enhances Mitotic Phase Arrest in TNBC Cells
3.5. Dox-NP Pretreatment with [125I]-PARPi-01 Leads Responsive Cells to Apoptosis
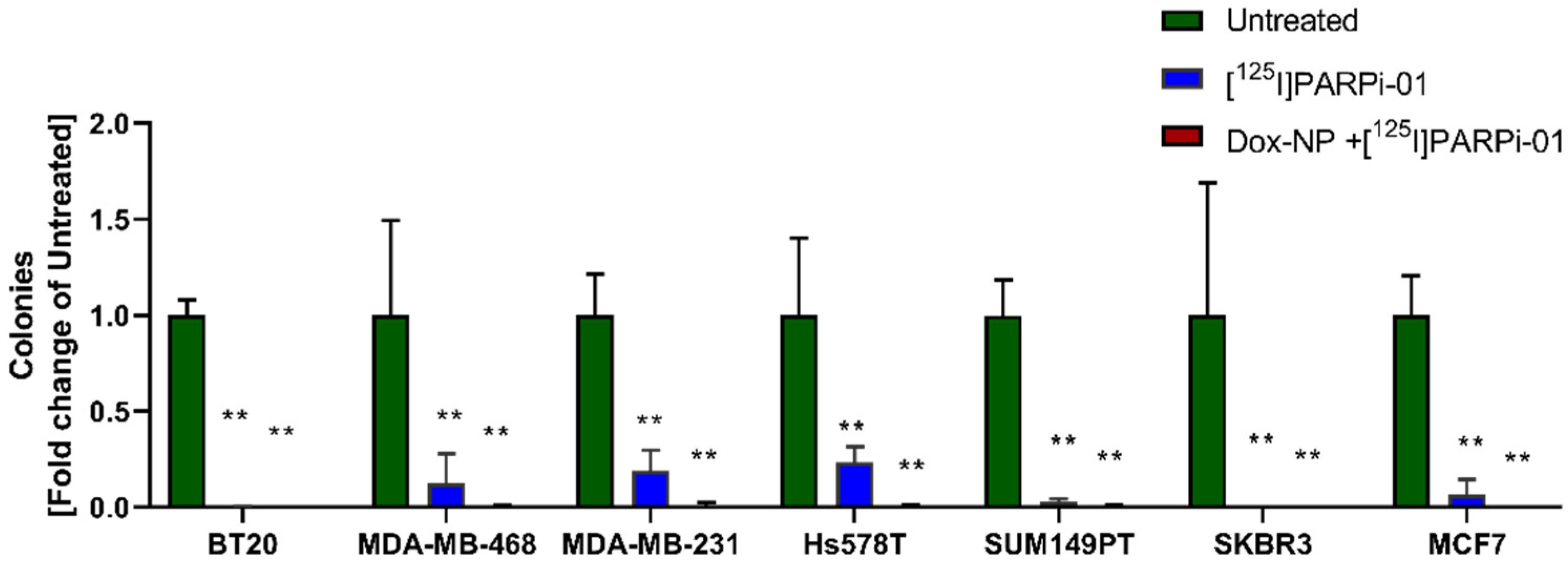
3.6. [125I]-PARPi-01 Significantly Reduces Tumorigenicity in All Colony Forming Cancer Cell Lines
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics. CA Cancer J. Clin. 2021, 71, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef]
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Holgado, E.; et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): A randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef]
- Bardia, A.; Mayer, I.A.; Vahdat, L.T.; Tolaney, S.M.; Isakoff, S.J.; Diamond, J.R.; O’Shaughnessy, J.; Moroose, R.L.; Santin, A.D.; Abramson, V.G.; et al. Sacituzumab Govitecan-hziy in Refractory Metastatic Triple-Negative Breast Cancer. N. Engl. J. Med. 2019, 380, 741–751. [Google Scholar] [CrossRef] [PubMed]
- Seligson, J.M.; Patron, A.M.; Berger, M.J.; Harvey, R.D.; Seligson, N.D. Sacituzumab Govitecan-hziy: An Antibody-Drug Conjugate for the Treatment of Refractory, Metastatic, Triple-Negative Breast Cancer. Ann. Pharmacother. 2020, 55, 921–931. [Google Scholar] [CrossRef]
- Robson, M.; Im, S.-A.; Senkus, E.; Xu, B.; Domchek, S.M.; Masuda, N.; Delaloge, S.; Li, W.; Tung, N.; Armstrong, A.; et al. Olaparib for Metastatic Breast Cancer in Patients with a Germline BRCA Mutation. N. Engl. J. Med. 2017, 377, 523–533. [Google Scholar] [CrossRef] [PubMed]
- Litton, J.K.; Rugo, H.S.; Ettl, J.; Hurvitz, S.A.; Gonçalves, A.; Lee, K.-H.; Fehrenbacher, L.; Yerushalmi, R.; Mina, L.A.; Martin, M.; et al. Talazoparib in Patients with Advanced Breast Cancer and a Germline BRCA Mutation. N. Engl. J. Med. 2018, 379, 753–763. [Google Scholar] [CrossRef]
- Lyons, T.G. Targeted Therapies for Triple-Negative Breast Cancer. Curr. Treat. Options Oncol. 2019, 20, 82. [Google Scholar] [CrossRef] [PubMed]
- Gupta, G.K.; Collier, A.L.; Lee, D.; Hoefer, R.A.; Zheleva, V.; van Reesema, L.L.S.; Tang-Tan, A.M.; Guye, M.L.; Chang, D.Z.; Winston, J.S.; et al. Perspectives on triple-negative breast cancer: Current treatment strategies, unmet needs, and potential targets for future therapies. Cancers 2020, 12, 2392. [Google Scholar] [CrossRef]
- Liedtke, C.; Mazouni, C.; Hess, K.R.; André, F.; Tordai, A.; Mejia, J.A.; Symmans, W.F.; Gonzalez-Angulo, A.M.; Hennessy, B.; Green, M.; et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 2008, 26, 1275–1281. [Google Scholar] [CrossRef]
- Pistelli, M.; Pagliacci, A.; Battelli, N.; Santinelli, A.; Biscotti, T.; Ballatore, Z.; Berardi, R.; Cascinu, S. Prognostic factors in early-stage triple-negative breast cancer: Lessons and limits from clinical practice. Anticancer Res. 2013, 33, 2737–2742. [Google Scholar]
- Kalimutho, M.; Parsons, K.; Mittal, D.; López, J.A.; Srihari, S.; Khanna, K.K. Targeted Therapies for Triple-Negative Breast Cancer: Combating a Stubborn Disease. Trends Pharmacol. Sci. 2015, 36, 822–846. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, A.R.; Nussenzweig, A. The multifaceted roles of PARP1 in DNA repair and chromatin remodelling. Nat. Rev. Mol. Cell Biol. 2017, 18, 610–621. [Google Scholar] [CrossRef] [PubMed]
- Chalakur-Ramireddy, N.K.R.; Pakala, S.B. Combined drug therapeutic strategies for the effective treatment of Triple Negative Breast Cancer. Biosci. Rep. 2018, 38, BSR20171357. [Google Scholar] [CrossRef] [PubMed]
- Pogoda, K.; Niwińska, A.; Sarnowska, E.; Nowakowska, D.; Jagiełło-Gruszfeld, A.; Siedlecki, J.; Nowecki, Z. Effects of BRCA Germline Mutations on Triple-Negative Breast Cancer Prognosis. J. Oncol. 2020, 2020, 8545643. [Google Scholar] [CrossRef] [PubMed]
- Anders, C.K.; Winer, E.P.; Ford, J.M.; Dent, R.; Silver, D.P.; Sledge, G.W.; Carey, L.A. Poly(ADP-ribose) polymerase inhibition: “Targeted” therapy for triple-negative breast cancer. Clin. Cancer Res. 2010, 16, 4702–4710. [Google Scholar] [CrossRef] [PubMed]
- Beniey, M.; Haque, T.; Hassan, S. Translating the role of PARP inhibitors in triple-negative breast cancer. Oncoscience 2019, 6, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Gelmon, K.A.; Tischkowitz, M.; Mackay, H.; Swenerton, K.; Robidoux, A.; Tonkin, K.; Hirte, H.; Huntsman, D.; Clemons, M.; Gilks, B.; et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: A phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011, 12, 852–861. [Google Scholar] [CrossRef]
- Adelstein, S.J.; Kassis, A.I. Strand breaks in plasmid DNA following positional changes of Auger-electron-emitting radionuclides. Acta Oncol. 1996, 35, 797–801. [Google Scholar] [CrossRef]
- Unverricht-Yeboah, M.; Holtmann, K.; Kriehuber, R. Comet Assay analysis of DNA strand breaks after exposure to the DNA-incorporated Auger Electron Emitter Iodine-125. Int. J. Radiat. Biol. 2020, 1–9. [Google Scholar] [CrossRef]
- Balagurumoorthy, P.; Xu, X.; Wang, K.; Adelstein, S.J.; Kassis, A.I.; Balagurumoorthy, P. Effect of distance between decaying 125 I and DNA on Auger-electron induced double-strand break yield. Int. J. Radiat. Biol. 2012, 88, 998–1008. [Google Scholar] [CrossRef]
- Ku, A.; Facca, V.J.; Cai, Z.; Reilly, R.M. Auger electrons for cancer therapy—A review. EJNMMI Radiopharm. Chem. 2019, 4, 27. [Google Scholar] [CrossRef]
- Terraneo, N.; Jacob, F.; Dubrovska, A.; Grünberg, J. Novel Therapeutic Strategies for Ovarian Cancer Stem Cells. Front. Oncol. 2020, 10, 319. [Google Scholar] [CrossRef]
- Wilson, T.C.; Jannetti, S.A.; Guru, N.; Pillarsetty, N.; Reiner, T.; Pirovano, G. Improved radiosynthesis of 123I-MAPi, an Auger theranostic agent. Int. J. Radiat. Biol. 2020, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Murai, J.; Zhang, Y.; Morris, J.; Ji, J.; Takeda, S.; Doroshow, J.H.; Pommier, Y. Rationale for poly(ADP-ribose) polymerase (PARP) inhibitors in combination therapy with camptothecins or temozolomide based on PARP trapping versus catalytic inhibition. J. Pharmacol. Exp. Ther. 2014, 349, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Del Conte, G.; Sessa, C.; von Moos, R.; Viganò, L.; Digena, T.; Locatelli, A.; Gallerani, E.; Fasolo, A.; Tessari, A.; Cathomas, R.; et al. Phase I study of Olaparib in combination with liposomal doxorubicin in patients with advanced solid tumours. Br. J. Cancer 2014, 111, 651–659. [Google Scholar] [CrossRef] [PubMed]
- Carey, J.P.W.; Karakas, C.; Bui, T.; Chen, X.; Vijayaraghavan, S.; Zhao, Y.; Wang, J.; Mikule, K.; Litton, J.K.; Hunt, K.K.; et al. Synthetic Lethality of PARP Inhibitors in Combination with MYC Blockade Is Independent of BRCA Status in Triple-Negative Breast Cancer. Cancer Res. 2017, 78, 742–757. [Google Scholar] [CrossRef]
- Xiao, G.; Lundine, D.; Annor, G.K.; Canar, J.; Ellison, V.; Polotskaia, A.; Donabedian, P.L.; Reiner, T.; Khramtsova, G.F.; Olopade, O.I.; et al. Gain-of-function mutant p53 R273H interacts with replicating DNA and PARP1 in breast cancer. Cancer Res. 2020, 80, 394–405. [Google Scholar] [CrossRef] [PubMed]
- Węsierska-Gądek, J.; Mauritz, M.; Mitulovic, G.; Cupo, M. Differential Potential of Pharmacological PARP Inhibitors for Inhibiting Cell Proliferation and Inducing Apoptosis in Human Breast Cancer Cells. J. Cell. Biochem. 2015, 116, 2824–2839. [Google Scholar] [CrossRef]
- Keung, M.Y.; Wu, Y.; Badar, F.; Vadgama, J.V. Response of Breast Cancer Cells to PARP Inhibitors Is Independent of BRCA Status. J. Clin. Med. 2020, 9, 940. [Google Scholar] [CrossRef]
- Minami, A.; Nakanishi, A.; Ogura, Y.; Kitagishi, Y.; Matsuda, S. Connection between tumor suppressor BRCA1 and PTEN in damaged DNA repair. Front. Oncol. 2014, 4, 318. [Google Scholar] [CrossRef]
- Wojdacz, T.K.; Dobrovic, A. Methylation-sensitive high resolution melting (MS-HRM): A new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007, 35, e41. [Google Scholar] [CrossRef]
- Paranjpe, A.; Bailey, N.I.; Konduri, S.; Bobustuc, G.C.; Ali-Osman, F.; Yusuf, M.A.; Punganuru, S.R.; Madala, H.R.; Basak, D.; Mostofa, A.G.M.; et al. New insights into estrogenic regulation of O6-methylguanine DNA-methyltransferase (MGMT) in human breast cancer cells: Co-degradation of ER-α and MGMT proteins by fulvestrant or O6-benzylguanine indicates fresh avenues for therapy. J. Biomed. Res. 2016, 30, 393–410. [Google Scholar] [CrossRef]
- Clements, K.E.; Thakar, T.; Nicolae, C.M.; Liang, X.; Wang, H.G.; Moldovan, G.L. Loss of E2F7 confers resistance to poly-ADP-ribose polymerase (PARP) inhibitors in BRCA2-deficient cells. Nucleic Acids Res. 2018, 46, 8898–8907. [Google Scholar] [CrossRef] [PubMed]
- Kodaz, H.; Hastanesi, A.E.; Klinigi, T.O.; Kostek, O.; Bekir Hacioglu, M.; Erdogan, B.; Elpen Kodaz, C.; Hacibekiroglu, I.; Turkmen, E.; Uzunoglu, S.; et al. Frequency of Ras Mutations (Kras, Nras, Hras) in Human Solid Cancer. EJMO 2017, 1, 1–7. [Google Scholar] [CrossRef]
- Patra, S.; Young, V.; Llewellyn, L.; Senapati, J.N.; Mathew, J. BRAF, KRAS and PIK3CA mutation and sensitivity to Trastuzumab in breast cancer cell line model. Asian Pac. J. Cancer Prev. 2017, 18, 2209–2213. [Google Scholar] [CrossRef] [PubMed]
- Koh, M.S.; Moon, A. Activation of H-Ras and Rac1 correlates with epidermal growth factor-induced invasion in Hs578T and MDA-MB-231 breast carcinoma cells. Biochem. Biophys. Res. Commun. 2011, 406, 25–29. [Google Scholar] [CrossRef]
- Wu, X.; Zahari, M.S.; Renuse, S.; Kelkar, D.S.; Bharbuiya, M.A.; Rojas, P.L.; Stearns, V.; Gabrielson, E.; Malla, P.; Sukumar, S.; et al. The non-receptor tyrosine kinase TNK2/ACK1 is a novel therapeutic target in triple negative breast cancer. Oncotarget 2017, 8, 2971–2983. [Google Scholar] [CrossRef] [PubMed]
- Tassone, P.; Tagliaferri, P.; Perricelli, A.; Blotta, S.; Quaresima, B.; Martelli, M.L.; Goel, A.; Barbieri, V.; Costanzo, F.; Boland, C.R.; et al. BRCA1 expression modulates chemosensitivity of BRCA1-defective HCC1937 human breast cancer cells. Br. J. Cancer 2003, 88, 1285–1291. [Google Scholar] [CrossRef]
- Sachdev, E.; Tabatabai, R.; Roy, V.; Rimel, B.J.; Mita, M.M. PARP Inhibition in Cancer: An Update on Clinical Development. Target. Oncol. 2019, 14, 657–679. [Google Scholar] [CrossRef]
- McGrail, D.J.; Lin, C.C.-J.; Garnett, J.; Liu, Q.; Mo, W.; Dai, H.; Lu, Y.; Yu, Q.; Ju, Z.; Yin, J.; et al. Improved prediction of PARP inhibitor response and identification of synergizing agents through use of a novel gene expression signature generation algorithm. NPJ Syst. Biol. Appl. 2017, 3, 1–12. [Google Scholar] [CrossRef]
- Shen, C.J.; Minn, I.; Hobbs, R.F.; Chen, Y.; Josefsson, A.; Brummet, M.; Banerjee, S.R.; Brayton, C.F.; Mease, R.C.; Pomper, M.G.; et al. Auger radiopharmaceutical therapy targeting prostate-specific membrane antigen in a micrometastatic model of prostate cancer. Theranostics 2020, 10, 2888–2896. [Google Scholar] [CrossRef] [PubMed]
- Rosenkranz, A.A.; Slastnikova, T.A.; Karmakova, T.A.; Vorontsova, M.S.; Morozova, N.B.; Petriev, V.M.; Abrosimov, A.S.; Khramtsov, Y.V.; Lupanova, T.N.; Ulasov, A.V.; et al. Antitumor activity of Auger electron emitter 111In delivered by modular nanotransporter for treatment of bladder cancer with EGFR overexpression. Front. Pharmacol. 2018, 9, 1331. [Google Scholar] [CrossRef] [PubMed]
- Osytek, K.M.; Blower, P.J.; Costa, I.M.; Smith, G.E.; Abbate, V.; Terry, S.Y.A. In vitro proof of concept studies of radiotoxicity from Auger electron-emitter thallium-201. EJNMMI Res. 2021, 11, 63. [Google Scholar] [CrossRef] [PubMed]
- Pirovano, G.; Jannetti, S.A.; Carter, L.M.; Sadique, A.; Kossatz, S.; Guru, N.; Demétrio De Souza França, P.; Maeda, M.; Zeglis, B.M.; Lewis, J.S.; et al. Targeted brain tumor radiotherapy using an Auger emitter. Clin. Cancer Res. 2020, 26, 2871–2881. [Google Scholar] [CrossRef]
- Riad, A.; Gitto, S.B.; Lee, H.; Winters, H.D.; Martorano, P.M.; Hsieh, C.-J.; Xu, K.; Omran, D.K.; Powell, D.J.; Mach, R.H.; et al. PARP Theranostic Auger Emitters Are Cytotoxic in BRCA Mutant Ovarian Cancer and Viable Tumors from Ovarian Cancer Patients Enable Ex-Vivo Screening of Tumor Response. Molecules 2020, 25, 6029. [Google Scholar] [CrossRef]
- Manjunath, M.; Choudhary, B. Triple-negative breast cancer: A run-through of features, classification and current therapies. Oncol. Lett. 2021, 22, 512. [Google Scholar] [CrossRef] [PubMed]
- Varanda, A.B.; Martins-Logrado, A.; Ferreira, M.G.; Fior, R. Zebrafish xenografts unveil sensitivity to Olaparib beyond BRCA status. Cancers 2020, 12, 1769. [Google Scholar] [CrossRef]
- Rajkumar-Calkins, A.S.; Szalat, R.; Dreze, M.; Khan, I.; Frazier, Z.; Reznichenkov, E.; Schnorenberg, M.R.; Tsai, Y.-F.; Nguyen, H.; Kochupurakkal, B.; et al. Functional Profiling of Nucleotide Excision Repair in Breast Cancer. DNA Repair 2019, 82, 102697. [Google Scholar] [CrossRef]
- Birkbak, N.J.; Wang, Z.C.; Kim, J.-Y.; Eklund, A.C.; Li, Q.; Tian, R.; Bowman-Colin, C.; Li, Y.; Greene-Colozzi, A.; Iglehart, J.D.; et al. Telomeric allelic imbalance indicates defective DNA repair and sensitivity to DNA-damaging agents. Cancer Discov. 2012, 2, 366–375. [Google Scholar] [CrossRef]
- Mansour, W.Y.; Tennstedt, P.; Volquardsen, J.; Oing, C.; Kluth, M.; Hube-Magg, C.; Borgmann, K.; Simon, R.; Petersen, C.; Dikomey, E.; et al. Loss of PTEN-assisted G2/M checkpoint impedes homologous recombination repair and enhances radio-curability and PARP inhibitor treatment response in prostate cancer. Sci. Rep. 2018, 8, 3947. [Google Scholar] [CrossRef]
- Mendes-Pereira, A.M.; Martin, S.A.; Brough, R.; McCarthy, A.; Taylor, J.R.; Kim, J.S.; Waldman, T.; Lord, C.J.; Ashworth, A. Synthetic lethal targeting of PTEN mutant cells with PARP inhibitors. EMBO Mol. Med. 2009, 1, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Holme, H.; Gulati, A.; Brough, R.; Fleuren, E.D.G.; Bajrami, I.; Campbell, J.; Chong, I.Y.; Costa-Cabral, S.; Elliott, R.; Fenton, T.; et al. Chemosensitivity profiling of osteosarcoma tumour cell lines identifies a model of BRCAness. Sci. Rep. 2018, 8, 10614. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Chapman, J.H.; Custance, M.F.; Tricola, G.M.; Jones, C.E.; Furano, A.V. Perturbation of base excision repair sensitizes breast cancer cells to APOBEC3 deaminase-mediated mutations. Elife 2020, 9, e51605. [Google Scholar] [CrossRef] [PubMed]
- Teraoka, S.; Muguruma, M.; Takano, N.; Miyahara, K.; Kawate, T.; Kaise, H.; Yamada, K.; Miyazawa, K.; Ishikawa, T. Association of BRCA Mutations and BRCAness Status With Anticancer Drug Sensitivities in Triple-Negative Breast Cancer Cell Lines. J. Surg. Res. 2020, 250, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Gu, Y.; Helenius, M.; Väänänen, K.; Bulanova, D.; Saarela, J.; Sokolenko, A.; Martens, J.; Imyanitov, E.; Kuznetsov, S. BRCA1-deficient breast cancer cell lines are resistant to MEK inhibitors and show distinct sensitivities to 6-thioguanine OPEN. Nat. Publ. Gr. 2016, 6, 28217. [Google Scholar] [CrossRef]
- Gu, Y.; Wang, C.; Zhu, R.; Yang, J.; Yuan, W.; Zhu, Y.; Zhou, Y.; Qin, N.; Shen, H.; Ma, H.; et al. The cancer-testis gene, MEIOB, sensitizes triple-negative breast cancer to PARP1 inhibitors by inducing homologous recombination deficiency. Cancer Biol. Med. 2021, 18, 74. [Google Scholar] [CrossRef]
- Eikesdal, H.P.; Yndestad, S.; Elzawahry, A.; Llop-Guevara, A.; Gilje, B.; Blix, E.S.; Espelid, H.; Lundgren, S.; Geisler, J.; Vagstad, G.; et al. Olaparib monotherapy as primary treatment in unselected triple negative breast cancer. Ann. Oncol. 2021, 32, 240–249. [Google Scholar] [CrossRef]
- Wagener-Ryczek, S.; Merkelbach-Bruse, S.; Siemanowski, J. Biomarkers for homologous recombination deficiency in cancer. J. Pers. Med. 2021, 11, 612. [Google Scholar] [CrossRef]
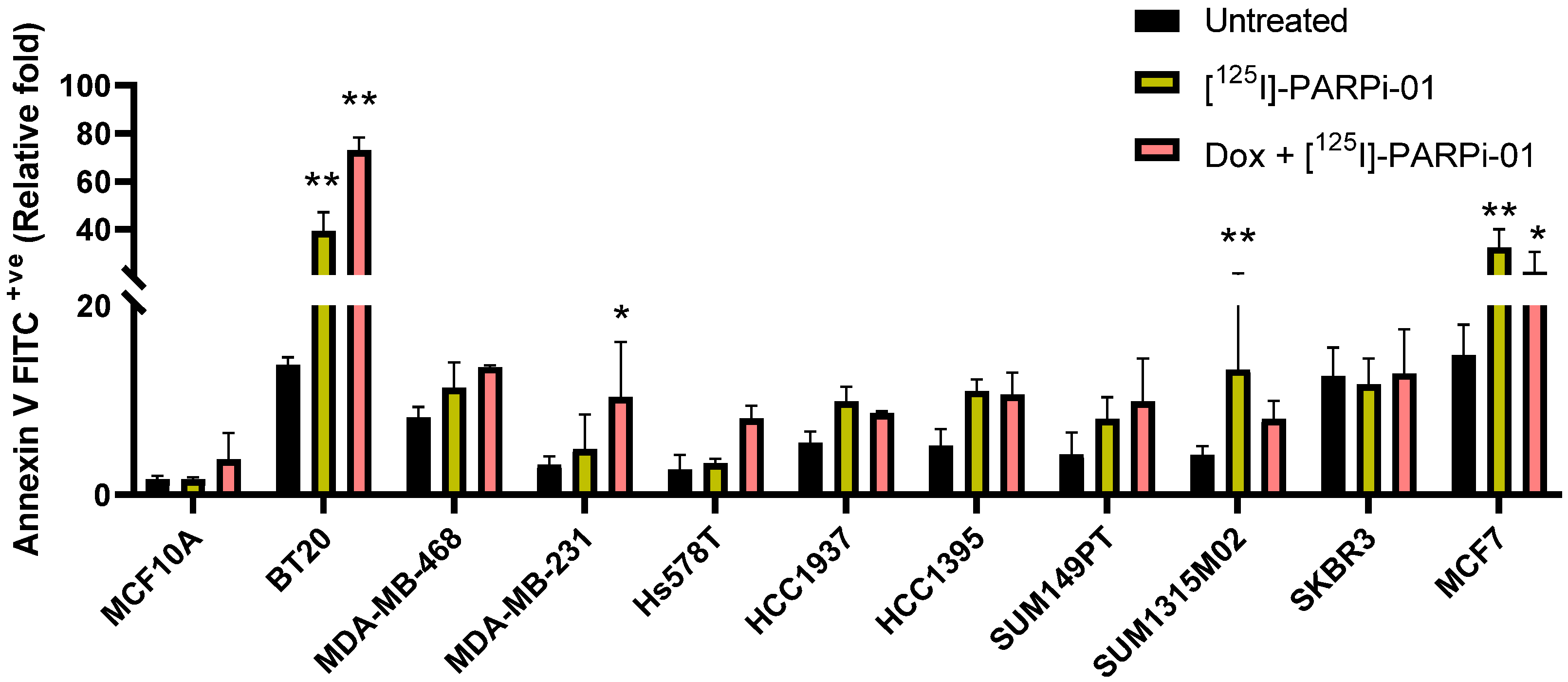
| [125I]-PARPi-01 | Dox+[125I]-PARPi-01 | ||||
|---|---|---|---|---|---|
| G2-M Arrest | Annexin V + ve | G2-M Arrest | Annexin V + ve | ||
| Control | MCF10A | 1.5 ± 0.4 | 1 ± 0.12 | 1.6 ± 0.5 | 2.27 ± 1.68 |
| TNBC BRCAwt | BT20 | 2.3 ± 0.3 | 2.86 ± 0.57 | 3.1 ± 0.4 | 5.30 ± 0.40 |
| MDA-MB-468 | 1.5 ± 0.1 | 1.38 ± 0.33 | 1.9 ± 0.4 | 1.65 ± 0.02 | |
| MDA-MB-231 | 1.4 ± 0.1 | 1.53 ± 1.14 | 1.6 ± 0.1 | 3.24 ± 1.84 | |
| Hs578T | 1.2 ± 0.1 | 1.25 ± 0.16 | 1.8 ± 0.4 | 3 ± 0.5 | |
| TNBC BRCAmut | HCC1937 | 1.6 ± 0.3 | 1.79 ± 0.27 | 1.8 ± 0.2 | 1.56 ± 0.03 |
| HCC1395 | 1.1 ± 0.7 | 2.09 ± 0.31 | 1.6 ± 0.1 | 1.88 ± 0.39 | |
| SUM149PT | 1.1 ± 0.3 | 1.88 ± 0.54 | 1.3 ± 0.5 | 2.31 ± 1.06 | |
| SUM1315M02 | 1.0 ± 0.2 | 3.16 ± 2.02 | 1.0 ± 0.1 | 1.91 ± 0.46 | |
| HER2+ve BRCAwt | SKBR3 | 0.9 ± 0.1 | 0.93 ± 0.22 | 0.9 ± 0.3 | 1.02 ± 0.37 |
| MCF7 | 0.8 ± 0.1 | 2.15 ± 0.70 | 0.8 ± 0.3 | 1.23 ± 0.36 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sankaranarayanan, R.A.; Peil, J.; Vogg, A.T.J.; Bolm, C.; Terhorst, S.; Classen, A.; Bauwens, M.; Maurer, J.; Mottaghy, F.; Morgenroth, A. Auger Emitter Conjugated PARP Inhibitor for Therapy in Triple Negative Breast Cancers: A Comparative In-Vitro Study. Cancers 2022, 14, 230. https://doi.org/10.3390/cancers14010230
Sankaranarayanan RA, Peil J, Vogg ATJ, Bolm C, Terhorst S, Classen A, Bauwens M, Maurer J, Mottaghy F, Morgenroth A. Auger Emitter Conjugated PARP Inhibitor for Therapy in Triple Negative Breast Cancers: A Comparative In-Vitro Study. Cancers. 2022; 14(1):230. https://doi.org/10.3390/cancers14010230
Chicago/Turabian StyleSankaranarayanan, Ramya Ambur, Jennifer Peil, Andreas T. J. Vogg, Carsten Bolm, Steven Terhorst, Arno Classen, Matthias Bauwens, Jochen Maurer, Felix Mottaghy, and Agnieszka Morgenroth. 2022. "Auger Emitter Conjugated PARP Inhibitor for Therapy in Triple Negative Breast Cancers: A Comparative In-Vitro Study" Cancers 14, no. 1: 230. https://doi.org/10.3390/cancers14010230
APA StyleSankaranarayanan, R. A., Peil, J., Vogg, A. T. J., Bolm, C., Terhorst, S., Classen, A., Bauwens, M., Maurer, J., Mottaghy, F., & Morgenroth, A. (2022). Auger Emitter Conjugated PARP Inhibitor for Therapy in Triple Negative Breast Cancers: A Comparative In-Vitro Study. Cancers, 14(1), 230. https://doi.org/10.3390/cancers14010230







