Novel Benzenesulfonate Scaffolds with a High Anticancer Activity and G2/M Cell Cycle Arrest
Abstract
Simple Summary
Abstract
1. Introduction
2. Results and Discussion
2.1. Synthesis of Sulfonic Styrylquinazolines
2.2. Sulfonates Demonstrate Antiproliferative Activity toward Cancer Cell Lines
2.3. Sulfonates Modulate Tyrosine Kinases Activity
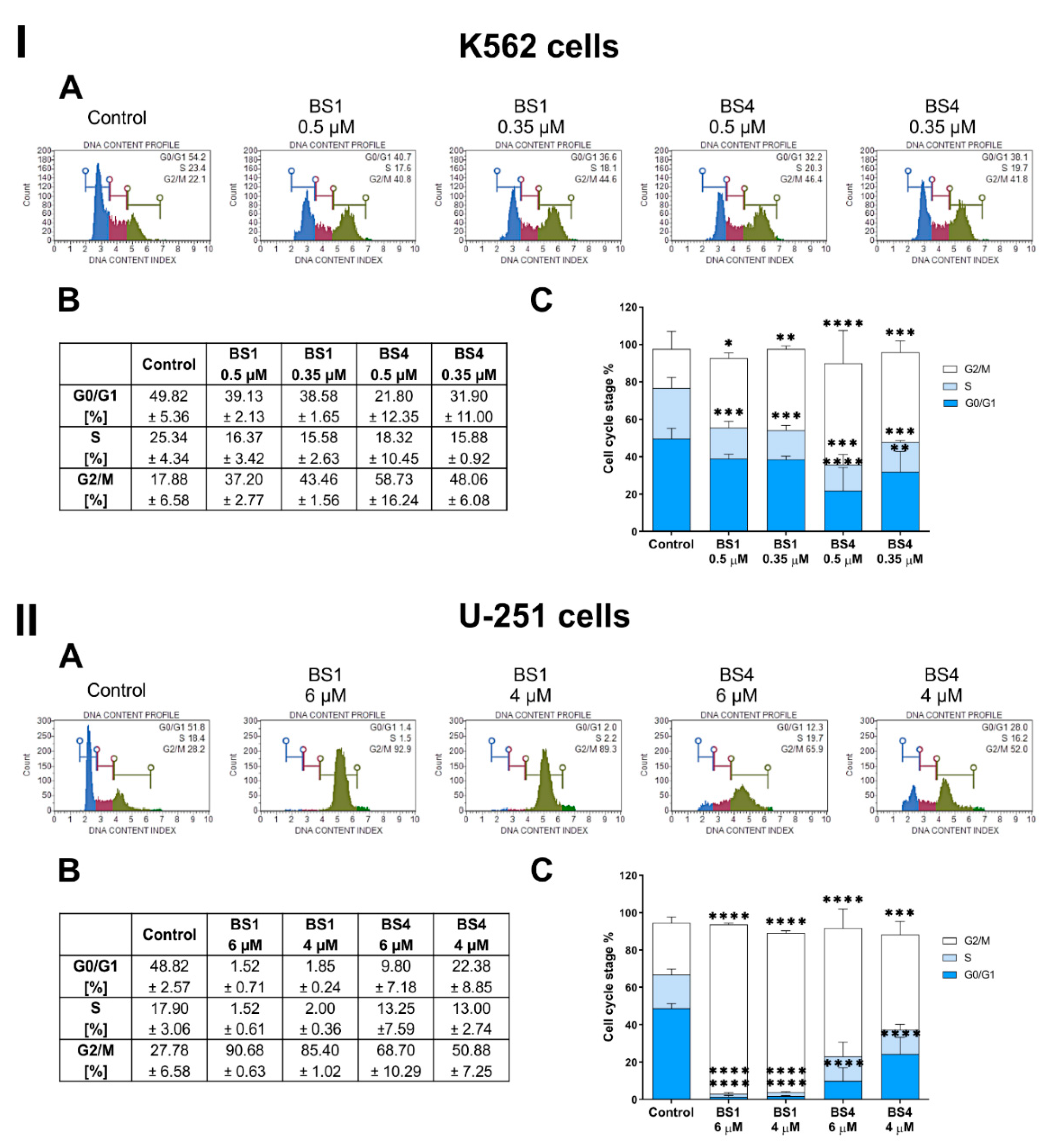
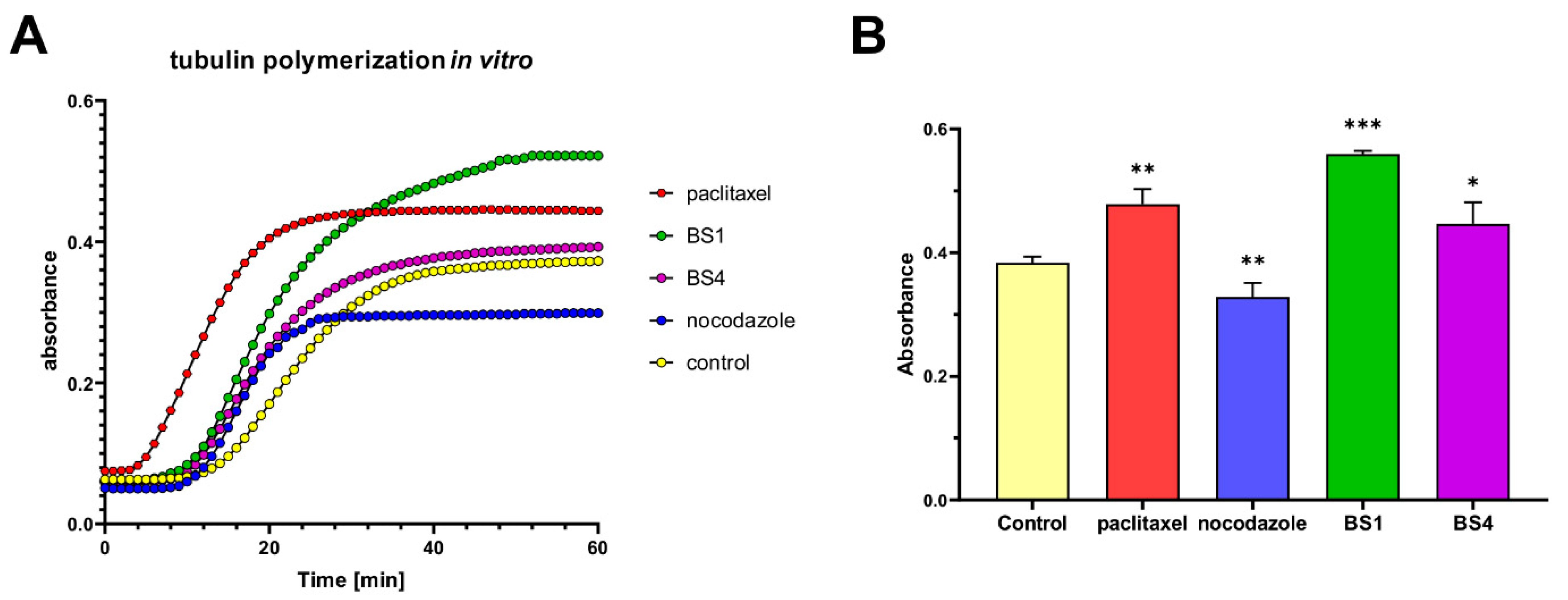
2.4. Sulfonates Inhibit Cell Cycle Progression
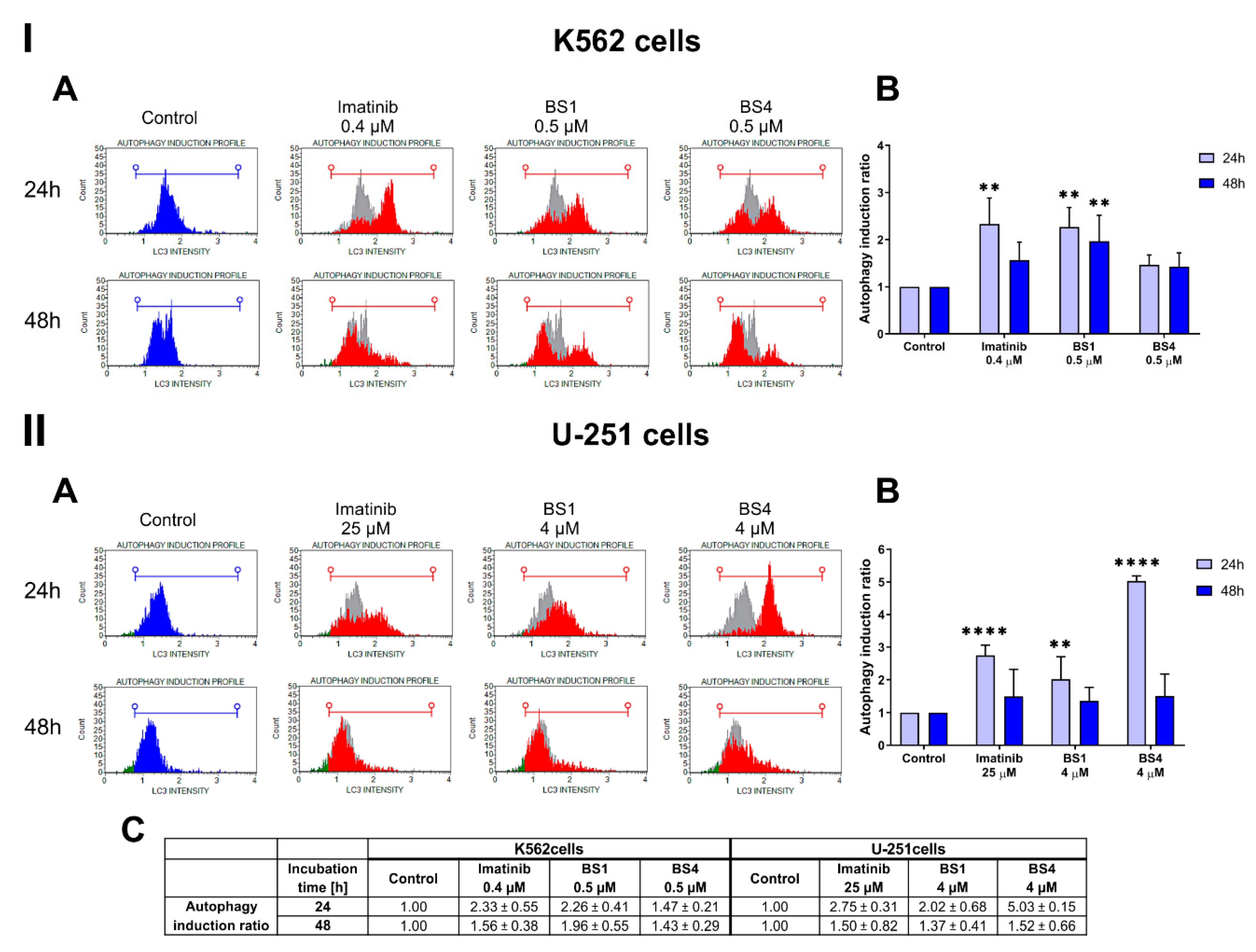
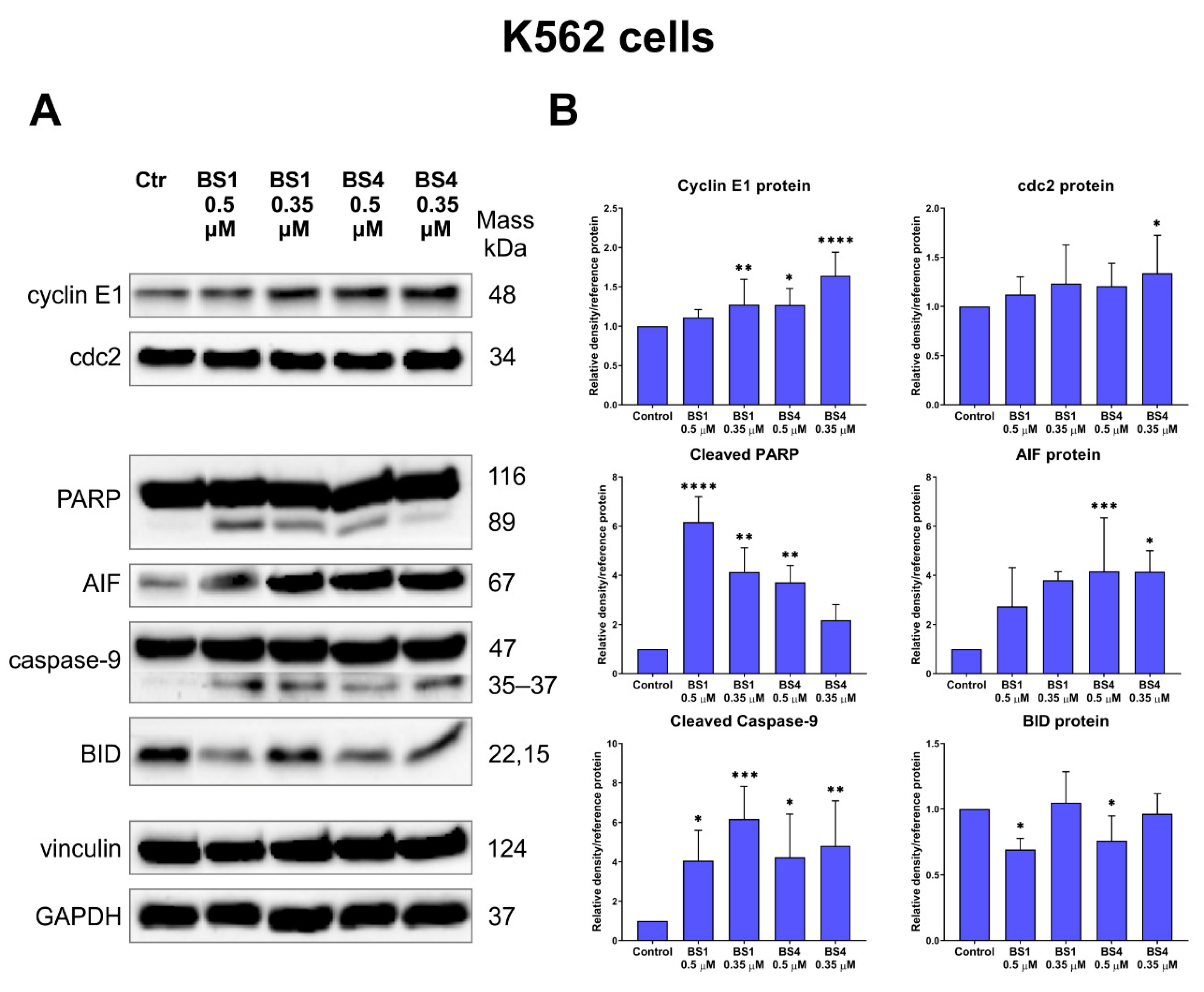
2.5. Sulfonates Induce Apoptosis and Autophagy
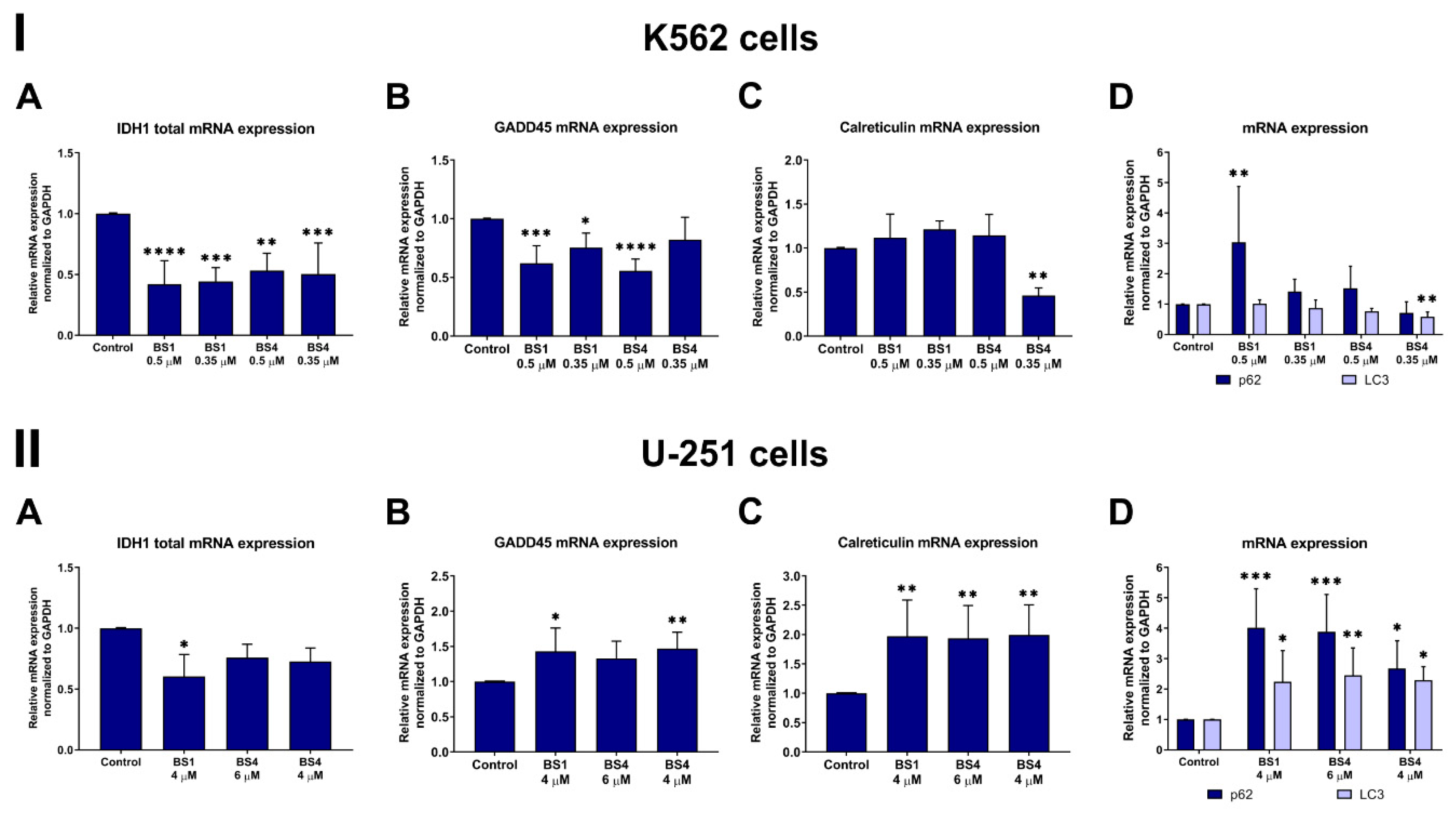
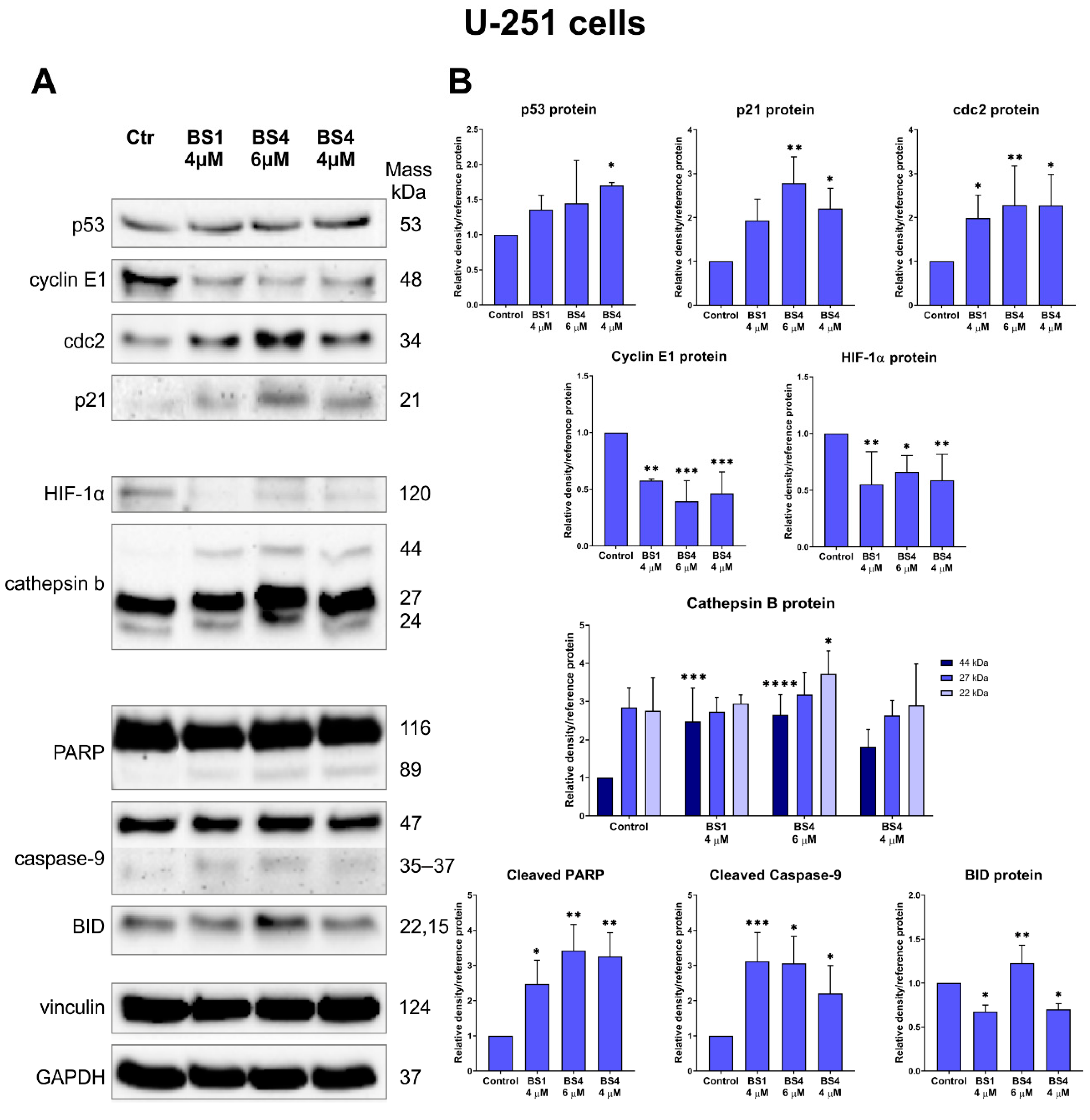
2.6. Sulfonates Change Expression of Genes and Proteins Associated with Cell Metabolism, Cell Cycle and Cell Death
3. Materials and Methods
3.1. Synthesis
- (BS1)
- 2-((E)-2-(2-methoxyphenyl)ethenyl)quinazolin-4-yl 1,3-benzodioxole-5-sulfonate 1H NMR (500 MHz, (CD3)2SO) δ 8.26 (d, J = 16.2 Hz, 1H), 8.15 (dd, J = 8.0, 1.5 Hz, 1H), 7.89 (ddd, J = 8.5, 7.2, 1.6 Hz, 1H), 7.73 (dd, J = 8.3, 1.1 Hz, 1H), 7.63 (dd, J = 7.8, 1.7 Hz, 1H), 7.56 (ddd, J = 8.1, 7.1, 1.1 Hz, 1H), 7.47 (ddd, J = 8.8, 7.4, 1.7 Hz, 1H), 7.17 (d, J = 8.4 Hz, 1H), 7.15 (d, J = 16.3 Hz, 1H), 7.13 (dd, J = 8.0, 1.7 Hz, 1H), 7.08 (td, J = 7.5, 1.0 Hz, 1H), 7.04 (d, J = 1.6 Hz, 1H), 6.83 (d, J = 8.0 Hz, 1H), 6.01 (s, 2H), 3.94 (s, 3H); 13C NMR (126 MHz, (CD3)2SO) δ 161.16, 158.94, 154.16, 147.71, 146.93, 143.74, 143.06, 139.23, 136.01, 133.18, 129.90, 127.84, 126.88, 123.54, 123.01, 121.53, 120.59, 119.88, 117.78, 112.61, 107.61, 106.72, 101.55, 56.28; HRMS (ESI) calcd for C24H18N2O6S [M − H]− 461.0813; found 461.0821; m.p. 223–225 °C.
- (BS2)
- 2-((E)-2-(2-methoxyphenyl)ethenyl)quinazolin-4-yl 6-bromo-1,3-benzodioxole-5-sulfonate 1H NMR (500 MHz, (CD3)2SO) δ 8.18 (ddd, J = 8.3, 1.5, 0.7 Hz, 1H), 8.08 (ddd, J = 8.4, 6.9, 1.4 Hz, 1H), 8.01 (appdt, J = 8.4, 1.0 Hz, 1H), 7.92 (d, J = 16.1 Hz, 1H), 7.86 (s, 1H), 7.78 (ddd, J = 8.1, 6.9, 1.2 Hz, 1H), 7.70 (dd, J = 7.7, 1.7 Hz, 1H), 7.62 (s, 1H), 7.41 (ddd, J = 8.9, 7.3, 1.7 Hz, 1H), 7.26 (d, J = 16.0 Hz, 1H), 7.13 (dd, J = 8.4, 1.1 Hz, 1H), 7.03 (apptd, J = 7.5, 1.1 Hz, 1H), 6.12 (s, 2H), 3.95 (s, 3H); 13C NMR (126 MHz, DMSO) δ 162.00, 158.43, 152.80, 148.39, 146.40, 141.67, 135.23, 132.10, 128.99, 126.86, 126.46, 123.66, 121.39, 121.27, 113.80, 112.41, 111.15, 109.47, 102.45, 56.16; HRMS (ESI) calcd for C24H17BrN2O6S [M + H]− 541.0063; found 541.0079; m.p. 149–151 °C.
- (BS3)
- 7-chloro-2-((E)-2-(2-methoxyphenyl)ethenyl)quinazolin-4-yl 4-methylbenzenesulfonate 1H NMR (500 MHz, (CD3)2SO) δ 8.19 (d, J = 16.2 Hz, 1H), 8.09 (d, J = 8.5 Hz, 1H), 7.74 (d, J = 2.0 Hz, 1H), 7.61 (dd, J = 7.7, 1.9 Hz, 1H), 7.50 (dd, J = 8.5, 2.1 Hz, 1H), 7.49–7.46 (m, 2H), 7.43 (ddd, J = 8.9, 7.5, 1.8 Hz, 1H), 7.14 (d, J = 8.2 Hz, 1H), 7.13 –7.10 (m, 2H), 7.08 (d, J = 16.2 Hz, 1H), 7.04 (appt, J = 7.3 Hz, 1H), 3.92 (s, 2H), 2.29 (s, 3H); 13C NMR (126 MHz, (CD3)2SO) δ 161.64, 158.38, 153.86, 150.28, 146.25, 139.60, 138.04, 135.35, 132.05, 128.88, 128.51 (2H), 128.42, 126.76, 126.33, 125.97 (2H), 128.51, 123.65, 121.37, 121.18, 120.25, 112.38, 56.14, 21.25; HRMS (ESI) calcd for C24H19ClN2O4S [M − H]− 465.0681; found 465.0685; m.p. 196–197 °C.
- (BS4)
- 2-((E)-2-(2-methoxyphenyl)ethenyl)quinazolin-4-yl 4-methylbenzenesulfonate 1H NMR (500 MHz, (CD3)2SO) δ 1H NMR (500 MHz, DMSO) δ 8.26 (d, J = 16.3 Hz, 1H), 8.14 (dd, J = 8.0, 1.5 Hz, 1H), 7.88 (ddd, J = 8.5, 7.1, 1.6 Hz, 1H), 7.73 (dd, J = 8.3, 1.1 Hz, 1H), 7.63 (dd, J = 7.7, 1.7 Hz, 1H), 7.56 (ddd, J = 8.1, 7.1, 1.1 Hz, 1H), 7.49–7.47 (m, 2H), 7.47 (ddd, J = 6.9, 1.8 Hz, 1H), 7.17 (dd, J = 8.4, 1.1 Hz, 1H), 7.15 (d, J = 16.4 Hz, 1H), 7.13–7.11 (m, 1H), 7.08 (td, J = 7.5, 1.0 Hz, 1H), 3.94 (s, 3H), 2.29 (s, 3H); 13C NMR (126 MHz, (CD3)2SO) δ 161.40, 159.96, 158.05, 153.16, 146.79, 136.23, 134.66, 131.60, 130.57, 129.26, 128.99, 128.40, 128.10, 127.13, 124.08, 123.55, 121.34, 114.50, 112.24, 56.24, 21.70; HRMS (ESI) calcd for C24H21N2O4S [M − H]− 431.1070; found 431.1068; m.p. 168–169 °C.
- (BS5)
- 2-((E)-2-(4-methoxyphenyl)ethenyl)quinazolin-4-yl 4-methylbenzenesulfonate 1H NMR (500 MHz, (CD3)2SO) δ 8.16 (dd, J = 8.0, 1.5 Hz, 1H), 8.12 (d, J = 16.3 Hz, 1H), 7.92 (ddd, J = 8.5, 7.2, 1.5 Hz, 1H), 7.73 (d, J = 7.5 Hz, 1H), 7.69–7.64 (m, 2H), 7.60 (ddd, J = 8.1, 7.2, 1.1 Hz, 1H), 7.52–7.48 (m, 2H), 7.13 (dd, J = 8.5, 0.8 Hz, 2H), 7.10–7.06 (m, 2H), 6.91 (d, J = 16.3 Hz, 1H), 3.84 (s, 3H), 2.29 (s, 3H); 13C NMR (126 MHz, (CD3)2SO) δ 161.80, 160.20, 153.64, 145.17, 143.61, 142.35, 137.76, 135.42, 130.17, 127.95, 127.23, 126.56, 126.31, 125.36, 122.21, 119.72, 114.76, 113.19, 55.36, 20.56. HRMS (ESI) calcd for C24H20N2O4S [M + H]− 433.1217; found 433.1214; m.p. 176–178 °C
- (BS6)
- 2-((E)-2-(2,4-dimethoxyphenyl)ethenyl)quinazolin-4-yl 4-methylbenzenesulfonate 1H NMR (500 MHz, (CD3)2SO) δ 8.18–8.14 (m, 2H), 8.09 (ddd, J = 8.2, 1.4, 0.7 Hz, 1H), 8.03 (ddd, J = 8.2, 7.0, 1.4 Hz, 1H), 8.01 (d, J = 16.2 Hz, 1H), 7.95 (appdt, J = 8.5, 1.0 Hz, 1H), 7.72 (ddd, J = 8.2, 7.0, 1.2 Hz, 1H), 7.69 (d, J = 8.6 Hz, 1H), 7.53 (m, 2H), 7.15 (d, J = 16.0 Hz, 1H), 6.70 (d, J = 2.4 Hz, 1H), 6.65 (dd, J = 8.5, 2.4 Hz, 1H), 3.99 (s, 3H), 3.86 (s, 3H), 2.42 (s, 3H); 13C NMR (126 MHz, DMSO) δ 162.57, 161.29, 160.38, 159.53, 153.25, 146.74, 136.14, 134.75, 133.52, 130.55, 129.80, 129.23, 128.63, 127.95, 124.48, 123.52, 117.06, 114.32, 106.69, 98.96, 56.33, 55.96, 21.71; HRMS (ESI) calcd for C25H23N2O5S [M + H]− 463.1322; found 463.1325; m.p. 178–180 °C.
3.2. Cell Culture
3.3. Cytotoxicity Studies
3.4. Tyrosine Kinase Assay
3.5. Cell Cycle Assay
3.6. Tubulin Polymerization Assay
3.7. Annexin V Binding Assay
3.8. Autophagy Assay
3.9. Analysis of the mRNA Expression
3.10. Immunoblotting
3.11. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Khan, I.; Ibrar, A.; Abbas, N.; Saeed, A. Recent advances in the structural library of functionalized quinazoline and quinazolinone scaffolds: Synthetic approaches and multifarious applications. Eur. J. Med. Chem. 2014, 76, 193–244. [Google Scholar] [CrossRef]
- Selvam, T.; Kumar, P. Quinazoline Marketed drugs—A Review. Res. Pharm. 2011, 1, 1–21. [Google Scholar]
- Marzaro, G.; Guiotto, A.; Chilin, A. Quinazoline derivatives as potential anticancer agents: A patent review (2007–2010). Expert Opin. Ther. Pat. 2012, 22, 223–252. [Google Scholar] [CrossRef]
- Kaur, K.; Jain, M.; Kaur, T.; Jain, R. Antimalarials from nature. Bioorg. Med. Chem. 2009, 17, 3229–3256. [Google Scholar] [CrossRef] [PubMed]
- Muscia, G.C.; Cazorla, S.I.; Frank, F.M.; Borosky, G.L.; Buldain, G.Y.; Asis, S.E.; Malchiodi, E.L. Synthesis, trypanocidal activity and molecular modeling studies of 2-alkylaminomethylquinoline derivatives. Eur. J. Med. Chem. 2011, 46, 3696–3703. [Google Scholar] [CrossRef] [PubMed]
- Tobe, M.; Isobe, Y.; Tomizawa, H.; Nagasaki, T.; Takahashi, H.; Fukazawa, T.; Hayashi, H. Discovery of quinazolines as a novel structural class of potent inhibitors of NF-κB activation. Bioorg. Med. Chem. 2003, 11, 383–391. [Google Scholar] [CrossRef]
- Lee, J.Y.; Park, J.H.; Lee, S.J.; Park, H.; Lee, Y.S. Styrylquinazoline Derivatives as HIV-1 Integrase Inhibitors. Arch. Der Pharm. 2002, 335. [Google Scholar] [CrossRef]
- Muhsin, M.; Graham, J.; Kirkpatrick, P. Gefitinib. Nat. Rev. Drug Discov. 2003, 2, 515–516. [Google Scholar] [CrossRef]
- Boschelli, D.H. 4-anilino-3-quinolinecarbonitriles: An emerging class of kinase inhibitors. Curr. Top. Med. Chem. 2002, 2, 1051–1063. [Google Scholar] [CrossRef]
- Brown, C.J.; Cheok, C.F.; Verma, C.S.; Lane, D.P. Reactivation of p53: From peptides to small molecules. Trends Pharmacol. Sci. 2011, 32, 53–62. [Google Scholar] [CrossRef]
- Krapf, M.K.; Wiese, M. Synthesis and Biological Evaluation of 4-Anilino-quinazolines and -quinolines as Inhibitors of Breast Cancer Resistance Protein (ABCG2). J. Med. Chem. 2016, 59, 5449–5461. [Google Scholar] [CrossRef]
- Li, D.D.; Hou, Y.P.; Wang, W.; Zhu, H.L. Exploration of chemical space based on 4-anilinoquinazoline. Curr. Med. Chem. 2012, 19, 871–892. [Google Scholar] [CrossRef]
- Jampilek, J.; Musiol, R.; Finster, J.; Pesko, M.; Carroll, J.; Kralova, K.; Vejsova, M.; O’Mahony, J.; Coffey, A.; Dohnal, J.; et al. Investigating biological activity spectrum for novel styrylquinazoline analogues. Molecules 2009, 14, 4246–4265. [Google Scholar] [CrossRef]
- Mrozek-Wilczkiewicz, A.; Kalinowski, D.S.; Musiol, R.; Finster, J.; Szurko, A.; Serafin, K.; Knas, M.; Kamalapuram, S.K.; Kovacevic, Z.; Jampilek, J.; et al. Investigating the anti-proliferative activity of styrylazanaphthalenes and azanaphthalenediones. Bioorg. Med. Chem. 2010, 18, 2664–2671. [Google Scholar] [CrossRef]
- Mularski, J.; Malarz, K.; Pacholczyk, M.; Musiol, R. The p53 stabilizing agent CP-31398 and multi-kinase inhibitors. Designing, synthesizing and screening of styrylquinazoline series. Eur. J. Med. Chem. 2019, 163, 610–625. [Google Scholar] [CrossRef]
- Nishikawa, S.; Maki, S.; Nishikimi, Y.; Kumazawa, Z.; Kashimura, N. Cytokinin Activity of 4-Aminopyridopyrimidines toward the Growth of Tobacco Callus. Biosci. Biotechnol. Biochem. 2014, 58, 1709–1710. [Google Scholar] [CrossRef]
- Yadav, M.R.; Grande, F.; Chouhan, B.S.; Naik, P.P.; Giridhar, R.; Garofalo, A.; Neamati, N. Cytotoxic potential of novel 6,7-dimethoxyquinazolines. Eur. J. Med. Chem. 2012, 48, 231–243. [Google Scholar] [CrossRef]
- Zhang, H.Q.; Gong, F.H.; Li, C.G.; Zhang, C.; Wang, Y.J.; Xu, Y.G.; Sun, L.P. Design and discovery of 4-anilinoquinazoline-acylamino derivatives as EGFR and VEGFR-2 dual TK inhibitors. Eur. J. Med. Chem. 2016, 109, 371–379. [Google Scholar] [CrossRef]
- Jian, S.; Gao, Q.L.; Wu, B.W.; Li, D.; Shi, L.; Zhu, T.; Lou, J.F.; Jin, C.Y.; Zhang, Y.B.; Zhang, S.Y.; et al. Novel tertiary sulfonamide derivatives containing benzimidazole moiety as potent anti-gastric cancer agents: Design, synthesis and SAR studies. Eur. J. Med. Chem. 2019, 183, 111731. [Google Scholar] [CrossRef]
- Elder, D.P.; Delaney, E.; Teasdale, A.; Eyley, S.; Reif, V.D.; Jacq, K.; Facchine, K.L.; Oestrich, R.S.; Sandra, P.; David, F. The utility of sulfonate salts in drug development. J. Pharm. Sci. 2010, 99, 2948–2961. [Google Scholar] [CrossRef]
- Elder, D.P.; Snodin, D.J. Drug substances presented as sulfonic acid salts: Overview of utility, safety and regulation. J. Pharm. Pharmacol. 2009, 61, 269–278. [Google Scholar] [CrossRef] [PubMed]
- Gibson, N.W. Characterization of DNA Damage and Cytotoxicity Induced in Two Human Colon Carcinoma Cell Lines by Cyclodisone. Cancer Res. 1989, 49, 154–157. [Google Scholar] [PubMed]
- Umprayn, K.; Waugh, W.; Stella, V.J. Mechanism of degradation of the investigational cytotoxic agent, cyclodisone (NSC-348948). Int. J. Pharm. 1990, 66, 253–262. [Google Scholar] [CrossRef]
- Raju, T.N. The Nobel chronicles. 1939: Gerhard Domagk (1895-1964). Lancet 1999, 353, 681. [Google Scholar] [CrossRef]
- Hazem, R.M.; Mohamed, A.A.; Ghareb, N.; Mehanna, E.T.; Mesbah, N.M.; Abo-Elmatty, D.M.; Elgawish, M.S. Anti-cancer activity of two novel heterocyclic compounds through modulation of VEGFR and miR-122 in mice bearing Ehrlich ascites carcinoma. Eur. J. Pharmacol. 2021, 892, 173747. [Google Scholar] [CrossRef] [PubMed]
- Shao, T.; Wang, J.; Chen, J.G.; Wang, X.M.; Li, H.; Li, Y.P.; Li, Y.; Yang, G.D.; Mei, Q.B.; Zhang, S.Q. Discovery of 2-methoxy-3-phenylsulfonamino-5-(quinazolin-6-yl or quinolin-6-yl)benzamides as novel PI3K inhibitors and anticancer agents by bioisostere. Eur. J. Med. Chem. 2014, 75, 96–105. [Google Scholar] [CrossRef] [PubMed]
- Dittrich, C.; Zandvliet, A.S.; Gneist, M.; Huitema, A.D.; King, A.A.; Wanders, J. A phase I and pharmacokinetic study of indisulam in combination with carboplatin. Br. J. Cancer 2007, 96, 559–566. [Google Scholar] [CrossRef]
- Assi, R.; Kantarjian, H.M.; Kadia, T.M.; Pemmaraju, N.; Jabbour, E.; Jain, N.; Daver, N.; Estrov, Z.; Uehara, T.; Owa, T.; et al. Final results of a phase 2, open-label study of indisulam, idarubicin, and cytarabine in patients with relapsed or refractory acute myeloid leukemia and high-risk myelodysplastic syndrome. Cancer 2018, 124, 2758–2765. [Google Scholar] [CrossRef]
- Niu, F.; Liu, Y.; Jing, Z.; Han, G.; Sun, L.; Yan, L.; Zhou, L.; Wu, Y.; Xu, Y.; Hu, L.; et al. Novel carbazole sulfonamide microtubule-destabilizing agents exert potent antitumor activity against esophageal squamous cell carcinoma. Cancer Lett. 2018, 420, 60–71. [Google Scholar] [CrossRef]
- Tokumitsu, H.; Chijiwa, T.; Hagiwara, M.; Mizutani, A.; Terasawa, M.; Hidaka, H. KN-62, 1-[N,O-bis(5-isoquinolinesulfonyl)-N-methyl-L-tyrosyl]-4-phenylpiperazine, a specific inhibitor of Ca2+/calmodulin-dependent protein kinase II. J. Biol. Chem. 1990, 265, 4315–4320. [Google Scholar] [CrossRef]
- Wang, Y.Y.; Zhao, R.; Zhe, H. The emerging role of CaMKII in cancer. Oncotarget 2015, 6, 11725–11734. [Google Scholar] [CrossRef]
- Malarz, K.; Mularski, J.; Pacholczyk, M.; Musiol, R. The Landscape of the Anti-Kinase Activity of the IDH1 Inhibitors. Cancers 2020, 12, 536. [Google Scholar] [CrossRef]
- Woyach, J.A.; Bojnik, E.; Ruppert, A.S.; Stefanovski, M.R.; Goettl, V.M.; Smucker, K.A.; Smith, L.L.; Dubovsky, J.A.; Towns, W.H.; MacMurray, J.; et al. Bruton’s tyrosine kinase (BTK) function is important to the development and expansion of chronic lymphocytic leukemia (CLL). Blood 2014, 123, 1207–1213. [Google Scholar] [CrossRef]
- Hendriks, R.W.; Yuvaraj, S.; Kil, L.P. Targeting Bruton’s tyrosine kinase in B cell malignancies. Nat. Rev. Cancer 2014, 14, 219–232. [Google Scholar] [CrossRef]
- Kim, E.; Hurtz, C.; Koehrer, S.; Wang, Z.; Balasubramanian, S.; Chang, B.Y.; Muschen, M.; Davis, R.E.; Burger, J.A. Ibrutinib inhibits pre-BCR(+) B-cell acute lymphoblastic leukemia progression by targeting BTK and BLK. Blood 2017, 129, 1155–1165. [Google Scholar] [CrossRef]
- Wang, J.; Liu, X.; Hong, Y.; Wang, S.; Chen, P.; Gu, A.; Guo, X.; Zhao, P. Ibrutinib, a Bruton’s tyrosine kinase inhibitor, exhibits antitumoral activity and induces autophagy in glioblastoma. J. Exp. Clin. Cancer Res. 2017, 36, 96. [Google Scholar] [CrossRef]
- Yue, C.; Niu, M.; Shan, Q.Q.; Zhou, T.; Tu, Y.; Xie, P.; Hua, L.; Yu, R.; Liu, X. High expression of Bruton’s tyrosine kinase (BTK) is required for EGFR-induced NF-kappaB activation and predicts poor prognosis in human glioma. J. Exp. Clin. Cancer Res. 2017, 36, 132. [Google Scholar] [CrossRef] [PubMed]
- Zepecki, J.P.; Snyder, K.M.; Moreno, M.M.; Fajardo, E.; Fiser, A.; Ness, J.; Sarkar, A.; Toms, S.A.; Tapinos, N. Regulation of human glioma cell migration, tumor growth, and stemness gene expression using a Lck targeted inhibitor. Oncogene 2019, 38, 1734–1750. [Google Scholar] [CrossRef]
- Peerzada, M.N.; Khan, P.; Ahmad, K.; Hassan, M.I.; Azam, A. Synthesis, characterization and biological evaluation of tertiary sulfonamide derivatives of pyridyl-indole based heteroaryl chalcone as potential carbonic anhydrase IX inhibitors and anticancer agents. Eur. J. Med. Chem. 2018, 155, 13–23. [Google Scholar] [CrossRef]
- Yin, S.; Kaluz, S.; Devi, N.S.; Jabbar, A.A.; de Noronha, R.G.; Mun, J.; Zhang, Z.; Boreddy, P.R.; Wang, W.; Wang, Z.; et al. Arylsulfonamide KCN1 inhibits in vivo glioma growth and interferes with HIF signaling by disrupting HIF-1α interaction with cofactors p300/CBP. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 6623–6633. [Google Scholar] [CrossRef]
- Gwaltney, S.L., II; Imade, H.M.; Li, Q.; Gehrke, L.; Credo, R.B.; Warner, R.B.; Lee, J.Y.; Kovar, P.; Frost, D.; Ng, S.C.; et al. Novel sulfonate derivatives: Potent antimitotic agents. Bioorganic Med. Chem. Lett. 2001, 11, 1671–1673. [Google Scholar] [CrossRef]
- Kakadiya, R.; Wu, Y.C.; Dong, H.; Kuo, H.H.; Yih, L.H.; Chou, T.C.; Su, T.L. Novel 2-substituted quinolin-4-yl-benzenesulfonate derivatives: Synthesis, antiproliferative activity, and inhibition of cellular tubulin polymerization. ChemMedChem 2011, 6, 1119–1129. [Google Scholar] [CrossRef]
- Shaik, T.B.; Malik, M.S.; Routhu, S.R.; Seddigi, Z.S.; Althagafi, I.I.; Kamal, A. Evaluation of Anticancer and Anti-Mitotic Properties of Quinazoline and Quinazolino-Benzothiadiazine Derivatives. Anticancer Agents Med. Chem. 2020, 20, 599–611. [Google Scholar] [CrossRef]
- Demidenko, Z.N.; Kalurupalle, S.; Hanko, C.; Lim, C.U.; Broude, E.; Blagosklonny, M.V. Mechanism of G1-like arrest by low concentrations of paclitaxel: Next cell cycle p53-dependent arrest with sub G1 DNA content mediated by prolonged mitosis. Oncogene 2008, 27, 4402–4410. [Google Scholar] [CrossRef]
- Mukhopadhyay, S.; Saqcena, M.; Foster, D.A. Synthetic lethality in KRas-driven cancer cells created by glutamine deprivation. Oncoscience 2015, 2, 807–808. [Google Scholar] [CrossRef] [PubMed]
- Shafer, A.; Zhou, C.; Gehrig, P.A.; Boggess, J.F.; Bae-Jump, V.L. Rapamycin potentiates the effects of paclitaxel in endometrial cancer cells through inhibition of cell proliferation and induction of apoptosis. Int. J. Cancer 2010, 126, 1144–1154. [Google Scholar] [CrossRef]
- Topham, C.H.; Taylor, S.S. Mitosis and apoptosis: How is the balance set? Curr. Opin. Cell Biol. 2013, 25, 780–785. [Google Scholar] [CrossRef] [PubMed]
- Gul, H.I.; Yamali, C.; Bulbuller, M.; Kirmizibayrak, P.B.; Gul, M.; Angeli, A.; Bua, S.; Supuran, C.T. Anticancer effects of new dibenzenesulfonamides by inducing apoptosis and autophagy pathways and their carbonic anhydrase inhibitory effects on hCA I, hCA II, hCA IX, hCA XII isoenzymes. Bioorg. Chem. 2018, 78, 290–297. [Google Scholar] [CrossRef] [PubMed]
- Alaoui, S.; Dufies, M.; Driowya, M.; Demange, L.; Bougrin, K.; Robert, G.; Auberger, P.; Pages, G.; Benhida, R. Synthesis and anti-cancer activities of new sulfonamides 4-substituted-triazolyl nucleosides. Bioorg. Med. Chem. Lett. 2017, 27, 1989–1992. [Google Scholar] [CrossRef] [PubMed]
- Bhat, P.; Kriel, J.; Priya, B.S.; Shivananju, N.S.; Loos, B. Modulating autophagy in cancer therapy: Advancements and challenges for cancer cell death sensitization. Biochem. Pharmacol. 2018, 147, 170–182. [Google Scholar] [CrossRef]
- Malarz, K.; Zych, D.; Gawecki, R.; Kuczak, M.; Musioł, R.; Mrozek-Wilczkiewicz, A. New derivatives of 4′-phenyl-2,2′:6′,2″-terpyridine as promising anticancer agents. Eur. J. Med. Chem. 2020, 212, 113032. [Google Scholar] [CrossRef]
- Zhang, J.; Ng, S.; Wang, J.; Zhou, J.; Tan, S.-H.; Yang, N.; Lin, Q.; Xia, D.; Shen, H.-M. Histone deacetylase inhibitors induce autophagy through FOXO1-dependent pathways. Autophagy 2015, 11, 629–642. [Google Scholar] [CrossRef]
- Chen, J.-H.; Zhang, P.; Chen, W.-D.; Li, D.-D.; Wu, X.-Q.; Deng, R.; Jiao, L.; Li, X.; Ji, J.; Feng, G.-K.; et al. ATM-mediated PTEN phosphorylation promotes PTEN nuclear translocation and autophagy in response to DNA-damaging agents in cancer cells. Autophagy 2015, 11, 239–252. [Google Scholar] [CrossRef]
- Dalby, K.; Tekedereli, I.; Lopez-Berestein, G.; Ozpolat, B. Targeting the pro-death and pro-survival functions of autophagy as novel therapeutic strategies in cancer. Autophagy 2010, 6, 322–329. [Google Scholar] [CrossRef]
- Pal, S.; Salunke-Gawalib, S.; Konkimallaa, V.B. Induction of Autophagic Cell Death in Apoptosis-resistant Pancreatic Cancer Cells using Benzo[α]phenoxazines Derivatives, 10-methyl-benzo[α]phenoxazine-5-one and benzo[α]phenoxazine-5-one. Anticancer Agents Med. Chem. 2017, 17, 115–125. [Google Scholar] [PubMed]
- Qu, H.; Song, X.; Song, Z.; Jiang, X.; Gao, X.; Bai, L.; Wu, J.; Na, L.; Yao, Z. Berberine reduces temozolomide resistance by inducing autophagy via the ERK1/2 signaling pathway in glioblastoma. Cancer Cell Int. 2020, 20. [Google Scholar] [CrossRef] [PubMed]
- Catalano, M.; D’Alessandro, G.; Lepore, F.; Corazzari, M.; Caldarola, S.; Valacca, C.; Faienza, F.; Esposito, V.; Limatola, C.; Cecconi, F.; et al. Autophagy induction impairs migration and invasion by reversing EMT in glioblastoma cells. Mol. Oncol. 2015, 9, 1612–1625. [Google Scholar] [CrossRef] [PubMed]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. In Autophagosome and Phagosome; Deretic, V., Ed.; Humana Press: Totowa, NJ, USA, 2008; pp. 77–88. [Google Scholar] [CrossRef]
- Chitneni, S.K.; Reitman, Z.J.; Gooden, D.M.; Yan, H.; Zalutsky, M.R. Radiolabeled inhibitors as probes for imaging mutant IDH1 expression in gliomas: Synthesis and preliminary evaluation of labeled butyl-phenyl sulfonamide analogs. Eur. J. Med. Chem. 2016, 119, 218–230. [Google Scholar] [CrossRef] [PubMed]
- Law, J.M.; Stark, S.C.; Liu, K.; Liang, N.E.; Hussain, M.M.; Leiendecker, M.; Ito, D.; Verho, O.; Stern, A.M.; Johnston, S.E.; et al. Discovery of 8-Membered Ring Sulfonamides as Inhibitors of Oncogenic Mutant Isocitrate Dehydrogenase 1. ACS Med. Chem. Lett. 2016, 7, 944–949. [Google Scholar] [CrossRef] [PubMed]
- Parker, S.J.; Metallo, C.M. Metabolic consequences of oncogenic IDH mutations. Pharmacol. Ther. 2015, 152, 54–62. [Google Scholar] [CrossRef] [PubMed]
- Ohgaki, H.; Kleihues, P. Genetic Pathways to Primary and Secondary Glioblastoma. Am. J. Pathol. 2007, 170, 1445–1453. [Google Scholar] [CrossRef]
- Zeng, W.F.; Navaratne, K.; Prayson, R.A.; Weil, R.J. Aurora B expression correlates with aggressive behaviour in glioblastoma multiforme. J. Clin. Pathol. 2006, 60, 218–221. [Google Scholar] [CrossRef] [PubMed]
- Brázdová, M.; Quante, T.; Tögel, L.; Walter, K.; Loscher, C.; Tichý, V.; Činčárová, L.; Deppert, W.; Tolstonog, G.V. Modulation of gene expression in U251 glioblastoma cells by binding of mutant p53 R273H to intronic and intergenic sequences. Nucleic Acids Res. 2009, 37, 1486–1500. [Google Scholar] [CrossRef] [PubMed]
- Law, J.C.; Ritke, M.K.; Yalowich, J.C.; Leder, G.H.; Ferrell, R.E. Mutational inactivation of the p53 gene in the human erythroid leukemic K562 cell line. Leuk. Res. 1993, 17, 1045–1050. [Google Scholar] [CrossRef]
- Stuppia, L.; Calabrese, G.; Peila, R.; Guanciali-Franchi, P.; Morizio, E.; Spadano, A.; Palka, G. p53 loss and point mutations are associated with suppression of apoptosis and progression of CML into myeloid blastic crisis. Cancer Genet. Cytogenet. 1997, 98, 28–35. [Google Scholar] [CrossRef]
- Wischhusen, J.; Naumann, U.; Ohgaki, H.; Rastinejad, F.; Weller, M. CP-31398, a novel p53-stabilizing agent, induces p53-dependent and p53-independent glioma cell death. Oncogene 2003, 22, 8233–8245. [Google Scholar] [CrossRef]
- Jin, S.; Tong, T.; Fan, W.; Fan, F.; Antinore, M.J.; Zhu, X.; Mazzacurati, L.; Li, X.; Petrik, K.L.; Rajasekaran, B.; et al. GADD45-induced cell cycle G2-M arrest associates with altered subcellular distribution of cyclin B1 and is independent of p38 kinase activity. Oncogene 2002, 21, 8696–8704. [Google Scholar] [CrossRef] [PubMed]
- Chadebech, P.; Truchet, I.; Brichese, L.; Valette, A. Up-regulation of cdc2 protein during paclitaxel-induced apoptosis. Int. J. Cancer 2000, 87, 779–786. [Google Scholar] [CrossRef]
- Choi, H.J.; Fukui, M.; Zhu, B.T. Role of Cyclin B1/Cdc2 Up-Regulation in the Development of Mitotic Prometaphase Arrest in Human Breast Cancer Cells Treated with Nocodazole. PLoS ONE 2011, 6, e24312. [Google Scholar] [CrossRef]
- Malarz, K.; Mrozek-Wilczkiewicz, A.; Serda, M.; Rejmund, M.; Polanski, J.; Musiol, R. The role of oxidative stress in activity of anticancer thiosemicarbazones. Oncotarget 2018, 9, 17689–17710. [Google Scholar] [CrossRef]
- Mrozek-Wilczkiewicz, A.; Malarz, K.; Rejmund, M.; Polanski, J.; Musiol, R. Anticancer activity of the thiosemicarbazones that are based on di-2-pyridine ketone and quinoline moiety. Eur. J. Med. Chem. 2019, 171, 180–194. [Google Scholar] [CrossRef] [PubMed]
- Zhong, B.; Shingyoji, M.; Hanazono, M.; Nguyễn, T.T.T.; Morinaga, T.; Tada, Y.; Hiroshima, K.; Shimada, H.; Tagawa, M. A p53-stabilizing agent, CP-31398, induces p21 expression with increased G2/M phase through the YY1 transcription factor in esophageal carcinoma defective of the p53 pathway. Am. J. Cancer Res. 2019, 9, 79–93. [Google Scholar] [PubMed]
- Mazumder, S.; Gong, B.; Almasan, A. Cyclin E induction by genotoxic stress leads to apoptosis of hematopoietic cells. Oncogene 2000, 19, 2828–2835. [Google Scholar] [CrossRef][Green Version]
- Tsuchihara, K.; Fujii, S.; Esumi, H. Autophagy and cancer: Dynamism of the metabolism of tumor cells and tissues. Cancer Lett. 2009, 278, 130–138. [Google Scholar] [CrossRef]
- Giansanti, V.; Torriglia, A.; Scovassi, A.I. Conversation between apoptosis and autophagy: “Is it your turn or mine?”. Apoptosis 2011, 16, 321–333. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Ma, F.; Liu, Z.; Su, Q.; Liu, Y.; Liu, Z.; Li, Y. The ER-localized Ca(2+)-binding protein calreticulin couples ER stress to autophagy by associating with microtubule-associated protein 1A/1B light chain 3. J. Biol. Chem. 2019, 294, 772–782. [Google Scholar] [CrossRef]
- Zhou, X.-Y.; Luo, Y.; Zhu, Y.-M.; Liu, Z.-H.; Kent, T.A.; Rong, J.-G.; Li, W.; Qiao, S.-G.; Li, M.; Ni, Y.; et al. Inhibition of autophagy blocks cathepsins-tBid-mitochondrial apoptotic signaling pathway via stabilization of lysosomal membrane in ischemic astrocytes. Cell Death Dis. 2017, 8, e2618. [Google Scholar] [CrossRef]
- Lockman, J.W.; Klimova, Y.; Anderson, M.B.; Willardsen, J.A. Synthesis of Substituted Quinazolines: Application to the Synthesis of Verubulin. Synth. Commun. 2012, 42, 1715–1723. [Google Scholar] [CrossRef]
- Serda, M.; Malarz, K.; Mrozek-Wilczkiewicz, A.; Wojtyniak, M.; Musiol, R.; Curley, S.A. Glycofullerenes as non-receptor tyrosine kinase inhibitors-towards better nanotherapeutics for pancreatic cancer treatment. Sci. Rep. 2020, 10, 260. [Google Scholar] [CrossRef]
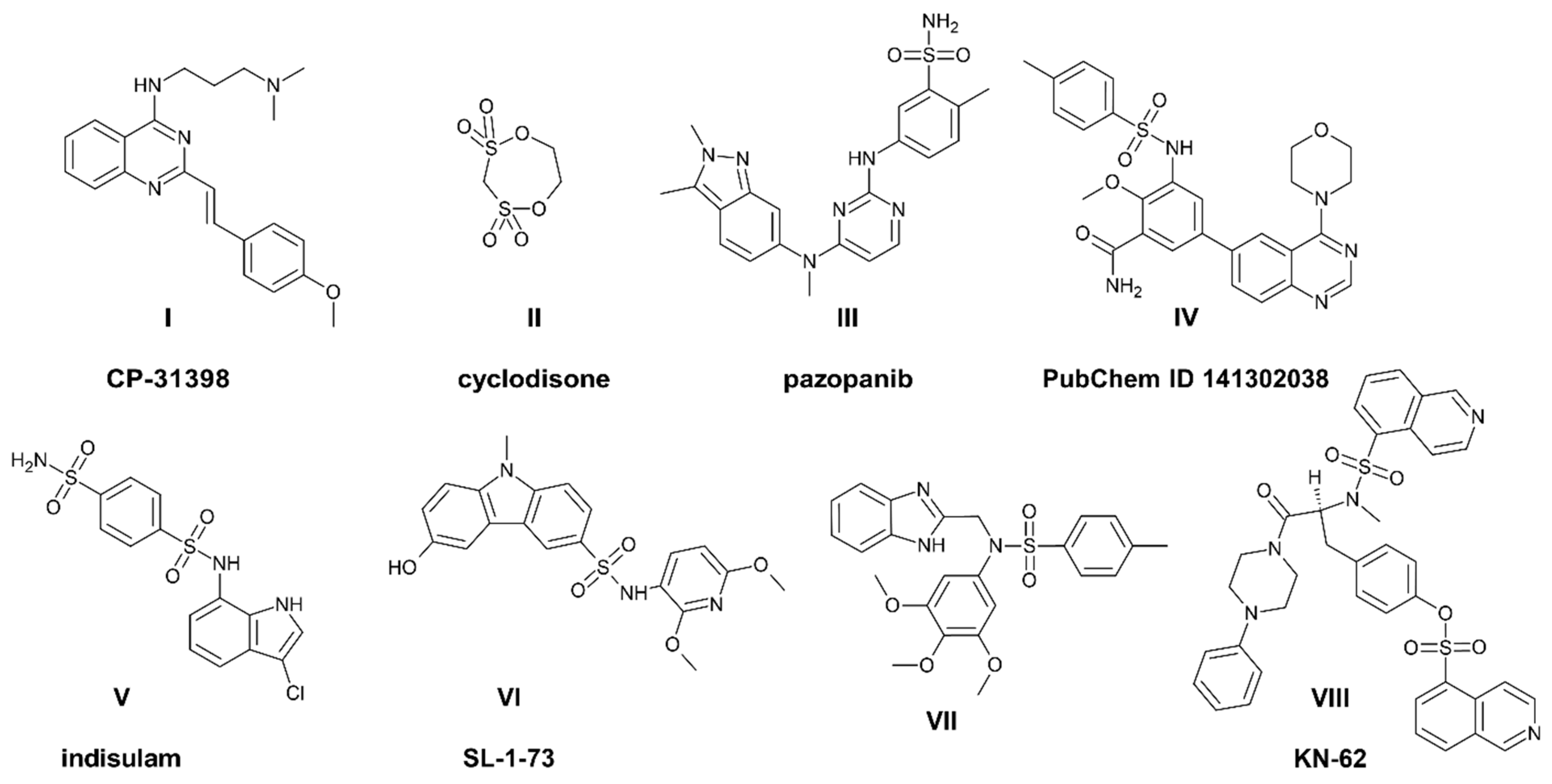



| No. | Activity IC50 Value [μM] | |||||||
|---|---|---|---|---|---|---|---|---|
| K562 | HCT 116 p53+/+ | HCT 116 p53−/− | MCF-7 | A549 | U-251 | PANC-1 | NHDF | |
| BS1 | 0.172 ± 0.034 | 0.880 ± 0.086 | 0.563 ± 0.121 | 8.325 ± 1.940 | >25 | 1.897 ± 0.649 | 3.981 ± 0.597 | 12.540 ± 0.855 |
| BS2 | 0.246 ± 0.055 | 1.200 ± 0.132 | 1.312 ± 0.290 | 16.760 ± 2.060 | >25 | 2.303 ± 0.234 | 2.905 ± 0.622 | 15.670 ± 1.410 |
| BS3 | 0.078 ± 0.027 | 0.363 ± 0.028 | 0.239 ± 0.030 | 4.599 ± 1.022 | 7.652 ± 0.987 | 1.757 ± 0.388 | 0.097 ± 0.030 | 9.415 ± 1.652 |
| BS4 | 0.173 ± 0.031 | 1.567 ± 0.357 | 3.724 ± 0.487 | 9.128 ± 2.053 | 11.340 ± 1.760 | 1.907 ± 0.214 | 0.235 ± 0.042 | >25 |
| BS5 | 10.190 ± 0.819 | >25 | >25 | >25 | >25 | >25 | >25 | >25 |
| BS6 | 2.699 ± 0.519 | >25 | 21.670 ± 1.185 | 9.791 ± 1.232 | >25 | >25 | >25 | >25 |
| CP-31398 | 3.087 ± 0.360 | 18.63 ± 0.92 | 26.28 ± 1.41 | 26.96 ± 2.10 | >25 | 18.77 ± 1.65 | >25 | 12.26 ± 0.54 |
| Imatinib | 0.133 ± 0.030 | 44.55 ± 2.41 | 51.21 ± 4.09 | >25 | >25 | >25 | >25 | >25 |
| No. | Inhibitory Effect of the Tyrosine Kinase Activity [%] at 0.5 μM | |||||||
|---|---|---|---|---|---|---|---|---|
| ABL1 | BRK | BTK | CSK | Fyn A | Lck | Lyn B | Src | |
| BS1 | 28.39 | 15.30 | 18.80 | 0 | 23.68 | 25.78 | 0 | 0.19 |
| BS2 | 0 | 9.33 | 40.18 | 34.25 | 0 | 9.93 | 37.96 | 26.32 |
| BS3 | 0 | 26.46 | 30.66 | 0 | 34.07 | 42.65 | 0 | 2.73 |
| BS4 | 0 | 35.45 | 26.89 | 28.30 | 3.51 | 19.50 | 0 | 0 |
| BS5 | 0 | 20.23 | 8.59 | 20.10 | 0 | 11.20 | 0 | 0 |
| BS6 | 0 | 0 | 17.95 | 21.14 | 62.60 | 37.26 | 0 | 0 |
| CP-31398 | 10.02 | 34.70 | 36.83 | 0.98 | 30.65 | 27.87 | 47.10 | 15.73 |
| Imatinib | 77.17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Malarz, K.; Mularski, J.; Kuczak, M.; Mrozek-Wilczkiewicz, A.; Musiol, R. Novel Benzenesulfonate Scaffolds with a High Anticancer Activity and G2/M Cell Cycle Arrest. Cancers 2021, 13, 1790. https://doi.org/10.3390/cancers13081790
Malarz K, Mularski J, Kuczak M, Mrozek-Wilczkiewicz A, Musiol R. Novel Benzenesulfonate Scaffolds with a High Anticancer Activity and G2/M Cell Cycle Arrest. Cancers. 2021; 13(8):1790. https://doi.org/10.3390/cancers13081790
Chicago/Turabian StyleMalarz, Katarzyna, Jacek Mularski, Michał Kuczak, Anna Mrozek-Wilczkiewicz, and Robert Musiol. 2021. "Novel Benzenesulfonate Scaffolds with a High Anticancer Activity and G2/M Cell Cycle Arrest" Cancers 13, no. 8: 1790. https://doi.org/10.3390/cancers13081790
APA StyleMalarz, K., Mularski, J., Kuczak, M., Mrozek-Wilczkiewicz, A., & Musiol, R. (2021). Novel Benzenesulfonate Scaffolds with a High Anticancer Activity and G2/M Cell Cycle Arrest. Cancers, 13(8), 1790. https://doi.org/10.3390/cancers13081790








