Malignant Pleural Mesothelioma Interactome with 364 Novel Protein-Protein Interactions
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
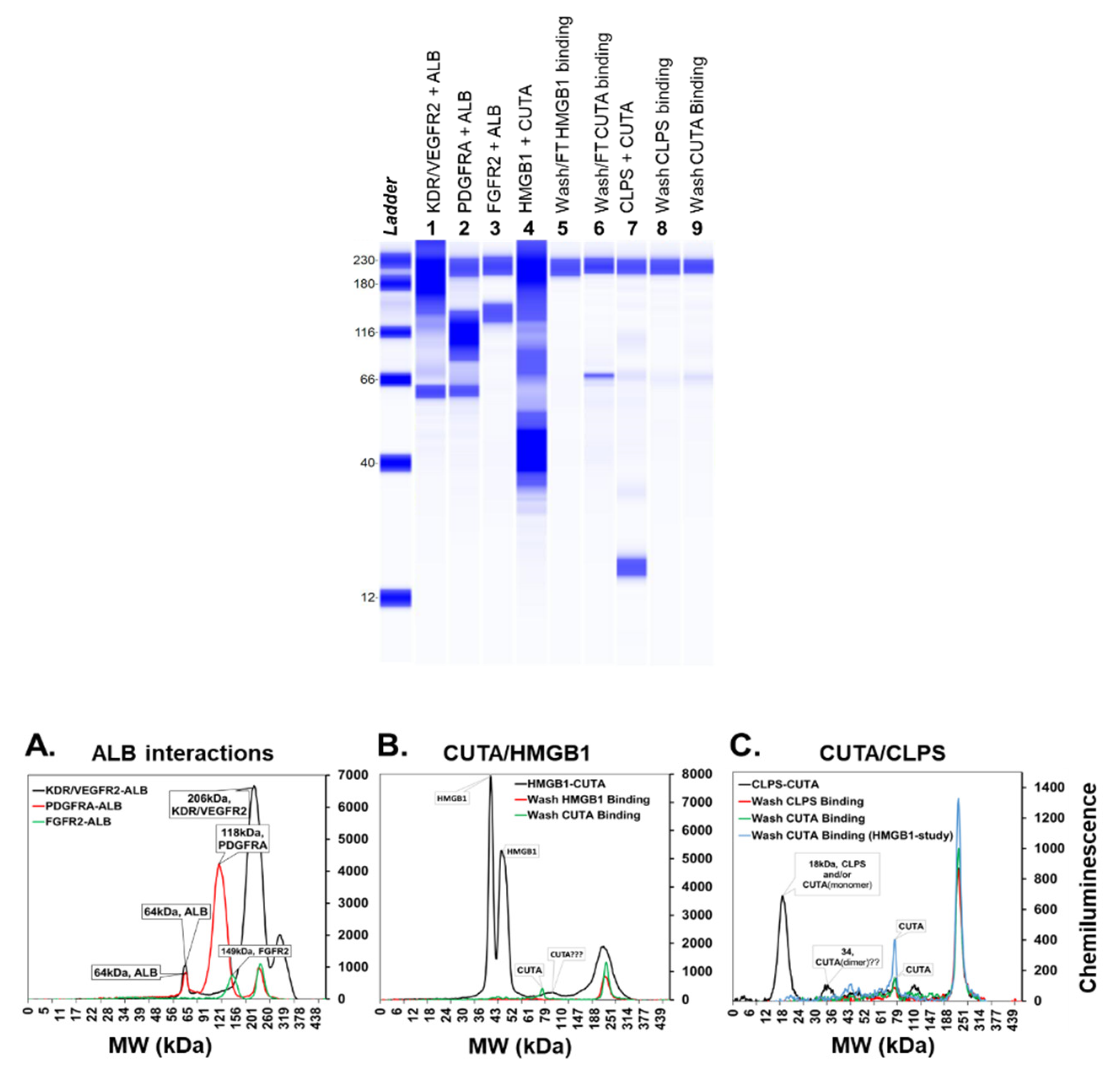
2.1. Experimental Validation of Selected Protein-Protein Interactions (PPIs)
2.2. Functional Interactions of Malignant Pleural Mesothelioma (MPM) Genes with Predicted Novel Interactors
2.3. Web Server
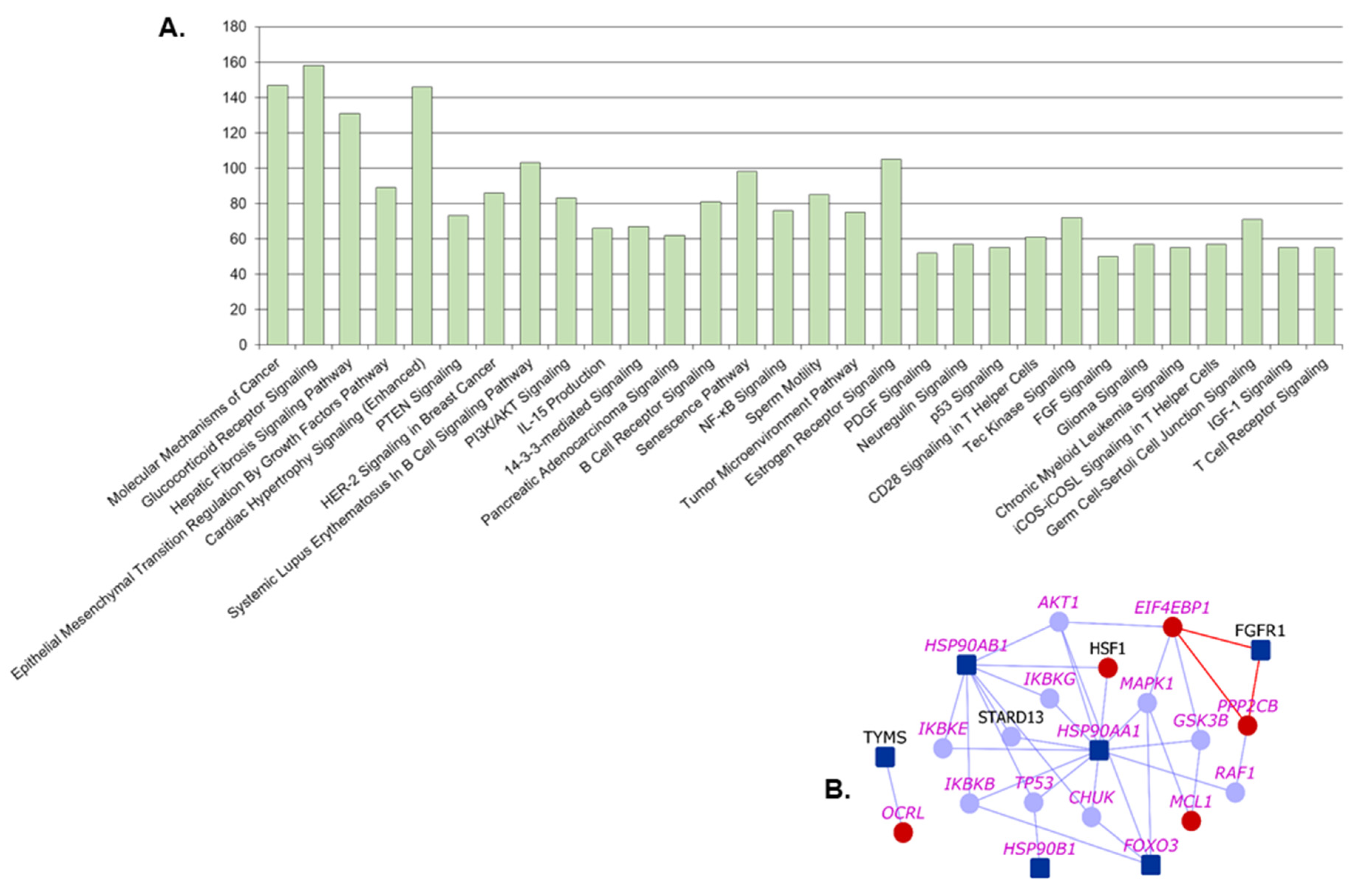
2.4. Pathway Analysis
2.5. Potentially Repurposable Drugs
2.6. Analysis with Other High-Throughput Data
3. Discussion
4. Methods
4.1. Data Collection
4.2. High-Precision Protein-Protein Interaction Prediction (HiPPIP) Model
4.3. Evaluation of PPI Prediction Model
4.4. Novel PPIs in the MPM Interactome
4.5. Previously Known PPIs in the MPM Interactome
4.6. In Vitro Pull-Down Assays
4.7. Protein Identification Methods
4.8. Ingenuity Pathway Analysis
4.9. Analysis of Differential Gene Expression in Pleural Mesothelioma Tumors and Lungs of Asbestos-Exposed Mice Versus Normal Tissue in Lungs
4.10. Analysis of DNA Methylation in MPM Tumors
4.11. Correlating Expression of MPM Genes with Lung Cancer Prognosis
4.12. Identification of Repurposable Drugs in the MPM Drug-Protein Interactome
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Repurposable Drugs for Treatment of Malignant Pleural Mesothelioma (MPM)
Appendix A.1. Repurposable Drugs Already Tested in Non-Small Cell Lung Cancer
Appendix A.2. Repurposable Drugs Targeting MPM Genes and Novel Interactors
Appendix A.3. Repurposable Drugs Targeting Known Interactors
References
- Mutsaers, S.E. The mesothelial cell. Int. J. Biochem. Cell Biol. 2004, 36, 9–16. [Google Scholar] [CrossRef]
- Carbone, M.; Adusumilli, P.S.; Alexander, H.R., Jr.; Baas, P.; Bardelli, F.; Bononi, A.; Bueno, R.; Felley-Bosco, E.; Galateau-Salle, F.; Jablons, D.; et al. Mesothelioma: Scientific clues for prevention, diagnosis, and therapy. CA Cancer J. Clin. 2019, 69, 402–429. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.J.; Reddy, G.P.; Gotway, M.B.; Higgins, C.B.; Jablons, D.M.; Ramaswamy, M.; Hawkins, R.A.; Webb, W.R. Malignant Pleural Mesothelioma: Evaluation with CT, MR Imaging, and PET. Radiographics 2004, 24, 105–119. [Google Scholar] [CrossRef] [PubMed]
- Lang-Lazdunski, L. Malignant pleural mesothelioma: Some progress, but still a long way from cure. J. Thorac. Dis. 2018, 10, 1172–1177. [Google Scholar] [CrossRef]
- Bueno, R.; Stawiski, E.W.; Goldstein, L.D.; Durinck, S.; De Rienzo, A.; Modrusan, Z.; Gnad, F.; Nguyen, T.T.; Jaiswal, B.S.; Chirieac, L.R.; et al. Comprehensive genomic analysis of malignant pleural mesothelioma identifies recurrent mutations, gene fusions and splicing alterations. Nat. Genet. 2016, 48, 407–416. [Google Scholar] [CrossRef]
- Bott, M.; Brevet, M.; Taylor, B.S.; Shimizu, S.; Ito, T.; Wang, L.; Creaney, J.; Lake, R.A.; Zakowski, M.F.; Reva, B.; et al. The nuclear deubiquitinase BAP1 is commonly inactivated by somatic mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat. Genet. 2011, 43, 668–672. [Google Scholar] [CrossRef]
- Jensen, D.E.; Proctor, M.; Marquis, S.T.; Gardner, H.P.; Ha, S.I.; Chodosh, L.A.; Ishov, A.M.; Tommerup, N.; Vissing, H.; Sekido, Y.; et al. BAP1: A novel ubiquitin hydrolase which binds to the BRCA1 RING finger and enhances BRCA1-mediated cell growth suppression. Oncogene 1998, 16, 1097–1112. [Google Scholar] [CrossRef] [PubMed]
- Hakiri, S.; Osada, H.; Ishiguro, F.; Murakami, H.; Murakami-Tonami, Y.; Yokoi, K.; Sekido, Y. Functional differences between wild-type and mutant-type BRCA1-associated protein 1 tumor suppressor against malignant mesothelioma cells. Cancer Sci. 2015, 106, 990–999. [Google Scholar] [CrossRef]
- Zauderer, M.G.; Kass, S.L.; Woo, K.; Sima, C.S.; Ginsberg, M.S.; Krug, L.M. Vinorelbine and gemcitabine as second- or third-line therapy for malignant pleural mesothelioma. Lung Cancer 2014, 84, 271–274. [Google Scholar] [CrossRef]
- Zucali, P.; Perrino, M.; Lorenzi, E.; Ceresoli, G.; De Vincenzo, F.; Simonelli, M.; Gianoncelli, L.; De Sanctis, R.; Giordano, L.; Santoro, A. Vinorelbine in pemetrexed-pretreated patients with malignant pleural mesothelioma. Lung Cancer 2014, 84, 265–270. [Google Scholar] [CrossRef]
- Zauderer, M.G.; Bott, M.; McMillan, R.; Sima, C.S.; Rusch, V.; Krug, L.M.; Ladanyi, M. Clinical characteristics of patients with malignant pleural mesothelioma harboring somatic BAP1 mutations. J. Thorac. Oncol. 2013, 8, 1430–1433. [Google Scholar] [CrossRef]
- Busacca, S.; Sheaff, M.; Arthur, K.; Gray, S.G.; O’Byrne, K.J.; Richard, D.J.; Soltermann, A.; Opitz, I.; Pass, H.; Harkin, D.P.; et al. BRCA1 is an essential mediator of vinorelbine-induced apoptosis in mesothelioma. J. Pathol. 2012, 227, 200–208. [Google Scholar] [CrossRef]
- Toyokawa, G.; Takenoyama, M.; Hirai, F.; Toyozawa, R.; Inamasu, E.; Kojo, M.; Morodomi, Y.; Shiraishi, Y.; Takenaka, T.; Yamaguchi, M.; et al. Gemcitabine and vinorelbine as second-line or beyond treatment in patients with malignant pleural mesothelioma pretreated with platinum plus pemetrexed chemotherapy. Int. J. Clin. Oncol. 2013, 19, 601–606. [Google Scholar] [CrossRef]
- Sahni, N.; Yi, S.; Taipale, M.; Bass, J.I.F.; Coulombe-Huntington, J.; Yang, F.; Peng, J.; Weile, J.; Karras, G.I.; Wang, Y.; et al. Widespread Macromolecular Interaction Perturbations in Human Genetic Disorders. Cell 2015, 161, 647–660. [Google Scholar] [CrossRef]
- Rolland, T.; Taşan, M.; Charloteaux, B.; Pevzner, S.J.; Zhong, Q.; Sahni, N.; Yi, S.; Lemmens, I.; Fontanillo, C.; Mosca, R.; et al. A Proteome-Scale Map of the Human Interactome Network. Cell 2014, 159, 1212–1226. [Google Scholar] [CrossRef]
- Ganapathiraju, M.K.; Thahir, M.; Handen, A.; Sarkar, S.N.; Sweet, R.A.; Nimgaonkar, V.L.; Loscher, C.E.; Bauer, E.M.; Chaparala, S. Schizophrenia interactome with 504 novel protein–protein interactions. Npj Schizophr. 2016, 2, 16012. [Google Scholar] [CrossRef]
- Zhu, J.; Zhang, Y.; Ghosh, A.; Cuevas, R.A.; Forero, A.; Dhar, J.; Ibsen, M.S.; Schmid-Burgk, J.L.; Schmidt, T.; Ganapathiraju, M.K.; et al. Antiviral Activity of Human OASL Protein Is Mediated by Enhancing Signaling of the RIG-I RNA Sensor. Immunity 2014, 40, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Klena, N.T.; Gabriel, G.C.; Liu, X.; Kim, A.J.; Lemke, K.; Chen, Y.; Chatterjee, B.; Devine, W.; Damerla, R.R.; et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nat. Cell Biol. 2015, 521, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yagi, H.; Saeed, S.; Bais, A.S.; Gabriel, G.C.; Chen, Z.; Peterson, K.; Li, Y.; Schwartz, M.C.; Reynolds, W.T.; et al. The complex genetics of hypoplastic left heart syndrome. Nat. Genet. 2017, 49, 1152–1159. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, K.B.; Chaparala, S.; Ganapathiraju, M.K. Potentially repurposable drugs for schizophrenia identified from its interactome. Sci. Rep. 2019, 9, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Cedrés, S.; Montero, M.; Martinez, P.; Rodríguez-Freixinós, V.; Torrejon, D.; Gabaldon, A.; Salcedo, M.; Cajal, S.R.Y.; Felip, E. Exploratory analysis of activation of PTEN–PI3K pathway and downstream proteins in malignant pleural mesothelioma (MPM). Lung Cancer 2012, 77, 192–198. [Google Scholar] [CrossRef] [PubMed]
- Prasad, T.S.K.; Goel, R.; Kandasamy, K.; Keerthikumar, S.; Kumar, S.; Mathivanan, S.; Telikicherla, D.; Raju, R.; Shafreen, B.; Venugopal, A.; et al. Human Protein Reference Database—2009 update. Nucleic Acids Res. 2008, 37, D767–D772. [Google Scholar] [CrossRef] [PubMed]
- Stark, C.; Breitkreutz, B.-J.; Reguly, T.; Boucher, L.; Breitkreutz, A.; Tyers, M. BioGRID: A general repository for interaction datasets. Nucleic Acids Res. 2006, 34, D535–D539. [Google Scholar] [CrossRef] [PubMed]
- Karunakaran, K.B.; Yanamala, N.; Boyce, G.; Ganapathiraju, M.K. Mesothelioma Interactome with 367 Novel Protein-Protein Interactions. bioRxiv 2018, 459065. [Google Scholar] [CrossRef]
- Wu, G.; Dawson, E.; Duong, A.; Haw, R.; Stein, L. ReactomeFIViz: A Cytoscape app for pathway and network-based data analysis. F1000Research 2014, 3, 146. [Google Scholar] [CrossRef]
- Orii, N.; Ganapathiraju, M.K. Wiki-Pi: A Web-Server of Annotated Human Protein-Protein Interactions to Aid in Discovery of Protein Function. PLoS ONE 2012, 7, e49029. [Google Scholar] [CrossRef]
- Amin, W.; Parwani, A.V.; Melamed, J.; Flores, R.M.; Pennathur, A.; Valdivieso, F.A.; Whelan, N.B.; Landreneau, R.; Luketich, J.D.; Feldman, M.; et al. National Mesothelioma Virtual Bank: A Platform for Collaborative Research and Mesothelioma Biobanking Resource to Support Translational Research. Lung Cancer Int. 2013, 2013, 1–9. [Google Scholar] [CrossRef]
- Amin, W.; Singh, H.; Pople, A.K.; Winters, S.; Dhir, R.; Parwani, A.V.; Becich, M.J. A decade of experience in the development and implementation of tissue banking informatics tools for intra and inter-institutional translational research. J. Pathol. Inform. 2010, 1, 12. [Google Scholar] [CrossRef]
- Krämer, A.; Green, J.; Pollard, J.; Tugendreich, S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014, 30, 523–530. [Google Scholar] [CrossRef]
- LoPiccolo, J.; Granville, C.A.; Gills, J.J.; Dennis, P.A. Targeting Akt in cancer therapy. Anti-Cancer Drugs 2007, 18, 861–874. [Google Scholar] [CrossRef]
- Kupershmidt, I.; Su, Q.J.; Grewal, A.; Sundaresh, S.; Halperin, I.; Flynn, J.; Shekar, M.; Wang, H.; Park, J.; Cui, W.; et al. Ontology-Based Meta-Analysis of Global Collections of High-Throughput Public Data. PLoS ONE 2010, 5, e13066. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Ganapathiraju, M.K. Demonstration Study: A Protocol to Combine Online Tools and Databases for Identifying Potentially Repurposable Drugs. Data 2017, 2, 15. [Google Scholar] [CrossRef]
- Suraokar, M.B.; Nunez, M.I.; Diao, L.; Chow, C.W.; Kim, D.; Behrens, C.; Lin, H.; Lee, S.; Raso, G.; Moran, C.; et al. Expression profiling stratifies mesothelioma tumors and signifies deregulation of spindle checkpoint pathway and microtubule network with therapeutic implications. Ann. Oncol. 2014, 25, 1184–1192. [Google Scholar] [CrossRef] [PubMed]
- Gordon, G.J.; Rockwell, G.N.; Jensen, R.V.; Rheinwald, J.G.; Glickman, J.N.; Aronson, J.P.; Pottorf, B.J.; Nitz, M.D.; Richards, W.G.; Sugarbaker, D.J.; et al. Identification of Novel Candidate Oncogenes and Tumor Suppressors in Malignant Pleural Mesothelioma Using Large-Scale Transcriptional Profiling. Am. J. Pathol. 2005, 166, 1827–1840. [Google Scholar] [CrossRef]
- Heintz, N.H.; Janssen-Heininger, Y.M.; Mossman, B.T. Asbestos, lung cancers, and mesotheliomas: From molecular approaches to targeting tumor survival pathways. Am. J. Respir. Cell Mol. Biol. 2010, 42, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Wishart, D.S.; Knox, C.; Guo, A.C.; Cheng, D.; Shrivastava, S.; Tzur, D.; Gautam, B.; Hassanali, M. DrugBank: A knowledgebase for drugs, drug actions and drug targets. Nuc. Acids Res. 2008, 36, D901–D906. [Google Scholar] [CrossRef] [PubMed]
- Kotsakis, A.; Matikas, A.; Koinis, F.; Kentepozidis, N.; Varthalitis, I.I.; Karavassilis, V.; Samantas, E.; Katsaounis, P.; Dermitzaki, E.K.; Hatzidaki, D.; et al. A multicentre phase II trial of cabazitaxel in patients with advanced non-small-cell lung cancer progressing after docetaxel-based chemotherapy. Br. J. Cancer 2016, 115, 784–788. [Google Scholar] [CrossRef] [PubMed]
- Johnson, F.M.; Bekele, B.N.; Feng, L.; Wistuba, I.; Tang, X.M.; Tran, H.T.; Erasmus, J.J.; Hwang, L.-L.; Takebe, N.; Blumenschein, G.R.; et al. Phase II Study of Dasatinib in Patients with Advanced Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2010, 28, 4609–4615. [Google Scholar] [CrossRef]
- Tsao, A.S.; Lin, H.; Carter, B.W.; Lee, J.J.; Rice, D.; Vaporcyan, A.; Swisher, S.; Mehran, R.; Heymach, J.; Nilsson, M.; et al. Biomarker-Integrated Neoadjuvant Dasatinib Trial in Resectable Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2018, 13, 246–257. [Google Scholar] [CrossRef]
- Comer, A.M.; Goa, K.L. Docetaxel. A review of its use in non-small cell lung cancer. Drugs Aging 2000, 17, 53–80. [Google Scholar] [CrossRef]
- Belani, C.P.; Adak, S.; Aisner, S.; Stella, P.J.; Levitan, N.; Johnson, D.H. Docetaxel for malignant mesothelioma: Phase II study of the Eastern Cooperative Oncology Group. Clin. Lung Cancer 2004, 6, 43–47. [Google Scholar] [CrossRef]
- Ralli, M.; Tourkantonis, I.; Makrilia, N.; Gkini, E.; Kotteas, E.; Gkiozos, I.; Katirtzoglou, N.; Syrigos, K. Docetaxel plus gemcitabine as first-line treatment in malignant pleural mesothelioma: A single institution phase II study. Anticancer Res. 2009, 29, 3441–3444. [Google Scholar]
- Tourkantonis, I.; Makrilia, N.; Ralli, M.; Alamara, C.; Nikolaidis, I.; Tsimpoukis, S.; Charpidou, A.; Kotanidou, A.; Syrigos, K. Phase II study of gemcitabine plus docetaxel as second-line treatment in malignant pleural mesothelioma: A single institution study. Am. J. Clin. Oncol. 2011, 34, 38–42. [Google Scholar] [CrossRef]
- Manegold, C. Gemcitabine (Gemzar®) in non-small cell lung cancer. Expert Rev. Anticancer Ther. 2004, 4, 345–360. [Google Scholar] [CrossRef]
- Kindler, H.L.; van Meerbeeck, J.P. The role of gemcitabine in the treatment of malignant mesothelioma. Semin. Oncol. 2002, 29, 70–76. [Google Scholar] [CrossRef]
- Malhotra, J.; Jabbour, S.K.; Aisner, J. Current state of immunotherapy for non-small cell lung cancer. Transl. Lung Cancer Res. 2007, 6, 196–211. [Google Scholar] [CrossRef]
- Scherpereel, A.; Mazieres, J.; Greillier, L.; Dô, P.; Bylicki, O.; Monnet, I.; Corre, R.; Audigier-Valette, C.; Locatelli-Sanchez, M.; Molinier, O. Second-or third-line nivolumab (Nivo) versus nivo plus ipilimumab (Ipi) in malignant pleural mesothelioma (MPM) patients: Results of the IFCT-1501 MAPS2 randomized phase II trial. Am. Soc. Clin. Oncol. 2017, 35, LBA8507. [Google Scholar] [CrossRef]
- Spigel, D.R.; Greco, F.A.; Waterhouse, D.M.; Shipley, D.L.; Zubkus, J.D.; Bury, M.J.; Webb, C.D.; Hart, L.L.; Gian, V.G.; Infante, J.R.; et al. Phase II trial of ixabepilone and carboplatin with or without bevacizumab in patients with previously untreated advanced non-small-cell lung cancer. Lung Cancer 2012, 78, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Puhalla, S.; Brufsky, A. Ixabepilone: A new chemotherapeutic option for refractory metastatic breast cancer. Biol. Targets Ther. 2008, 2, 505. [Google Scholar]
- Altorki, N.; Lane, M.E.; Bauer, T.; Lee, P.C.; Guarino, M.J.; Pass, H.; Felip, E.; Peylan-Ramu, N.; Gurpide, A.; Grannis, F.W.; et al. Phase II Proof-of-Concept Study of Pazopanib Monotherapy in Treatment-Naive Patients with Stage I/II Resectable Non–Small-Cell Lung Cancer. J. Clin. Oncol. 2010, 28, 3131–3137. [Google Scholar] [CrossRef] [PubMed]
- Hiddinga, B.I.; Rolfo, C.; Van Meerbeeck, J.P. Mesothelioma treatment: Are we on target? A review. J. Adv. Res. 2015, 6, 319–330. [Google Scholar] [CrossRef]
- Scagliotti, G.; Parikh, P.; Von Pawel, J.; Biesma, B.; Vansteenkiste, J.; Manegold, C.; Serwatowski, P.; Gatzemeier, U.; Digumarti, R.; Zukin, M.; et al. Phase III Study Comparing Cisplatin Plus Gemcitabine with Cisplatin Plus Pemetrexed in Chemotherapy-Naive Patients with Advanced-Stage Non–Small-Cell Lung Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. 2008, 26, 3543–3551. [Google Scholar] [CrossRef]
- Vogelzang, N.J.; Rusthoven, J.J.; Symanowski, J.; Denham, C.; Kaukel, E.; Ruffie, P.; Gatzemeier, U.; Boyer, M.; Emri, S.; Manegold, C.; et al. Phase III Study of Pemetrexed in Combination with Cisplatin Versus Cisplatin Alone in Patients with Malignant Pleural Mesothelioma. J. Clin. Oncol. 2003, 21, 2636–2644. [Google Scholar] [CrossRef]
- Jamil, M.O.; Jerome, M.S.; Miley, D.; Selander, K.S.; Robert, F. A pilot study of zoledronic acid in the treatment of patients with advanced malignant pleural mesothelioma. Lung Cancer Targets Ther. 2017, 8, 39–44. [Google Scholar] [CrossRef][Green Version]
- Okamoto, S.; Kawamura, K.; Li, Q.; Yamanaka, M.; Yang, S.; Fukamachi, T.; Tada, Y.; Tatsumi, K.; Shimada, H.; Hiroshima, K.; et al. Zoledronic Acid Produces Antitumor Effects on Mesothelioma Through Apoptosis and S-Phase Arrest in p53-Independent and Ras prenylation-Independent Manners. J. Thorac. Oncol. 2012, 7, 873–882. [Google Scholar] [CrossRef]
- Hartman, M.-L.; Esposito, J.M.; Yeap, B.Y.; Sugarbaker, D.J. Combined treatment with cisplatin and sirolimus to enhance cell death in human mesothelioma. J. Thorac. Cardiovasc. Surg. 2010, 139, 1233–1240. [Google Scholar] [CrossRef][Green Version]
- Tomasetti, M.; Gellert, N.; Procopio, A.; Neuzil, J. A vitamin E analogue suppresses malignant mesothelioma in a preclinical model: A future drug against a fatal neoplastic disease? Int. J. Cancer 2004, 109, 641–642. [Google Scholar] [CrossRef] [PubMed]
- Kovarova, J.; Bajzikova, M.; Vondrusová, M.; Stursa, J.; Goodwin, J.; Nguyen, M.; Zobalova, R.; Pesdar, E.A.; Truksa, J.; Tomasetti, M.; et al. Mitochondrial targeting of α-tocopheryl succinate enhances its anti-mesothelioma efficacy. Redox Rep. 2014, 19, 16–25. [Google Scholar] [CrossRef] [PubMed]
- Sato, A.; Virgona, N.; Sekine, Y.; Yano, T. The evidence to date: A redox-inactive analogue of tocotrienol as a new anti-mesothelioma agent. J. Rare Dis. Res. Treat. 2016, 2, 38–42. [Google Scholar]
- Karantza, V. Keratins in health and cancer: More than mere epithelial cell markers. Oncogene 2010, 30, 127–138. [Google Scholar] [CrossRef]
- Røe, O.D.; Szulkin, A.; Anderssen, E.; Flatberg, A.; Sandeck, H.; Amundsen, T.; Erlandsen, S.E.; Dobra, K.; Sundstrøm, S.H. Molecular resistance fingerprint of pemetrexed and platinum in a long-term survivor of mesothelioma. PLoS ONE 2012, 7, e40521. [Google Scholar] [CrossRef] [PubMed]
- Miettinen, M.; Limon, J.; Niezabitowski, A.; Lasota, J. Calretinin and other mesothelioma markers in synovial sarcoma: Analysis of antigenic similarities and differences with malignant mesothelioma. Am. J. Surg. Pathol. 2001, 25, 610–617. [Google Scholar] [CrossRef] [PubMed]
- Choi, A.-R.; Kim, J.-H.; Woo, Y.H.; Kim, H.S.; Yoon, S. Anti-malarial Drugs Primaquine and Chloroquine Have Different Sensitization Effects with Anti-mitotic Drugs in Resistant Cancer Cells. Anticancer Res. 2016, 36, 1641–1648. [Google Scholar] [CrossRef]
- Kimura, T.; Takagi, H.; Suzuma, K.; Kita, M.; Watanabe, D.; Yoshimura, N. Comparisons between the beneficial effects of different sulphonylurea treatments on ischemia-induced retinal neovascularization. Free Radic. Biol. Med. 2007, 43, 454–461. [Google Scholar] [CrossRef]
- Yasumitsu, A.; Tabata, C.; Tabata, R.; Hirayama, N.; Murakami, A.; Yamada, S.; Terada, T.; Iida, S.; Tamura, K.; Fukuoka, K.; et al. Clinical Significance of Serum Vascular Endothelial Growth Factor in Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2010, 5, 479–483. [Google Scholar] [CrossRef]
- Hirayama, N.; Tabata, C.; Tabata, R.; Maeda, R.; Yasumitsu, A.; Yamada, S.; Kuribayashi, K.; Fukuoka, K.; Nakano, T. Pleural effusion VEGF levels as a prognostic factor of malignant pleural mesothelioma. Respir. Med. 2011, 105, 137–142. [Google Scholar] [CrossRef]
- Pasello, G.; Urso, L.; Conte, P.; Favaretto, A. Effects of Sulfonylureas on Tumor Growth: A Review of the Literature. Oncologist 2013, 18, 1118–1125. [Google Scholar] [CrossRef]
- Krug, L.M.; Heelan, R.T.; Kris, M.G.; Venkatraman, E.; Sirotnak, F.M. Phase II Trial of Pralatrexate (10-Propargyl-10-deazaaminopterin, PDX) in Patients with Unresectable Malignant Pleural Mesothelioma. J. Thorac. Oncol. 2007, 2, 317–320. [Google Scholar] [CrossRef] [PubMed]
- Christensen, B.C.; Marsit, C.J.; Houseman, E.A.; Godleski, J.J.; Longacker, J.L.; Zheng, S.; Yeh, R.-F.; Wrensch, M.R.; Wiemels, J.L.; Karagas, M.R.; et al. Differentiation of Lung Adenocarcinoma, Pleural Mesothelioma, and Nonmalignant Pulmonary Tissues Using DNA Methylation Profiles. Cancer Res. 2009, 69, 6315–6321. [Google Scholar] [CrossRef]
- Wang, S.; Jiang, L.; Han, Y.; Chew, S.H.; Ohara, Y.; Akatsuka, S.; Weng, L.; Kawaguchi, K.; Fukui, T.; Sekido, Y.; et al. Urokinase-type plasminogen activator receptor promotes proliferation and invasion with reduced cisplatin sensitivity in malignant mesothelioma. Oncotarget 2016, 7, 69565–69578. [Google Scholar] [CrossRef] [PubMed]
- Marek, L.A.; Hinz, T.K.; Von Mässenhausen, A.; Olszewski, K.A.; Kleczko, E.K.; Boehm, D.; Weiser-Evans, M.C.; Nemenoff, R.A.; Hoffmann, H.; Warth, A.; et al. Nonamplified FGFR1 Is a Growth Driver in Malignant Pleural Mesothelioma. Mol. Cancer Res. 2014, 12, 1460–1469. [Google Scholar] [CrossRef]
- Wilson, T.R.; Lee, D.Y.; Berry, L.; Shames, D.S.; Settleman, J. Neuregulin-1-Mediated Autocrine Signaling Underlies Sensitivity to HER2 Kinase Inhibitors in a Subset of Human Cancers. Cancer Cell 2011, 20, 158–172. [Google Scholar] [CrossRef] [PubMed]
- Kaarteenaho-Wiik, R.; Soini, Y.; Pöllänen, R.; Pääkkö, P.; Kinnula, V. Over-expression of tenascin-C in malignant pleural mesothelioma. Histopathology 2003, 42, 280–291. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.-C.; Chen, L.-C.; Tseng, V.S.; Yan, J.-J.; Lai, W.-W.; Su, W.-P.; Huang, C.-Y.F. Malignant pleural effusion cells show aberrant glucose metabolism gene expression. Eur. Respir. J. 2010, 37, 1453–1465. [Google Scholar] [CrossRef] [PubMed]
- Crispi, S.; Calogero, R.A.; Santini, M.; Mellone, P.; Vincenzi, B.; Citro, G.; Vicidomini, G.; Fasano, S.; Meccariello, R.; Cobellis, G.; et al. Global Gene Expression Profiling of Human Pleural Mesotheliomas: Identification of Matrix Metalloproteinase 14 (MMP-14) as Potential Tumour Target. PLoS ONE 2009, 4, e7016. [Google Scholar] [CrossRef]
- De Rienzo, A.; Richards, W.G.; Yeap, B.Y.; Coleman, M.H.; Sugarbaker, P.E.; Chirieac, L.R.; Wang, Y.E.; Quackenbush, J.; Jensen, R.V.; Bueno, R. Sequential Binary Gene Ratio Tests Define a Novel Molecular Diagnostic Strategy for Malignant Pleural Mesothelioma. Clin. Cancer Res. 2013, 19, 2493–2502. [Google Scholar] [CrossRef]
- Consortium, G. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science 2015, 348, 648–660. [Google Scholar] [CrossRef] [PubMed]
- Yanamala, N.; Kisin, E.R.; Gutkin, D.W.; Shurin, M.R.; Harper, M.; Shvedova, A.A. Characterization of pulmonary responses in mice to asbestos/asbestiform fibers using gene expression profiles. J. Toxicol. Environ. Health Part A 2017, 81, 60–79. [Google Scholar] [CrossRef] [PubMed]
- Maxim, L.D.; McConnell, E.E. A Review of the Toxicology and Epidemiology of Wollastonite. Inhal. Toxicol. 2005, 17, 451–466. [Google Scholar] [CrossRef]
- Uhlén, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F.; et al. A pathology atlas of the human cancer transcriptome. Science 2017, 357, eaan2507. [Google Scholar] [CrossRef]
- Strizzi, L.; Catalano, A.; Vianale, G.; Orecchia, S.; Casalini, A.; Tassi, G.; Puntoni, R.; Mutti, L.; Procopio, A. Vascular endothelial growth factor is an autocrine growth factor in human malignant mesothelioma. J. Pathol. 2001, 193, 468–475. [Google Scholar] [CrossRef]
- Seto, T.; Higashiyama, M.; Funai, H.; Imamura, F.; Uematsu, K.; Seki, N.; Eguchi, K.; Yamanaka, T.; Ichinose, Y. Prognostic value of expression of vascular endothelial growth factor and its flt-1 and KDR receptors in stage I non-small-cell lung cancer. Lung Cancer 2006, 53, 91–96. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Ji, H.; Chen, M.; Robinson, B.W.S.; Dick, I.M.; Creaney, J.; Simpson, R.J. Secreted primary human malignant mesothelioma exosome signature reflects oncogenic cargo. Sci. Rep. 2016, 6, 32643. [Google Scholar] [CrossRef]
- Großerueschkamp, F.; Bracht, T.; Diehl, H.C.; Kuepper, C.; Ahrens, M.; Kallenbach-Thieltges, A.; Mosig, A.; Eisenacher, M.; Marcus, K.; Behrens, T.; et al. Spatial and molecular resolution of diffuse malignant mesothelioma heterogeneity by integrating label-free FTIR imaging, laser capture microdissection and proteomics. Sci. Rep. 2017, 7, srep44829. [Google Scholar] [CrossRef]
- Cigognetti, M.; Lonardi, S.; Fisogni, S.; Balzarini, P.; Pellegrini, V.; Tironi, A.; Bercich, L.; Bugatti, M.; De Rossi, G.; Murer, B.; et al. BAP1 (BRCA1-associated protein 1) is a highly specific marker for differentiating mesothelioma from reactive mesothelial proliferations. Mod. Pathol. 2015, 28, 1043–1057. [Google Scholar] [CrossRef]
- Lupo, B.; Trusolino, L. Inhibition of poly(ADP-ribosyl)ation in cancer: Old and new paradigms revisited. Biochim. Biophys. Acta (BBA) Bioenerg. 2014, 1846, 201–215. [Google Scholar] [CrossRef]
- Nasu, M. Identification of bap1 as a Predisposing Gene for Malignant Mesothelioma; University of Hawaii: Manoa, HI, USA, 2012. [Google Scholar]
- Yao, Z.-H.; Tian, G.-Y.; Yang, S.-X.; Wan, Y.-Y.; Kang, Y.-M.; Liu, Q.-H.; Yao, F.; Lin, D.-J. Serum albumin as a significant prognostic factor in patients with malignant pleural mesothelioma. Tumor Biol. 2014, 35, 6839–6845. [Google Scholar] [CrossRef] [PubMed]
- Iacono, M.L.; Monica, V.; Righi, L.; Grosso, F.; Libener, R.; Vatrano, S.; Bironzo, P.; Novello, S.; Musmeci, L.; Volante, M.; et al. Targeted Next-Generation Sequencing of Cancer Genes in Advanced Stage Malignant Pleural Mesothelioma: A Retrospective Study. J. Thorac. Oncol. 2015, 10, 492–499. [Google Scholar] [CrossRef]
- Liu, Y.; Zou, X.; Sun, G.; Bao, Y. Codonopsis lanceolata polysaccharide CLPS inhibits melanoma metastasis via regulating integrin signaling. Int. J. Biol. Macromol. 2017, 103, 435–440. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Iwata, S.; Yamazaki, H.; Hatano, R.; Komiya, E.; Dang, N.H.; Ohnuma, K.; Morimoto, C. CD9 Negatively Regulates CD26 Expression and Inhibits CD26-Mediated Enhancement of Invasive Potential of Malignant Mesothelioma Cells. PLoS ONE 2014, 9, e86671. [Google Scholar] [CrossRef] [PubMed]
- Pereira, S.; Lowell, C. The Lyn Tyrosine Kinase Negatively Regulates Neutrophil Integrin Signaling. J. Immunol. 2003, 171, 1319–1327. [Google Scholar] [CrossRef] [PubMed]
- Orlova, V.V.; Choi, E.Y.; Xie, C.; Chavakis, E.; Bierhaus, A.; Ihanus, E.; Ballantyne, C.M.; Gahmberg, C.G.; Bianchi, M.E.; Nawroth, P.P.; et al. A novel pathway of HMGB1-mediated inflammatory cell recruitment that requires Mac-1-integrin. EMBO J. 2007, 26, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Podar, K.; Tai, Y.-T.; Lin, B.K.; Narsimhan, R.P.; Sattler, M.; Kijima, T.; Salgia, R.; Gupta, D.; Chauhan, D.; Anderson, K.C. Vascular Endothelial Growth Factor-induced Migration of Multiple Myeloma Cells Is Associated with β1 Integrin and Phosphatidylinositol 3-Kinase-dependent PKCα Activation. J. Biol. Chem. 2002, 277, 7875–7881. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yang, L.; Chen, W.; Kong, M. Identification of Potential Hub Genes and Therapeutic Drugs in Malignant Pleural Mesothelioma by Integrated Bioinformatics Analysis. Oncol. Res. Treat. 2020, 43, 656–671. [Google Scholar] [CrossRef]
- Cheresh, P.; Morales-Nebreda, L.; Kim, S.-J.; Yeldandi, A.; Williams, D.B.; Cheng, Y.; Mutlu, G.M.; Budinger, G.R.S.; Ridge, K.; Schumacker, P.T.; et al. Asbestos-Induced Pulmonary Fibrosis Is Augmented in 8-Oxoguanine DNA Glycosylase Knockout Mice. Am. J. Respir. Cell Mol. Biol. 2015, 52, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Bozelka, B.; Sestini, P.; Gaumer, H.; Hammad, Y.; Heather, C.; Salvaggio, J. A murine model of asbestosis. Am. J. Pathol. 1983, 112, 326. [Google Scholar]
- Rehrauer, H.; Wu, L.; Blum, W.; Pecze, L.; Henzi, T.; Serre-Beinier, V.; Aquino, C.; Vrugt, B.; De Perrot, M.; Schwaller, B.; et al. How asbestos drives the tissue towards tumors: YAP activation, macrophage and mesothelial precursor recruitment, RNA editing, and somatic mutations. Oncogene 2018, 37, 2645–2659. [Google Scholar] [CrossRef] [PubMed]
- Altomare, D.A.; Vaslet, C.A.; Skele, K.L.; De Rienzo, A.; Devarajan, K.; Jhanwar, S.C.; McClatchey, A.I.; Kane, A.B.; Testa, J.R. A Mouse Model Recapitulating Molecular Features of Human Mesothelioma. Cancer Res. 2005, 65, 8090–8095. [Google Scholar] [CrossRef] [PubMed]
- Breschi, A.; Gingeras, T.R.; Guigó, A.B.R. Comparative transcriptomics in human and mouse. Nat. Rev. Genet. 2017, 18, 425–440. [Google Scholar] [CrossRef]
- Thahir, M.; Sharma, T.; Ganapathiraju, M.K. An efficient heuristic method for active feature acquisition and its application to protein-protein interaction prediction. BMC Proc. 2012, 6, S2. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2013, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Tagawa, M.; Tada, Y.; Shimada, H.; Hiroshima, K. Gene therapy for malignant mesothelioma: Current prospects and challenges. Cancer Gene Ther. 2013, 20, 150–156. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, T.; Nakanishi, Y.; Nakagawa, Y.; Tsujino, I.; Takahashi, N.; Nemoto, N.; Hashimoto, S. Association between expression of thymidylate synthase, dihydrofolate reductase, and glycinamide ribonucleotide formyltransferase and efficacy of pemetrexed in advanced non-small cell lung cancer. Anticancer Res. 2012, 32, 4589–4596. [Google Scholar] [PubMed]
- Giammarioli, A.M.; Maselli, A.; Casagrande, A.; Gambardella, L.; Gallina, A.; Spada, M.; Giovannetti, A.; Proietti, E.; Malorni, W.; Pierdominici, M. Pyrimethamine Induces Apoptosis of Melanoma Cells via a Caspase and Cathepsin Double-Edged Mechanism. Cancer Res. 2008, 68, 5291–5300. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.W.; Saadalla, A.; Ewida, A.H.; Al-Katranji, K.; Al-Saoudi, G.; Giaccone, Z.T.; Gounari, F.; Zhang, M.; Frank, D.A.; Khazaie, K. The STAT3 inhibitor pyrimethamine displays anti-cancer and immune stimulatory effects in murine models of breast cancer. Cancer Immunol. Immunother. 2017, 67, 13–23. [Google Scholar] [CrossRef] [PubMed]
- Grose, W.E.; Bodey, G.P.; Rodriguez, V. Sulfamethoxazole-Trimethoprim for Infections in Cancer Patients. JAMA 1977, 237, 352–354. [Google Scholar] [CrossRef]
- Bodey, G.P.; Grose, W.E.; Keating, M.J. Use of Trimethoprim-Sulfamethoxazole for Treatment of Infections in Patients with Cancer. Clin. Infect. Dis. 1982, 4, 579–585. [Google Scholar] [CrossRef]
- Lu, S.-H.; Tsai, W.-S.; Chang, Y.-C.; Chou, T.-Y.; Pang, S.-T.; Lin, P.-H.; Tsai, C.-M. Identifying cancer origin using circulating tumor cells. Cancer Biol. Ther. 2016, 17, 430–438. [Google Scholar] [CrossRef]
- Nitadori, J.-i.; Ishii, G.; Tsuta, K.; Yokose, T.; Murata, Y.; Kodama, T.; Nagai, K.; Kato, H.; Ochiai, A. Immunohistochemical differential diagnosis between large cell neuroendocrine carcinoma and small cell carcinoma by tissue microarray analysis with a large antibody panel. Am. J. Clin. Pathol. 2006, 125, 682–692. [Google Scholar] [CrossRef]
- Camilo, R.; Capelozzi, V.L.; Siqueira, S.A.C.; Bernardi, F.D.C. Expression of p63, keratin 5/6, keratin 7, and surfactant-A in non–small cell lung carcinomas. Hum. Pathol. 2006, 37, 542–546. [Google Scholar] [CrossRef]
- Ordóñez, N.G. Value of cytokeratin 5/6 immunostaining in distinguishing epithelial mesothelioma of the pleura from lung adenocarcinoma. Am. J. Surg. Pathol. 1998, 22, 1215–1221. [Google Scholar] [CrossRef] [PubMed]
- Heard, C.; Monk, B.; Modley, A. Binding of primaquine to epidermal membranes and keratin. Int. J. Pharm. 2003, 257, 237–244. [Google Scholar] [CrossRef]
- Kimura, T.; Shirakawa, R.; Yaoita, N.; Hayashi, T.; Nagano, K.; Horiuchi, H. The antimalarial drugs chloroquine and primaquine inhibit pyridoxal kinase, an essential enzyme for vitamin B6 production. FEBS Lett. 2014, 588, 3673–3676. [Google Scholar] [CrossRef] [PubMed]
- Basso, L.G.; Rodrigues, R.Z.; Naal, R.M.; Costa-Filho, A.J. Effects of the antimalarial drug primaquine on the dynamic structure of lipid model membranes. Biochim. Biophys. Acta (BBA) Biomembr. 2011, 1808, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Gakhar, G.; Ohira, T.; Battina, S.; Hua, D.H.; Nguyen, T.A. Anti-Tumor Effect of Primaquine Compounds in Human Breast Cancer Cells. In Proceedings of the AACR Annual Meeting, Las Angeles, CA, USA, 14–18 April 2007; American Association for Cancer Research: Philadelphia, PA, USA, 2007. [Google Scholar]
- Ou, S.-H.I.; Moon, J.; Garland, L.L.; Mack, P.C.; Testa, J.R.; Tsao, A.S.; Wozniak, A.J.; Gandara, D.R. SWOG S0722: Phase II Study of mTOR Inhibitor Everolimus (RAD001) in Advanced Malignant Pleural Mesothelioma (MPM). J. Thorac. Oncol. 2015, 10, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Mamputu, J.-C.; Renier, G. Advanced glycation end products increase, through a protein kinase C-dependent pathway, vascular endothelial growth factor expression in retinal endothelial cells: Inhibitory effect of gliclazide. J. Diabetes Complicat. 2002, 16, 284–293. [Google Scholar] [CrossRef]
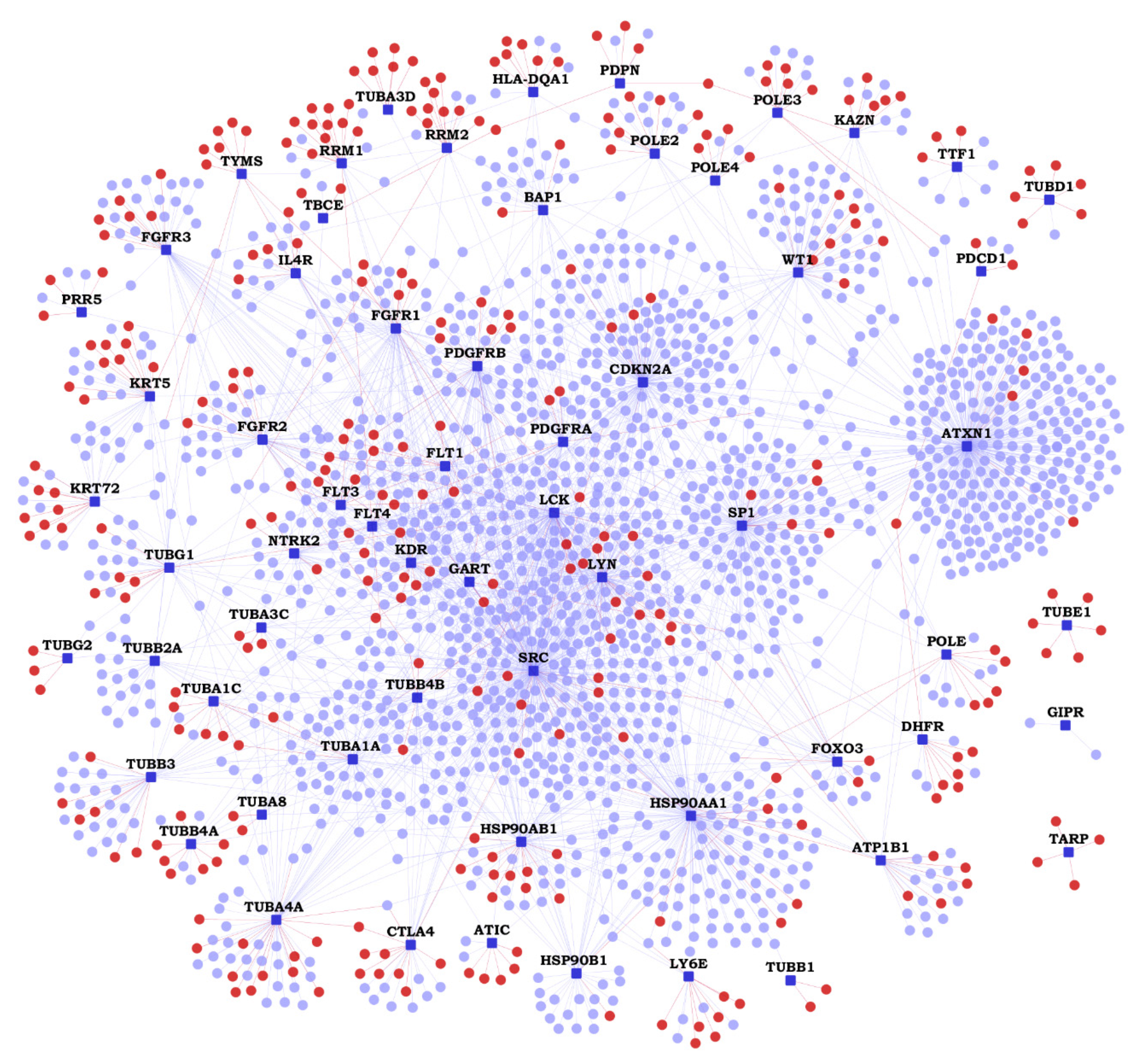
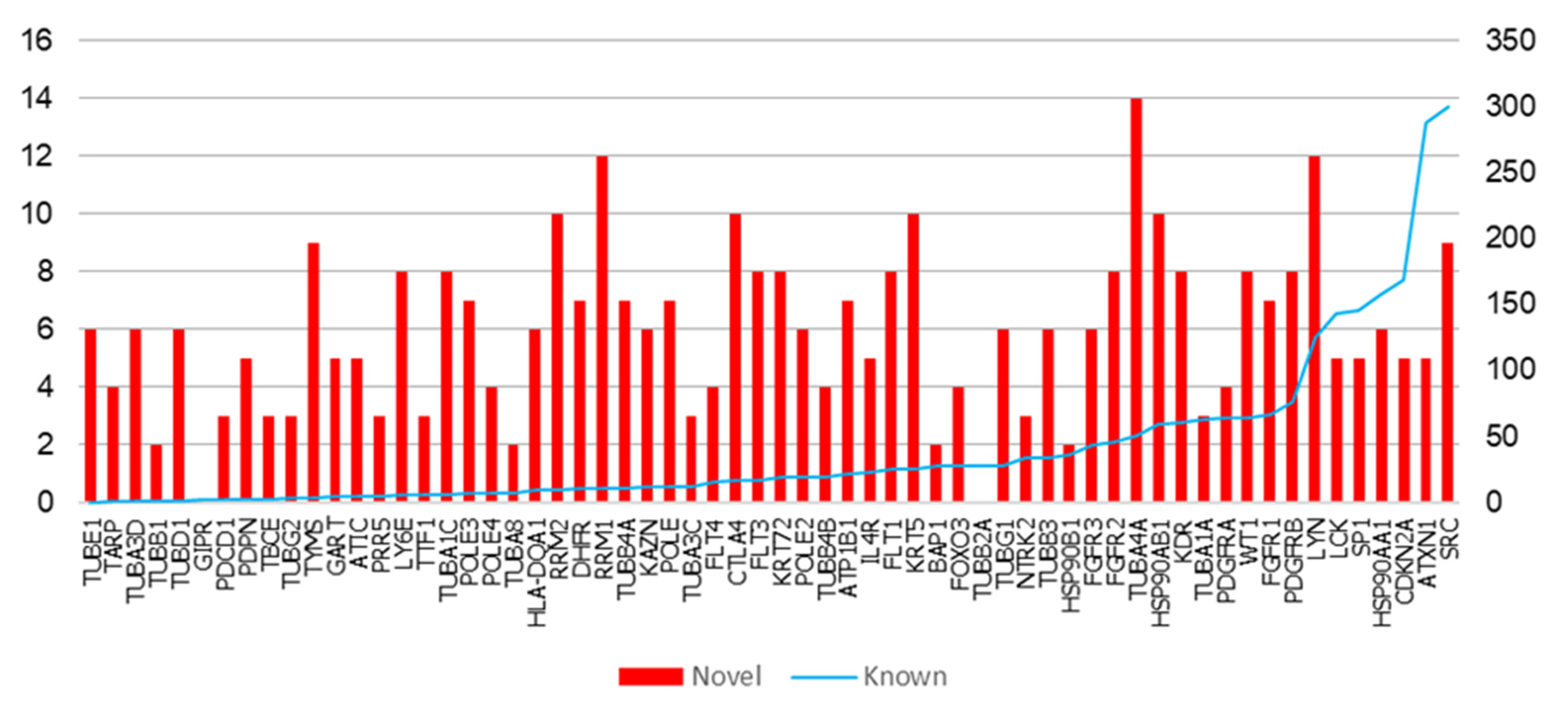

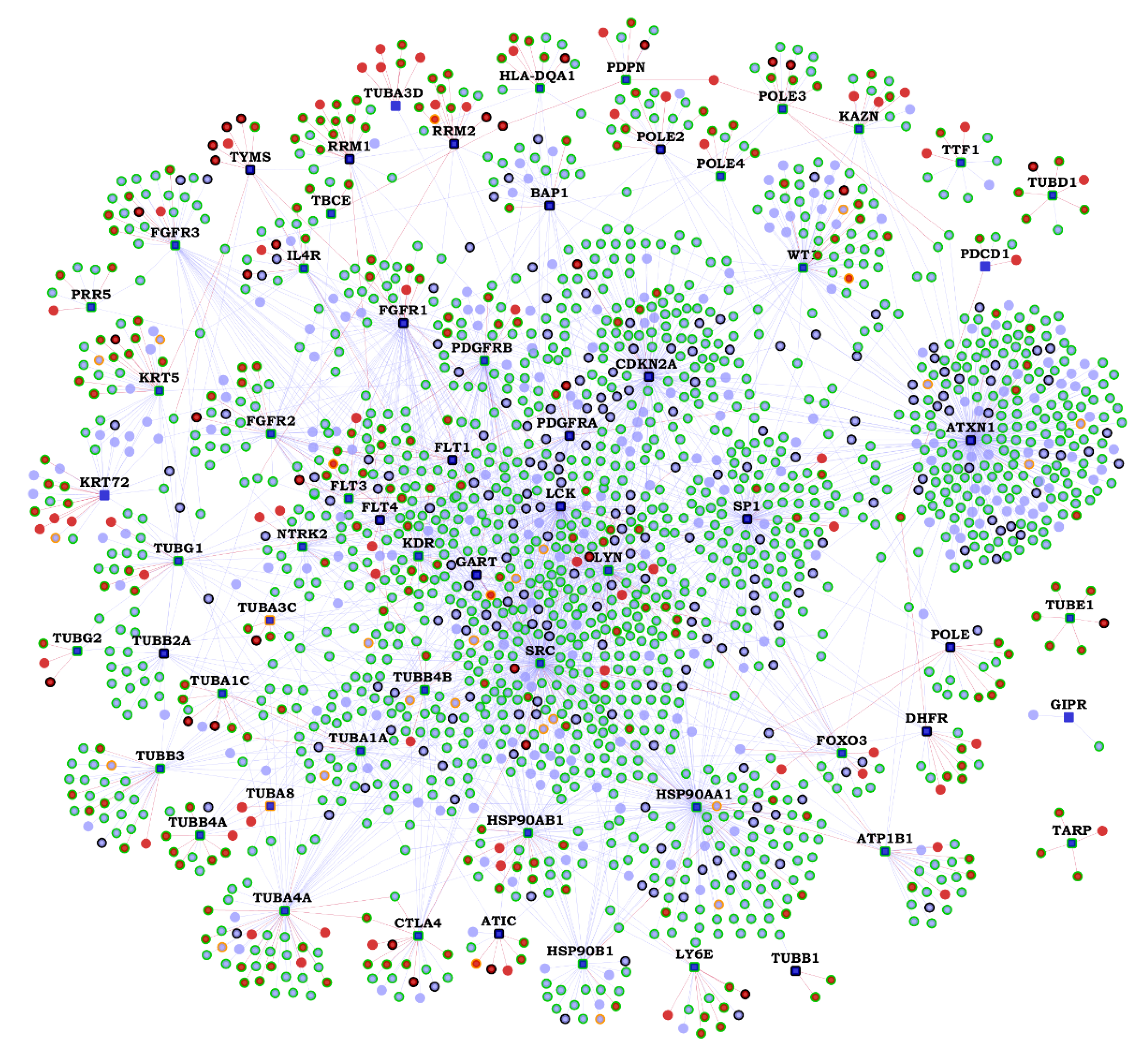
| Gene | K | N | Novel Interactors |
|---|---|---|---|
| ATP1B1 | 21 | 7 | HCRTR1, SERPINC1, TM4SF1, PRRX1, CD84, CREG1, THOC1 |
| ATIC | 5 | 5 | MAP3K7, CPS1, KIAA1524, VWC2L, DES |
| ATXN1 | 287 | 5 | CNOT6L, XPO7, C7, PITX3, RPL19 |
| BAP1 | 27 | 2 | PLN, PARP3 |
| CDKN2A | 168 | 5 | NFX1, DNAI1, GLIPR2, SIT1, CA9 |
| CTLA4 | 17 | 10 | PLCL1, DCTD, SKP1, GLP1R, AOX1, CD28, ATP5G3, CLK1, BCS1L, CDC26 |
| DHFR | 10 | 7 | RHOQ, SCZD1, TOMM7, EXOC4, DTYMK, COPS8, CRHBP |
| FGFR1 | 67 | 7 | ZFYVE1, NRG1, TPMT, OR51B4, SHB, PPP2CB, EIF4EBP1 |
| FGFR2 | 46 | 8 | PTPRE, OAT, PLXNA1, SEC23IP, MDM2, MGMT, PLSCR1, ELK4 |
| FGFR3 | 43 | 6 | GRK4, GMPS, STK32B, IDUA, IRF2BPL, ADD1 |
| FLT1 | 25 | 8 | MIPEP, RASSF9, HMGB1, FLT3, LATS2, ALOX5AP, ARL2BP, CDK8 |
| FLT3 | 17 | 8 | FMO1, SNRPA1, PNPLA3, NFIB, GPR12, SHC1, FLT1, CDK8 |
| FLT4 | 16 | 4 | NKX2-5, HNRNPH1, GRIA1, PNPLA8 |
| FOXO3 | 27 | 4 | GPR6, HDAC2, PRDM13, SIM1 |
| GART | 4 | 5 | TIAM1, NMI, TMPRSS15, JUN, IFNAR1 |
| GIPR | 2 | 0 | None |
| HLA-DQA1 | 9 | 6 | HLA-DQA2, KLHDC3, TAL2, NXF1, BRD2, HLA-DPB1 |
| HSP90AA1 | 158 | 6 | IGHA2, MED28, PHLDA2, TCIRG1, IGHD, USP13 |
| HSP90AB1 | 59 | 10 | SLC25A27, PENK, ZFP36L2, MTX2, TPSAB1, PROS1, GPRC5B, CCR7, GNPDA1, CETN3 |
| HSP90B1 | 36 | 2 | MMP17, EPB41L4B |
| IL4R | 23 | 5 | RBBP6, NPIPB5, SLC20A1, ERN2, HDGFRP3 |
| KAZN | 12 | 6 | KIF1B, NPPA, CELA2A, CELA2B, CTRC, FBLIM1 |
| KDR | 60 | 8 | UTP3, SRP72, SHOX2, KIT, ALB, CACNA1S, CHIC2, GSTA2 |
| KRT5 | 25 | 10 | SORD, KRT6A, NADSYN1, SAP18, KRT7, TARBP2, KRT6B, KRT4, DCTN1, GPD1 |
| KRT72 | 19 | 8 | SP7, KRT78, KRT80, LARP4, MYL6B, KRT74, BCDIN3D, GRASP |
| LCK | 143 | 5 | NCDN, ZSCAN20, YBX1, CITED4, CAMK1D |
| LY6E | 6 | 8 | PIP, GLI4, HSF1, AKR1B1, EIF3H, JRK, GML, GPAA1 |
| LYN | 125 | 12 | NEK7, SGK3, PDCD4, TRPA1, TERF1, PNMA2, IL7, CLCF1, AGXT, ARFGEF1, CRH, KLHL41 |
| NTRK2 | 34 | 3 | NXNL2, KCNS1, CDK20 |
| PDCD1 | 2 | 3 | COPS8, MCL1, OR6B3 |
| PDGFRA | 64 | 4 | SPOCK1, RAPGEF1, ALB, CD244 |
| PDGFRB | 76 | 8 | PLAUR, TUFM, CDX1, CHRM3, FAXDC2, ITK, CDK14, MITF |
| PDPN | 2 | 5 | PRDM2, PRMT1, ZBTB48, CELA2B, LHX1 |
| POLE | 12 | 7 | SCARB1, RAN, VSIG4, ULK1, EIF2B1, MMP17, NOS1 |
| POLE2 | 19 | 6 | SAV1, PYGL, NID2, PARK7, DRD3, ATOH1 |
| POLE3 | 7 | 7 | TNC, TRIM32, EIF4G2, ASTN2, GSN, CST3, ALAD |
| POLE4 | 7 | 4 | REG3G, SGOL1, EVA1A, B4GALT4 |
| PRR5 | 5 | 3 | WNT7B, TTC38, SCUBE1 |
| RRM1 | 10 | 12 | SLC22A18AS, SIRPA, SLC22A18, STIM1, SPINK1, ZFPM2, SH2D3A, PSMD13, RNH1, NUP98, CUZD1, RGS4 |
| RRM2 | 9 | 10 | TAF1B, ST3GAL3, NPBWR2, LPIN1, GCG, MGAT4A, BARX1, ASAP2, ITSN2, LAPTM4A |
| SP1 | 146 | 5 | HNRNPA1, REG1A, RAPGEF3, GRIN1, ENDOU |
| SRC | 300 | 9 | ZNF687, ENPP7, FMR1, PI3, PTPRT, CUL4B, DPYD, BARD1, PLTP |
| TARP | 1 | 4 | TBX20, GGCT, IL6, CPVL |
| TBCE | 2 | 3 | SERTAD3, EIF2B2, PRDM2 |
| TTF1 | 6 | 3 | AMPH, DFNB31, QRFP |
| TUBA1A | 63 | 3 | TUBA1C, AMHR2, ACVR1B |
| TUBA1C | 63 | 8 | PRKAG1, SHMT2, AMHR2, SCAF11, ACVR1B, AQP5, KMT2D, TUBA1A |
| TUBA3C | 12 | 3 | XPO4, EIF3FP2, PARP4 |
| TUBA3D | 1 | 6 | TUBA3E, WTH3DI, CCDC74B, FAM168B, LOC151121, IMP4 |
| TUBA4A | 51 | 14 | WNT6, ETV6, ATP5G3, CAPN2, CXCR1, SLC11A1, CDK5R2, ALPP, IL1RL1, NUPR1, HPCA, SKP1, DPYSL2, STK16 |
| TUBA8 | 7 | 2 | POTEH, CCT8L2 |
| TUBB1 | 1 | 2 | C20orf85, SLMO2 |
| TUBB2A | 27 | 0 | None |
| TUBB3 | 34 | 6 | PRDM7, SLC7A5, PIEZO1, MVD, TRAPPC2L, TCF25 |
| TUBB4A | 10 | 7 | UQCR11, APC2, ABCA7, PLIN3, KDM4B, SBNO2, HMG20B |
| TUBB4B | 19 | 4 | TSC1, NELFB, C9orf9, PTPRE |
| TUBD1 | 1 | 6 | TMED1, PTRH2, TRPV1, GJB3, EPX, RFX5 |
| TUBE1 | 0 | 6 | DPAGT1, NUDC, RPS20, CDC40, GOPC, C6orf203 |
| TUBG1 | 28 | 6 | WNT3, PHB, RND2, CTRL, SGCA, RARA |
| TUBG2 | 3 | 3 | NBR2, IKZF3, CLMP |
| TYMS | 3 | 9 | YES1, TAF3, ITGAM, NDUFV2, EPB41L3, SMCHD1, OCRL, THOC1, NAPG |
| WT1 | 64 | 8 | FJX1, PEX3, CAPRIN1, PAX6, BST2, B3GNT3, CALML5, HIPK3 |
| Pathway | p-Value | MPM Genes | Novel Interactors |
|---|---|---|---|
| Glucocorticoid Receptor Signaling | 6.13 × 10−56 | KRT72, HSP90B1, FGFR3, HSP90AB1, FGFR1, KRT5, FOXO3, FGFR2, HSP90AA1 | KRT74, HMGB1, PRKAG1, IL6, KRT6B, KRT78, KRT80, KRT7, KRT4, TAF3, NPPA, MAP3K7, KRT6A |
| Molecular Mechanisms of Cancer | 5.01 × 10−53 | CDKN2A, SRC, FGFR3, FGFR1, FGFR2 | CDK14, CDK20, CDKN2B, PRKAG1, WNT7B, RND2, WNT6, CDK8, RHOQ, RAPGEF3, MAP3K7, WNT3 |
| NF-κB Signaling | 1.26 × 10−39 | FGFR1, LCK, FLT1, KDR, PDGFRA, FGFR2, NTRK2, FGFR3, PDGFRB, FLT4 | MAP3K7 |
| Small Cell Lung Cancer Signaling | 2.00 × 10−37 | FGFR1, FGFR2, FGFR3 | CDKN2B |
| Axonal Guidance Signaling | 2.51 × 10−37 | TUBB1, TUBA1A, TUBA4A, TUBA8, TUBB2A, NTRK2, FGFR3, FGFR1, TUBB3, TUBG1, TUBA1C, TUBB4B, FGFR2, TUBB4A | MYL6B, DPYSL2, PRKAG1, PLCL1, WNT7B, WNT6, PLXNA1, TUBA3E, WNT3 |
| PI3K/AKT Signaling | 1.58 × 10−36 | HSP90B1, FOXO3, HSP90AA1, HSP90AB1 | OCRL, PPP2CB, MCL1, EIF4EBP1 |
| VEGF Signaling | 3.98 × 10−36 | FGFR1, FLT1, SRC, KDR, FOXO3, FGFR2, FGFR3, FLT4 | EIF2B1, EIF2B2 |
| Role of Macrophages, Fibroblasts and Endothelial Cells in Rheumatoid Arthritis | 6.31 × 10−36 | SRC, FGFR3, FGFR1, FGFR2 | IL1RL1, IL6, PLCL1, WNT7B, IL7, WNT6, CALML5, MAP3K7, WNT3, APC2 |
| Natural Killer Cell Signaling | 6.31 × 10−32 | FGFR1, LCK, FGFR2, FGFR3 | OCRL, CD244 |
| Actin Cytoskeleton Signaling | 1.58 × 10−30 | FGFR1, FGFR2, FGFR3 | MYL6B, GSN, APC2 |
| eNOS Signaling | 3.16 × 10−30 | FGFR1, FLT1, KDR, HSP90B1, FGFR2, HSP90AA1, FGFR3, FLT4, HSP90AB1 | PRKAG1, CALML5, AQP5, CHRM3 |
| Neuroinflammation Signaling Pathway | 3.98 × 10−30 | FGFR1, HLA-DQA1, FGFR2, FGFR3 | HMGB1, HLA-DQB1, ACVR1B, IL6, GRIN1, GRIA1 |
| Gap Junction Signaling | 1.00 × 10−29 | FGFR1, TUBB3, TUBG1, TUBB1, TUBA1C, TUBA1A, SRC, TUBB4B, TUBA4A, FGFR2, TUBA8, TUBB2A, FGFR3, SP1, TUBB4A | GJB3, PRKAG1, TUBA3E, PLCL1, GRIA1 |
| Integrin Signaling | 1.58 × 10−28 | FGFR1, SRC, FGFR2, FGFR3 | GSN, ITGAM, RHOQ, CAPN2, RND2 |
| IL-6 Signaling | 1.58 × 10−28 | FGFR1, FGFR2, FGFR3 | IL1RL1, MCL1, IL6, MAP3K7 |
| Drug Name & Score | Original Therapeutic Purpose(s) | Delivery | Half-Life | Toxicity | Targets |
|---|---|---|---|---|---|
| Pemetrexed negative 79 | Chemotherapeutic drug for pleural mesothelioma and non-small cell lung cancer | Powder for solution; Intravenous | 3.5 h | Data not available | ATIC, DHFR, GART, TYMS |
| Mitomycin negative 64 | Chemotherapeutic drug for breast, bladder, esophageal, stomach, pancreas, mesothelioma, lung and liver cancers | Injection, powder or lyophilized for solution; Intravenous | 8–48 min | Nausea and vomiting | - |
| Cabazitaxel negative 79 | Anti-neoplastic agent in hormone-refractory metastatic prostate cancer | Solution; Intravenous | Rapid initial-phase of 4 min, intermediate-phase of 2 h and prolonged terminal-phase of 95 h | Neutropenia, hypersensitivity reactions, gastrointestinal symptoms, renal failure | TUBB1, TUBA4A |
| Pyrimethamine negative 83 | Anti-parasitic agent in toxoplasmosis and acute malaria | Tablet; Oral | 4 days | Data not available | DHFR |
| Trimethoprim negative 63 | Anti-bacterial agent/antibiotic in urinary tract, respiratory tract and middle-ear infections and traveler’s diarrhea | Tablet/solution; Oral | 8 to 11 h | Oral toxicity in mice at LD50 = 4850 mg/kg | DHFR, TYMS |
| Primaquine negative 71 | Anti-malarial agent | Tablet; Oral | 3.7 to 7.4 h | Data not available | KRT7 |
| Gliclazide negative 56 | Anti-diabetic/hypoglycemic medication in type 2 diabetes mellitus | Tablet; Oral | 10.4 h | Oral toxicity in mice at LD50 = 3000 mg/kg, accumulation in people with severe hepatic and/or renal dysfunction, side-effects of hypoglycemia including dizziness, lack of energy, drowsiness, headache and sweating | VEGFA |
| A | B | C | D | E | F | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Novel Interactor | Differential Gene Expression | Exosome-Derived Proteins | Differential Protein Levels | Genetic Variants | Total | ||||||
| B1 | B2 | B3 | B4 | B5 | B6 | B7 | |||||
| CAPRIN1 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 6 | ||||
| RAN | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 6 | ||||
| TNC | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | 6 | ||||
| CUL4B | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| GMPS | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| IL6 | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| MGMT | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| NFIB | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| NUDC | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| PLAUR | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| PLIN3 | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| PLXNA1 | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| PRMT1 | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| RNH1 | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| SCARB1 | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| SLC7A5 | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| SMCHD1 | ✓ | ✓ | ✓ | ✓ | ✓ | 5 | |||||
| ASAP2 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| B4GALT4 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| CAPN2 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| CDC40 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| DTYMK | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| EIF3H | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| EPB41L3 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| EXOC4 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| GNPDA1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| HNRNPA1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| HNRNPH1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| LARP4 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| MGAT4A | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| MITF | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| NDUFV2 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| OAT | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| PHB | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| PHLDA2 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| PLCL1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| PRKAG1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| PROS1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| PTRH2 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| PYGL | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| RBBP6 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| SEC23IP | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| SGK3 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| SHMT2 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| SLC20A1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| TCIRG1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| XPO4 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
| YBX1 | ✓ | ✓ | ✓ | ✓ | 4 | ||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karunakaran, K.B.; Yanamala, N.; Boyce, G.; Becich, M.J.; Ganapathiraju, M.K. Malignant Pleural Mesothelioma Interactome with 364 Novel Protein-Protein Interactions. Cancers 2021, 13, 1660. https://doi.org/10.3390/cancers13071660
Karunakaran KB, Yanamala N, Boyce G, Becich MJ, Ganapathiraju MK. Malignant Pleural Mesothelioma Interactome with 364 Novel Protein-Protein Interactions. Cancers. 2021; 13(7):1660. https://doi.org/10.3390/cancers13071660
Chicago/Turabian StyleKarunakaran, Kalyani B., Naveena Yanamala, Gregory Boyce, Michael J. Becich, and Madhavi K. Ganapathiraju. 2021. "Malignant Pleural Mesothelioma Interactome with 364 Novel Protein-Protein Interactions" Cancers 13, no. 7: 1660. https://doi.org/10.3390/cancers13071660
APA StyleKarunakaran, K. B., Yanamala, N., Boyce, G., Becich, M. J., & Ganapathiraju, M. K. (2021). Malignant Pleural Mesothelioma Interactome with 364 Novel Protein-Protein Interactions. Cancers, 13(7), 1660. https://doi.org/10.3390/cancers13071660






