Clinical Implication of Liquid Biopsy in Colorectal Cancer Patients Treated with Metastasectomy
Abstract
Simple Summary
Abstract
1. Introduction
2. Patients and Methods
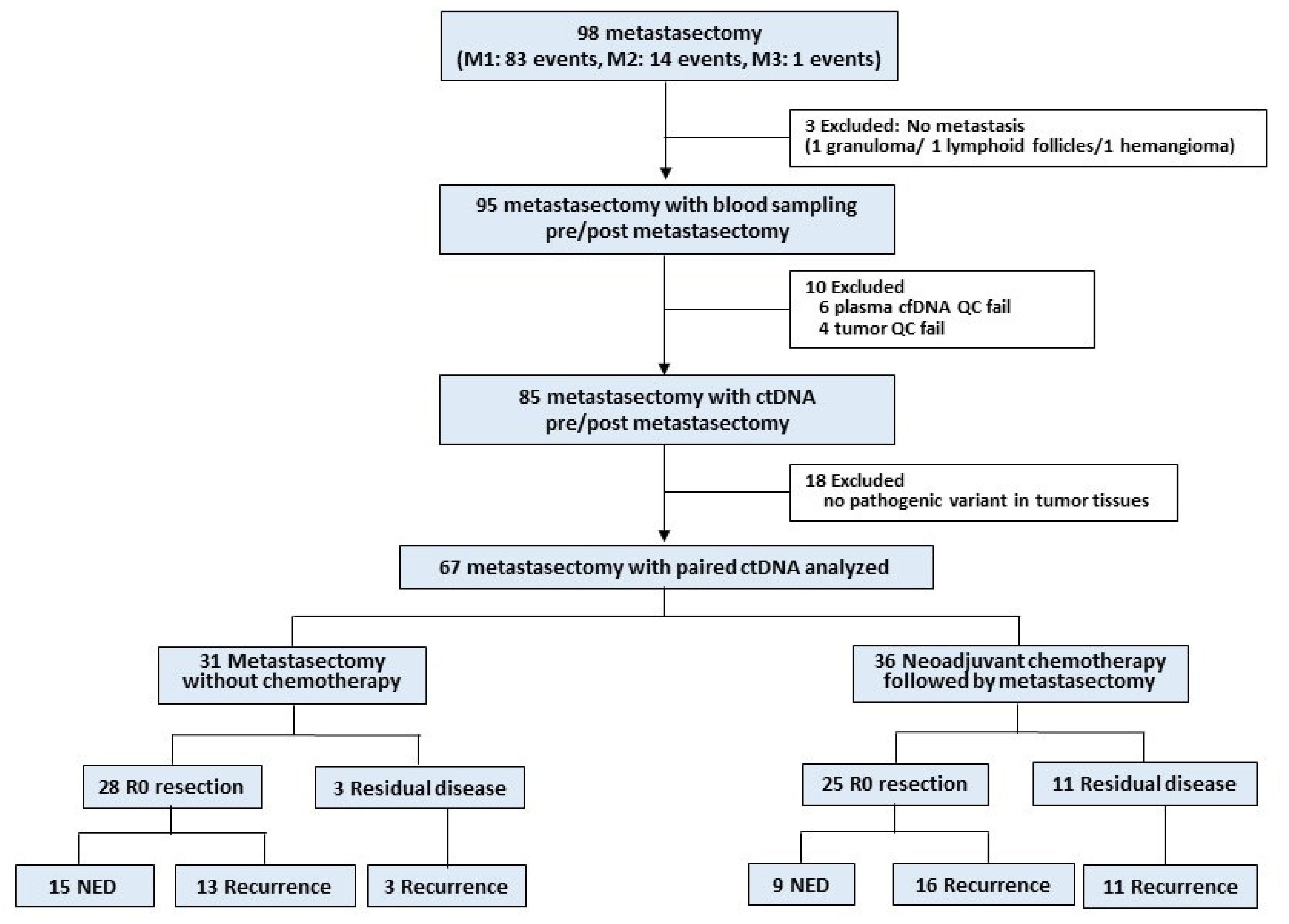
2.1. Study Design and Participants
2.2. Tumor Tissue and Blood Collection and Mutation Analysis by Next-Generation Sequencing
2.3. Tumor-Guided ctDNA Identification
2.4. Statistical Analysis
3. Results
3.1. Patient Characteristics
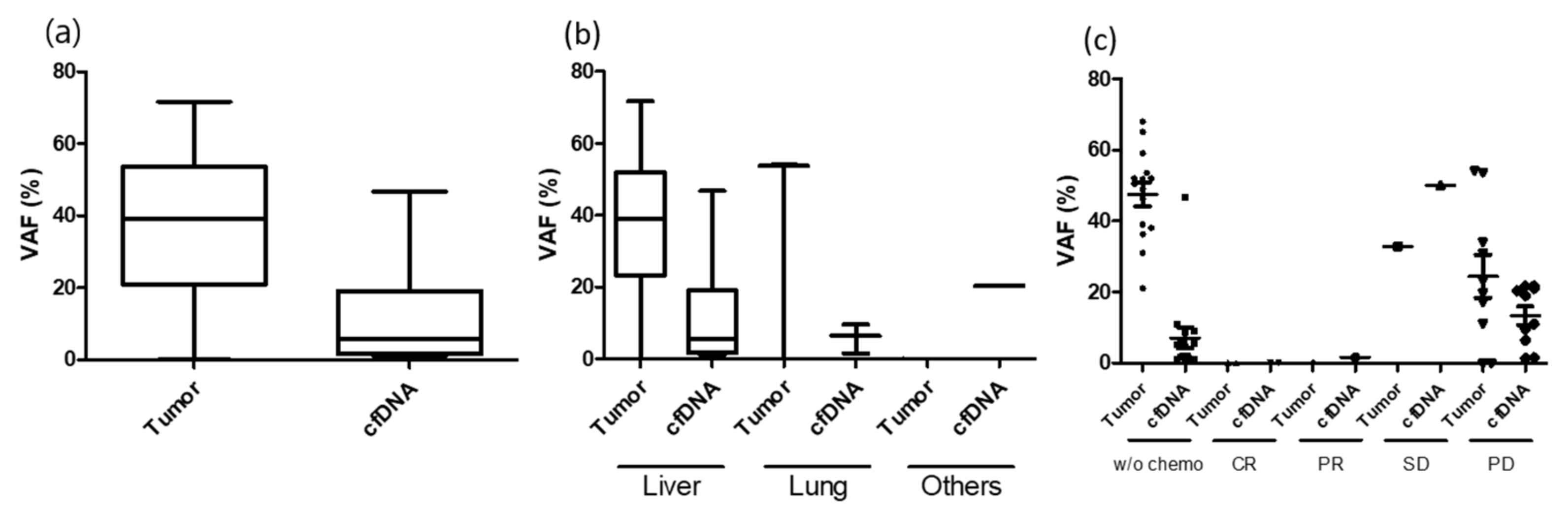
3.2. ctDNA Detection before and after Metastasectomy
3.3. Group 1: Upfront Metastasectomy without Neoadjuvant Chemotherapy (R0 Resection = 28 Cases)
3.4. Group 2: Upfront Metastasectomy without Neoadjuvant Chemotherapy (R0 Resection Failed = 3 Cases)
3.5. Group 3: Neoadjuvant Chemotherapy Followed by Metastasectomy (R0 Resection = 25 Cases)
3.6. Group 4: Neoadjuvant Chemotherapy Followed by Metastasectomy (R0 Resection Failed = 11)
3.7. Longitudinal Tracking with Serial ctDNA Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hong, S.; Won, Y.J.; Park, Y.R.; Jung, K.W.; Kong, H.J.; Lee, E.S. The Community of Population-Based Regional Cancer Registries. Cancer Statistics in Korea: Incidence, Mortality, Survival, and Prevalence in 2017. Cancer Res. Treat. 2020, 52, 335–350. [Google Scholar] [CrossRef]
- Van Cutsem, E.; Nordlinger, B.; Adam, R.; Köhne, C.-H.; Pozzo, C.; Poston, G.; Ychou, M.; Rougier, P. Towards a pan-European consensus on the treatment of patients with colorectal liver metastases. Eur. J. Cancer 2006, 42, 2212–2221. [Google Scholar] [CrossRef] [PubMed]
- Biasco, G.; Derenzini, E.; Grazi, G.; Ercolani, G.; Ravaioli, M.; Pantaleo, M.; Brandi, G. Treatment of hepatic metastases from colorectal cancer: Many doubts, some certainties. Cancer Treat. Rev. 2006, 32, 214–228. [Google Scholar] [CrossRef]
- Garden, O.J.; Rees, M.; Poston, G.J.; Mirza, D.; Saunders, M.; Ledermann, J.; Primrose, J.N.; Parks, R.W. Guidelines for resection of colorectal cancer liver metastases. Gut 2006, 55 (Suppl. 3), iii1–iii8. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.-S.; Yun, S.H.; Chun, H.-K.; Lee, W.-Y.; Yun, H.-R.; Kim, J.; Kim, K.; Shim, Y.M. Pulmonary resection for metastases from colorectal cancer: Prognostic factors and survival. Int. J. Colorectal Dis. 2007, 22, 699–704. [Google Scholar] [CrossRef]
- Kemeny, N.E.; Chou, J.F.; Boucher, T.M.; Capanu, M.; DeMatteo, R.P.; Jarnagin, W.R.; Allen, P.J.; Fong, Y.C.; Cercek, A.; D’Angelica, M.I. Updated long-term survival for patients with metastatic colorectal cancer treated with liver resection followed by hepatic arterial infusion and systemic chemotherapy. J. Surg. Oncol. 2016, 113, 477–484. [Google Scholar] [CrossRef]
- Penna, C.; Nordlinger, B. Colorectal metastasis (liver and lung). Surg. Clin. N. Am. 2002, 82, 1075–1090. [Google Scholar] [CrossRef]
- Merker, J.D.; Oxnard, G.R.; Compton, C.; Diehn, M.; Hurley, P.; Lazar, A.J.; Lindeman, N.; Lockwood, C.M.; Rai, A.J.; Schilsky, R.L. Circulating tumor DNA analysis in patients with cancer: American Society of Clinical Oncology and College of American Pathologists joint review. J. Clin. Oncol. 2018, 36, 1631–1641. [Google Scholar] [CrossRef] [PubMed]
- Bach, S.; Sluiter, N.R.; Beagan, J.J.; Mekke, J.M.; Ket, J.C.; van Grieken, N.C.; Steenbergen, R.D.; Ylstra, B.; Kazemier, G.; Tuynman, J.B. Circulating tumor DNA analysis: Clinical implications for colorectal cancer patients. A systematic review. JNCI Cancer Spectr. 2019, 3, pkz042. [Google Scholar] [CrossRef]
- Tie, J.; Kinde, I.; Wang, Y.; Wong, H.-L.; Roebert, J.; Christie, M.; Tacey, M.; Wong, R.; Singh, M.; Karapetis, C. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann. Oncol. 2015, 26, 1715–1722. [Google Scholar] [CrossRef]
- Wang, Y.; Li, L.; Cohen, J.D.; Kinde, I.; Ptak, J.; Popoli, M.; Schaefer, J.; Silliman, N.; Dobbyn, L.; Tie, J.; et al. Prognostic Potential of Circulating Tumor DNA Measurement in Postoperative Surveillance of Nonmetastatic Colorectal Cancer. JAMA Oncol. 2019, 5, 1118–1123. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2019, 68, 663–671. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Wang, Y.; Christie, M.; Simons, K.; Lee, M.; Wong, R.; Kosmider, S.; Ananda, S.; McKendrick, J.; et al. Circulating Tumor DNA Analyses as Markers of Recurrence Risk and Benefit of Adjuvant Therapy for Stage III Colon Cancer. JAMA Oncol. 2019, 5, 1710–1717. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Wang, Y.; Tomasetti, C.; Li, L.; Springer, S.; Kinde, I.; Silliman, N.; Tacey, M.; Wong, H.-L.; Christie, M. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci. Transl. Med. 2016, 8, 346ra392. [Google Scholar] [CrossRef]
- Carrato, A. Adjuvant treatment of colorectal cancer. Gastrointest. Cancer Res. 2008, 2, S42–S46. [Google Scholar]
- Grávalos, C.; García-Escobar, I.; García-Alfonso, P.; Cassinello, J.; Malón, D.; Carrato, A. Adjuvant chemotherapy for stages II, III and IV of colon cancer. Clin. Transl. Oncol. 2009, 11, 526–533. [Google Scholar] [CrossRef]
- Schøler, L.V.; Reinert, T.; Ørntoft, M.-B.W.; Kassentoft, C.G.; Árnadóttir, S.S.; Vang, S.; Nordentoft, I.; Knudsen, M.; Lamy, P.; Andreasen, D. Clinical implications of monitoring circulating tumor DNA in patients with colorectal cancer. Clin. Cancer Res. 2017, 23, 5437–5445. [Google Scholar] [CrossRef]
- Tie, J.; Cohen, J.D.; Lo, S.N.; Wang, Y.; Li, L.; Christie, M.; Lee, M.; Wong, R.; Kosmider, S.; Skinner, I.; et al. Prognostic significance of postsurgery circulating tumor DNA in nonmetastatic colorectal cancer: Individual patient pooled analysis of three cohort studies. Int. J. Cancer 2020. [Google Scholar] [CrossRef]
- Sung, J.S.; Chong, H.Y.; Kwon, N.J.; Kim, H.M.; Lee, J.W.; Kim, B.; Lee, S.B.; Park, C.W.; Choi, J.Y.; Chang, W.J.; et al. Detection of somatic variants and EGFR mutations in cell-free DNA from non-small cell lung cancer patients by ultra-deep sequencing using the ion ampliseq cancer hotspot panel and droplet digital polymerase chain reaction. Oncotarget 2017, 8, 106901–106912. [Google Scholar] [CrossRef]
- Genovese, G.; Kahler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef]
- Pecuchet, N.; Zonta, E.; Didelot, A.; Combe, P.; Thibault, C.; Gibault, L.; Lours, C.; Rozenholc, Y.; Taly, V.; Laurent-Puig, P.; et al. Base-Position Error Rate Analysis of Next-Generation Sequencing Applied to Circulating Tumor DNA in Non-Small Cell Lung Cancer: A Prospective Study. PLoS Med. 2016, 13, e1002199. [Google Scholar] [CrossRef] [PubMed]
- Santiago-Walker, A.; Gagnon, R.; Mazumdar, J.; Casey, M.; Long, G.V.; Schadendorf, D.; Flaherty, K.; Kefford, R.; Hauschild, A.; Hwu, P.; et al. Correlation of BRAF Mutation Status in Circulating-Free DNA and Tumor and Association with Clinical Outcome across Four BRAFi and MEKi Clinical Trials. Clin. Cancer Res. 2016, 22, 567–574. [Google Scholar] [CrossRef]
- Reinert, T.; Henriksen, T.V.; Christensen, E.; Sharma, S.; Salari, R.; Sethi, H.; Knudsen, M.; Nordentoft, I.; Wu, H.T.; Tin, A.S.; et al. Analysis of Plasma Cell-Free DNA by Ultradeep Sequencing in Patients With Stages I to III Colorectal Cancer. JAMA Oncol. 2019. [Google Scholar] [CrossRef]
- Levy, M.; Benesova, L.; Lipska, L.; Belsanova, B.; Minarikova, P.; Veprekova, G.; Zavoral, M.; Minarik, M. Utility of cell-free tumour DNA for post-surgical follow-up of colorectal cancer patients. Anticancer Res. 2012, 32, 1621–1626. [Google Scholar]
- Flamini, E.; Mercatali, L.; Nanni, O.; Calistri, D.; Nunziatini, R.; Zoli, W.; Rosetti, P.; Gardini, N.; Lattuneddu, A.; Verdecchia, G.M.; et al. Free DNA and carcinoembryonic antigen serum levels: An important combination for diagnosis of colorectal cancer. Clin. Cancer Res. 2006, 12, 6985–6988. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef] [PubMed]
- Avanzini, S.; Kurtz, D.M.; Chabon, J.J.; Moding, E.J.; Hori, S.S.; Gambhir, S.S.; Alizadeh, A.A.; Diehn, M.; Reiter, J.G. A mathematical model of ctDNA shedding predicts tumor detection size. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef]
- Oikkonen, J.; Zhang, K.; Salminen, L.; Schulman, I.; Lavikka, K.; Andersson, N.; Ojanpera, E.; Hietanen, S.; Grenman, S.; Lehtonen, R.; et al. Prospective Longitudinal ctDNA Workflow Reveals Clinically Actionable Alterations in Ovarian Cancer. JCO Precis. Oncol. 2019, 3, 1–12. [Google Scholar] [CrossRef]
- Garlan, F.; Laurent-Puig, P.; Sefrioui, D.; Siauve, N.; Didelot, A.; Sarafan-Vasseur, N.; Michel, P.; Perkins, G.; Mulot, C.; Blons, H.; et al. Early Evaluation of Circulating Tumor DNA as Marker of Therapeutic Efficacy in Metastatic Colorectal Cancer Patients (PLACOL Study). Clin. Cancer Res. 2017, 23, 5416–5425. [Google Scholar] [CrossRef]
- Katoh, M.; Unakami, M.; Hara, M.; Fukuchi, S. Bone metastasis from colorectal cancer in autopsy cases. J. Gastroenterol. 1995, 30, 615–618. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef]
| Clinical Variables | Npatient = 58 (%) | |
|---|---|---|
| Age years (median, range) | 56 (35–78) | |
| Gender | Male | 37 (63.8) |
| Female | 21 (36.2) | |
| Primary tumor site | Ascending colon | 8 (13.8) |
| Descending colon | 20 (34.5) | |
| Rectum | 30 (51.7) | |
| Primary tumor surgery | Yes | 57 (98.3%) |
| No | 1 (1.7%) | |
| Synchronicity of metastasis | Metachronous | 18 (31.1) |
| Synchronous | 40 (68.9) | |
| Metastasectomy organ | Liver | 38 (65.5) |
| Lung | 25 (43.1) | |
| Peritoneum | 4 (6.9) | |
| Lymph node | 2 (3.4) | |
| No | Synchronicity | Variant | Primary Tissue VAF | Metastatic Tissue VAF | Metastatic Site | PreM ctDNA VAF | PostM ctDNA VAF | Recurence | |
|---|---|---|---|---|---|---|---|---|---|
| No CTx | CTx | ||||||||
| ctDNA positive before metastasectomy | |||||||||
| 020b | M | TP53 R196 * | 38.5 | 65.1 | Liver 2.5 cm | 1.2 | 0.0 | N | |
| APC Q1294 * | 25.4 | 51.6 | 0.0 | 0.0 | |||||
| 037b | M | TP53 T211I | 14.9 | 59.1 | Liver 2.5 cm | 46.8 | 0.0 | N | |
| 073 | M | APC T1445Qfs | 21.0 | 26.0 | Liver 7.4 cm | 0.0 | 0.0 | N | |
| APC R876 * | 31.2 | 0.0 | 0.0 | 0.0 | |||||
| TP53 R248W | 23.0 | 21.0 | 1.1 | 0.0 | |||||
| 025a | M | FXW7 R465C | 33.0 | 33.4 | Liver 2.1 cm | 0.0 | 0.0 | N | |
| KRAS G12D | 23.8 | 28.4 | 0.0 | 0.0 | |||||
| TP53 R196 * | 35.9 | 51.8 | 2.1 | 0.0 | |||||
| APC L1488fs | 26.1 | 0 | 0.0 | 0.0 | |||||
| 025b | M | FXW7 R465C | 33.0 | 31.0 | Liver 1.5 cm | 1.1 | 0.0 | Liver LN | |
| KRAS G12D | 23.8 | 36.3 | 2.0 | 0.0 | |||||
| TP53 R196 * | 35.9 | 49.0 | 1.8 | 0.0 | |||||
| APC L1488fs | 26.1 | 0 | 0.0 | 0.0 | |||||
| 050 | M | APC Q1367 * | 32.6 | 34.0 | Liver 3.0 cm | 0.0 | 0.0 | Liver | |
| TP53 R237H | 26.2 | 39.0 | 1.0 | 0.0 | |||||
| BRAF G469E | 0.0 | 1.3 | 0.0 | 0.0 | |||||
| CTNNB1 G34E | 0.0 | 1.4 | 0.0 | 0.0 | |||||
| VHL R161 * | 0.0 | 1.4 | 0.0 | 0.0 | |||||
| 062 | M | APC R1450 * | 35.3 | 52.0 | Liver 3.2 cm | 5.9 | 0.0 | Lung | |
| KRAS G12S | 55.0 | 68.0 | 8.6 | 0.0 | |||||
| TP53 R342 * | 50.9 | 52.0 | 9.0 | 0.0 | |||||
| 092 | M | SMAD4 G419R | QC failed * | 53.6 | Liver 4.7 cm | 11.0 | 0.0 | Lung Liver | |
| ctDNA negative before metastasectomy | |||||||||
| 002 | M | NRAS G12D | 0.0 | 19.0 | Lung 0.9 cm | 0.0 | 0.0 | N | |
| 005 | M | TP53 R282W | 43.0 | 22.0 | Lung 0.8 cm | 0.0 | 0.0 | N | |
| APC R876 * | 34.0 | 14.0 | 0.0 | 0.0 | |||||
| 028b | M | KRAS G12V | 14.3 | 0.0 | PT 4.0 cm | 0.0 | 0.0 | N | |
| PIK3CA E545K | 14.8 | 0.0 | 0.0 | 0.0 | |||||
| SMAD4 R361H | 16.4 | 0.0 | 0.0 | 0.0 | |||||
| SMAD4 E330Q | 16.6 | 50.2 | 0.0 | 0.0 | |||||
| PIK3CA G914R | 14.9 | 0 | 0.0 | 0.0 | |||||
| 034 | M | TP53 R175H | 0.0 | 17.2 | LN 0.9 cm | 0.0 | 0.0 | N | |
| KRAS G12C | 0.0 | 18.0 | 0.0 | 0.0 | |||||
| PIK3CA E545V | 0.0 | 3.3 | 0.0 | 0.0 | |||||
| 041 | M | KRAS A59T | 59.0 | 44.0 | Lung 1.0 cm | 0.0 | 0.0 | N | |
| TP53 R342 * | 44.0 | 22.0 | 0.0 | 0.0 | |||||
| 056 | M | KRAS G12D | 24.0 | 0.0 | Liver 0.7 cm | 0.0 | 0.0 | N | |
| TP53 C238Y | 35.7 | 0.0 | 0.0 | 0.0 | |||||
| 060 | M | TP53 N239 * | 21.0 | 32.0 | Liver 2.0 cm | 0.0 | 0.0 | N | |
| 066 | M | TP53 R282W | 23.4 | 3.9 | Lung 0.4 cm | 0.0 | 0.0 | N | |
| APC R876 * | 31.2 | 0.0 | 0.0 | 0.0 | |||||
| APC E1379 * | 26.4 | 0.0 | 0.0 | 0.0 | |||||
| 068 | M | TP53 R213 * | 0.0 | 25.1 | Lung 1.7 cm | 0.0 | 0.0 | N | |
| 081 | S | KRAS G12S | 30.0 | 29.0 | Lung 1.2 cm | 0.0 | 0.0 | N | |
| APC Q1378 * | 23.0 | 25.0 | 0.0 | 0.0 | |||||
| 093 | M | PIK3CA V344M | BDL * | 17.4 | Lung 1.4 cm | 0.0 | 0.0 | N | |
| 098 | M | KRAS G13D | 0.0 | 21.1 | Liver 2.2 cm | 0.0 | 0.0 | N | |
| APC L1488fs | 22.9 | 0.0 | 0.0 | 0.0 | |||||
| APC E1494fs | 0.0 | 20.4 | 0.0 | 0.0 | |||||
| 007 | M | APC C1289 * | 6.9 | 20.8 | Lung 1.4 cm | 0.0 | 0.0 | PT | |
| PIK3CA E542K | 3.8 | 20.5 | 0.0 | 0.0 | |||||
| TP53 R282W | 2.6 | 14.0 | 0.0 | 0.0 | |||||
| TP53 S99fs * | 2.1 | 11.0 | 0.0 | 0.0 | |||||
| 031a | S | APC R1114 * | 8.1 | 29.4 | Lung 1.4 cm | 0.0 | 0.0 | Lung | |
| KRAS G13C | 13.4 | 42.6 | 0.0 | 0.0 | |||||
| APC R1463fs | 2.0 | 0.0 | 0.0 | 0.0 | |||||
| APC E1345 * | 8.1 | 30.7 | 0.0 | 0.0 | |||||
| 032 | M | KRAS G12D | 22.3 | 29.2 | Lung 1.2 cm | 0.0 | 0.0 | LN | |
| TP53 R282W | 0.0 | 34.6 | 0.0 | 0.0 | |||||
| TP53 S127F | 14.3 | 0.0 | 0.0 | 0.0 | |||||
| APC L1488fs | 13.2 | 0.0 | 0.0 | 0.0 | |||||
| 035 | M | KRAS G12C | 37.7 | 15.6 | Lung 0.5 cm | 0.0 | 0.0 | Lung | |
| TP53 I195T | 41.4 | 17.1 | 0.0 | 0.0 | |||||
| APC R1463fs | 1.9 | 0.0 | 0.0 | 0.0 | |||||
| APC S1501fs | 24.0 | 8.9 | 0.0 | 0.0 | |||||
| 040 | M | KRAS G12D | 3.4 | 18.5 | Lung 0.6 cm | 0.0 | 0.0 | Lung Bone | |
| TP53 S241P | 1.2 | 10.4 | 0.0 | 0.0 | |||||
| PTEN R173H | 1.0 | 0.0 | 0.0 | 0.0 | |||||
| 058 | M | NRAS G12D | 24.6 | 32.7 | Lung 1.0 cm | 0.0 | 0.0 | Lung | |
| TP53 H297fs | 26.2 | 14.6 | 0.0 | 0.0 | |||||
| TP53 R333fs | 22.5 | 10.0 | 0.0 | 0.0 | |||||
| 072 | M | APC R876 * | 17.8 | 10.6 | PT 2.7 cm | 0.0 | 0.0 | PT LN | |
| TP53 R248Q | 16.8 | 10.8 | 0.0 | 0.0 | |||||
| APC Q1303 * | 14.1 | 12.0 | 0.0 | 0.0 | |||||
| 078 | M | KRAS G12V | 41.0 | 12.9 | Lung 1.2 cm | 0.0 | 0.0 | Lung | |
| TP53 R175H | 54.1 | 20.8 | 0.0 | 0.0 | |||||
| No | Synchronicity | Variant | Primary Tissue VAF | Metastatic Tissue VAF | Metastatic Site | PreM ctDNA VAF | PostM ctDNA VAF | Residual | |
|---|---|---|---|---|---|---|---|---|---|
| No CTx | CTx | ||||||||
| ctDNA positive before metastasectomy | |||||||||
| 046 | M | KRAS G12D | 0 | 38.1 | Liver 2.1 cm | 4.4 | 0.0 | Lung <0.5 #4 Pelvic LNs | |
| TP53 G245C | 1.0 | 46.3 | 5.6 | 0.0 | |||||
| APC K1561 * | 0 | 50.5 | 5.0 | 0.0 | |||||
| ctDNA negative before metastasectomy | |||||||||
| 022 | M | TP53 V272M | 39.4 | 55.4 | Liver 3.0 cm | 0.0 | 0.0 | LN 2.2 cm | |
| SMAD4 | 7.4 | 0.0 | |||||||
| 061c | M | TP53 | 51.5 | 25.1 | Lung 2.1 cm | 0.0 | 0.0 | PT | |
| APC H1490fs | 30.2 | 16.2 | 0.0 | 0.0 | |||||
| No | Synchronicity | Variant | Primary Tissue VAF | Metastatic Tissue VAF | Meta Site | PreM ctDNA | PostM ctDNA | Recurence | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No CTx | CTx | Rx | CTx | Rx | |||||||
| ctNDA positive before metastasectomy | |||||||||||
| 001 | S | TP53 I255F | 49.8 | SD | 0.0 | PR | Liver 0.1 cm | 1.6 | 0.0 | N | |
| 036 | S | APC R1114 * | 44.2 | 31.0 | PD | Liver 2.5 cm | 21.0 | 0.0 | N | ||
| APC R1450 * | 21.5 | 17.0 | 11.0 | 0.0 | |||||||
| KRAS G12D | 46.5 | 34.0 | 19.0 | 0.0 | |||||||
| 008b | S | KRAS Q61R | 48.8 | 53.7 | PD | Lung 2.5 cm | 9.6 | 0.0 | Lung | ||
| TP53 R175H | 49.5 | 54.1 | 6.4 | 0.0 | |||||||
| 028a | S | KRAS G12V | 14.3 | 7.8 | PD | Liver 2.2 cm | 0.0 | 0.0 | PT | ||
| PIK3CA E545K | 14.8 | 11. 1 | 1.3 | 0.0 | |||||||
| PIK3CA G914R | 14.9 | 10.2 | 0.0 | 0.0 | |||||||
| SMAD4 E330Q | 16.6 | 8.9 | 0.0 | 0.0 | |||||||
| SMAD4 R361H/C | 16.4 | 1.7 | 0.0 | 0.0 | |||||||
| ctNDA negative before metastasectomy | |||||||||||
| 004 | S | TP53 R273C | 60.4 | 4.1 | PR | Liver 0.8 cm | 0.0 | 0.0 | N | ||
| APC K1308 * | 40.2 | 2.8 | 0.0 | 0.0 | |||||||
| 012 | S | KRAS G12V | 14.6 | 0.0 | CR | Liver 0 cm | 0.0 | 0.0 | N | ||
| PIK3CA H1047R | 20.6 | 0.0 | 0.0 | 0.0 | |||||||
| 031b | S | APC R1114 * | 8.1 | 11.6 | SD | Lung 1.4 cm | 0.0 | 0.0 | N | ||
| APC E1345 * | 8.1 | 15.4 | 0.0 | 0.0 | |||||||
| KRAS G13C | 13.4 | 9.1 | 0.0 | 0.0 | |||||||
| 042 | M | APC Q1378 * | 47.5 | 36.2 | PR | Liver 2.5 cm | 0.0 | 0.0 | N | ||
| KRAS G12D | 29.9 | 18.3 | 0.0 | 0.0 | |||||||
| TP53 R175H | 30.5 | 19.3 | 0.0 | 0.0 | |||||||
| 044 | S | CTNNB1 S45F | 36.8 | 6.5 | PR | Liver 2.2 cm | 0.0 | 0.0 | N | ||
| KRAS G12D | 17.3 | 0.0 | 0.0 | 0.0 | |||||||
| PTEN R335 * | 5.2 | 0.0 | 0.0 | 0.0 | |||||||
| 057 | S | TP53 C275Y | 26.5 | 36.9 | SD | Lung 1.6 cm | 0.0 | 0.0 | N | ||
| 091 | S | APC E1306 * | 18.0 | 0.0 | PR | Liver 0.8 cm | 0.0 | 0.0 | N | ||
| TP53 R175H | 40.2 | 6.2 | 0.0 | 0.0 | |||||||
| APC Q886 * | 26.9 | 4.6 | 0.0 | 0.0 | |||||||
| 006 | S | KRAS G13C | 37.2 | 10.7 | PR | Liver 1.6 cm | 0.0 | 0.0 | Bone | ||
| TP53 R306 * | 34.9 | 9.1 | 0.0 | 0.0 | |||||||
| 011 | M | APC R876 * | 30.6 | 0.0 | PD | Lung 1.0 cm | 0.0 | 0.0 | Lung | ||
| KRAS E545K | 34.3 | 2.2 | 0.0 | 0.0 | |||||||
| TP53 R342 * | 0.0 | 2.5 | 0.0 | 0.0 | |||||||
| SMAD4 R361H | 0.0 | 3.8 | 0.0 | 0.0 | |||||||
| SMAD4 A118V | 64.8 | 0.0 | 0.0 | 0.0 | |||||||
| 014 | S | KRAS Q61H | 25.3 | 5.6 | SD | Liver 2.3 cm | 0.0 | 0.0 | PT | ||
| SMAD4 R361C | 15.2 | 3.0 | 0.0 | 0.0 | |||||||
| 019 | S | PIK3CA G1049R | 3.1 | SD | 3.6 | SD | Liver 0.9 cm | 0.0 | 0.0 | Liver | |
| 020a | S | TP53 R196 * | 38.5 | SD | 21.2 | PR | Liver 2.5 cm | 0.0 | 0.0 | Liver | |
| APC Q1294 * | 25.4 | 13.8 | 0.0 | 0.0 | |||||||
| 027 | S | TP53 R175H | 6.1 | PR | 2.6 | PR | Liver 3.2 cm | 0.0 | 0.0 | Liver Lung | |
| PTEN R335 * | 1.1 | 0.0 | 0.0 | 0.0 | |||||||
| 037a | S | TP53 T211I | 14.9 | 1.0 | PR | Liver 0.5 cm | 0.0 | 0.0 | Liver | ||
| 048 | S | APC R1450 * | 1.5 | 0.0 | PR | Lung 0.6 cm | 0.0 | 0.0 | Lung | ||
| CTNNB1 S37Y | 0.0 | 21.2 | 0.0 | 0.0 | |||||||
| KRAS G12D | 1.4 | 0.0 | 0.0 | 0.0 | |||||||
| PIK3CA E545K | 2.0 | 0.0 | 0.0 | 0.0 | |||||||
| TP53 R209Kfs | 1.8 | 0 | 0.0 | 0.0 | |||||||
| 051b | M | KRAS G13S | 0.0 | 1.1 | PD | Lung 0.9 cm | 0.0 | 0.0 | Lung | ||
| PTEN R130Q | 0.0 | 1.0 | 0.0 | 0.0 | |||||||
| SMAD4 C115Y | 0.0 | 1.2 | 0.0 | 0.0 | |||||||
| TP53 R175H | 10.7 | 40.0 | 0.0 | 0.0 | |||||||
| 052 | S | RB1 A201fs | 0.0 | 5.5 | PR | Lung 0.4 cm | 0.0 | 0.0 | Lung | ||
| 055 | S | KRAS G13D | 60.3 | 16.6 | PR | Liver 3.5 cm | 0.0 | 0.0 | Liver | ||
| 061b | S | TP53? | 51.5 | SD | 27.1 | SD | Lung 0.5 cm | 0.0 | 0.0 | Lung | |
| APC H1490fs | 30.2 | 19.5 | 0.0 | 0.0 | |||||||
| 063 | S | KRAS G12V | BDL | 5.3 | PR | Liver 2.2 cm | 0.0 | 0.0 | Lung | ||
| KRAS A146T | BDL | 25.0 | 0.0 | 0.0 | |||||||
| TP53 V173L | BDL | 2.4 | 0.0 | 0.0 | |||||||
| APC P1381fs | BDL | 15.5 | 0.0 | 0.0 | |||||||
| 067 | S | TP53 M246R | 23.2 | SD | 39.3 | SD | Rec 3.4 cm | 0.0 | 0.0 | PT | |
| No | Synchronicity | Variant | Primary Tissue VAF | Metastatic Tissue VAF | Meta Site | PreM ctDNA VAF | PostM ctDNA VAF | Residual | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No CTx | CTx | Rx | CTx | Rx | |||||||
| ctDNA positive before metastasectomy | |||||||||||
| 053 | S | EBRR2 D769Y | 0 | 71.7 | PD | Liver #2 (2.3) | 32.9 | 50 | Liver multiple Bone | ||
| 076 | S | TP53 I195fs | 19.2 | 0 | PD | PT 4.1 cm | 20.4 | 16.9 | Huge liver | ||
| 079 | M | APC Q1378 * | BDL | 23.2 | PD | Liver #7 | 21.6 | 0 | Lung 0.9 cm | ||
| TP53 S215G | BDL | 19.8 | 21.7 | 0 | |||||||
| 096 | S | KRAS G13D | 31.4 | 8.6 | PR | Liver #9 | 0 | 0 | Liver #2 (<0.5) | ||
| TP53 V73fs | 50.8 | 0 | 0 | 0 | |||||||
| ctDNA negative before metastasectomy | |||||||||||
| 003 | S | TP53 R196 * | 30.2 | 22.8 | PD | Liver 2.3 cm | 0 | 0 | Liver 1.6 cm | ||
| 008a | S | KRAS Q61R | 48.8 | 10.4 | SD | Lung 0.6 cm | 0 0 | 0 0 | Lung 0.8 cm | ||
| TP53 R175H | 49.5 | 10.1 | |||||||||
| 026 | S | TP53 R306 * | 39.7 | 25.2 | PD | Liver #5 PT #5 | 0 | 0 | Liver 0.5 cm | ||
| 051a | S | TP53 R175H | 10.7 | 11.5 | PD | Liver #4 | 0 | 0 | Lung 0.5 cm | ||
| BRAF V471F | 0 | 12.3 | 0 | 0 | |||||||
| 054 | M | KRAS G12D | 45.5 | 49.9 | PD | Liver 1.6 cm PT 2.6 cm | 0 | 10.1 | Liver #6 (<1.0 cm) | ||
| 061a | S | TP53? | 51.5 | SD | 14.5 | PD | Lung 1.0 cm | 0 | 0 | Lung <0.5 cm #2 | |
| APC H1490fs | 30.2 | 12.4 | 0 | 0 | |||||||
| 065 | S | APC R1114 * | 37.5 | 37.5 | PR | Liver #4 | 0 | 3.6 | Liver 1.4 cm | ||
| TP53 G244S | 52.7 | 46.2 | 0 | 2.2 | |||||||
| APC S1356 * | 29.3 | 15.8 | 0 | 2.9 | |||||||
| Clinical Condition | ctDNA Positive (N = 16) | ctDNA Negative (N = 51) | p Value | |
|---|---|---|---|---|
| Neoadjuvant chemotherapy before R0 resection | Yes | 7 | 29 | 0.3587 |
| No | 9 | 22 | ||
| Metastasectomy organ | Liver | 14 | 21 | 0.0045 |
| Lung | 1 | 24 | ||
| Other | 1 | 6 | ||
| Tumor burden (tumor diameter) | >1 cm | 15 | 28 | 0.0183 |
| ≤1 cm | 1 | 23 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lee, S.; Park, Y.-S.; Chang, W.-J.; Choi, J.Y.; Lim, A.; Kim, B.; Lee, S.-B.; Lee, J.-W.; Kim, S.-H.; Kim, J.; et al. Clinical Implication of Liquid Biopsy in Colorectal Cancer Patients Treated with Metastasectomy. Cancers 2021, 13, 2231. https://doi.org/10.3390/cancers13092231
Lee S, Park Y-S, Chang W-J, Choi JY, Lim A, Kim B, Lee S-B, Lee J-W, Kim S-H, Kim J, et al. Clinical Implication of Liquid Biopsy in Colorectal Cancer Patients Treated with Metastasectomy. Cancers. 2021; 13(9):2231. https://doi.org/10.3390/cancers13092231
Chicago/Turabian StyleLee, Soohyeon, Young-Soo Park, Won-Jin Chang, Jung Yoon Choi, Ahreum Lim, Boyeon Kim, Saet-Byeol Lee, Jong-Won Lee, Seon-Hahn Kim, Jin Kim, and et al. 2021. "Clinical Implication of Liquid Biopsy in Colorectal Cancer Patients Treated with Metastasectomy" Cancers 13, no. 9: 2231. https://doi.org/10.3390/cancers13092231
APA StyleLee, S., Park, Y.-S., Chang, W.-J., Choi, J. Y., Lim, A., Kim, B., Lee, S.-B., Lee, J.-W., Kim, S.-H., Kim, J., Kwak, J.-M., Yoon, K.-C., Lee, S.-H., & Kim, Y. H. (2021). Clinical Implication of Liquid Biopsy in Colorectal Cancer Patients Treated with Metastasectomy. Cancers, 13(9), 2231. https://doi.org/10.3390/cancers13092231









