Simple Summary
This in silico study aimed to investigate associations between dipeptidyl peptidase (DPP) 9 mRNA expression, survival and gene signature in human hepatocellular carcinoma (HCC). DPP9 loss-of-function exonic variants were mostly associated with cancers. In HCC patients, DPP9 and the closely related genes DPP4 and DPP8 were upregulated in liver tumors. High co-expression of genes that were positively correlated with DPP4, DPP8 and DPP9 was associated with poor survival in HCC patients. These findings strongly implicate the DPP4 gene family, especially DPP9, in the pathogenesis of human HCC and therefore encourages future functional studies.
Abstract
Dipeptidyl peptidase (DPP) 9, DPP8, DPP4 and fibroblast activation protein (FAP) are the four enzymatically active members of the S9b protease family. Associations of DPP9 with human liver cancer, exonic single nucleotide polymorphisms (SNPs) in DPP9 and loss of function (LoF) variants have not been explored. Human genomic databases, including The Cancer Genome Atlas (TCGA), were interrogated to identify DPP9 LoF variants and associated cancers. Survival and gene signature analyses were performed on hepatocellular carcinoma (HCC) data. We found that DPP9 and DPP8 are intolerant to LoF variants. DPP9 exonic LoF variants were most often associated with uterine carcinoma and lung carcinoma. All four DPP4-like genes were overexpressed in liver tumors and their joint high expression was associated with poor survival in HCC. Increased DPP9 expression was associated with obesity in HCC patients. High expression of genes that positively correlated with overexpression of DPP4, DPP8, and DPP9 were associated with very poor survival in HCC. Enriched pathways analysis of these positively correlated genes featured Toll-like receptor and SUMOylation pathways. This comprehensive data mining suggests that DPP9 is important for survival and that the DPP4 protease family, particularly DPP9, is important in the pathogenesis of human HCC.
1. Introduction
Dipeptidyl peptidase (DPP) 9, DPP4, DPP8 and fibroblast activation protein (FAP) are the enzymatic members of the DPP4 family of serine proteases and have been implicated in cancer pathogenesis [1]. DPP9 is ubiquitously expressed in tissues [2] and has diverse roles in cell behaviors [3], immune regulation [4,5,6] and cancer [7,8,9]. DPP9 can interact with H-Ras, which is a key molecule of the epidermal growth factor receptor and PI3K/Akt signaling pathways; these pathways are important for cell survival, proliferation and apoptosis [3]. DPP9 inhibition stimulates the immune system by activating pyroptosis in monocytes and macrophages, in which inhibition of DPP9 causes formation of multiprotein complexes called inflammasomes, which activates caspase-1 and generates pyroptosis [3]. A study using a non-small-cell-lung cancer (NSCLC) model demonstrated that knockdown of DPP9 could inhibit lung cancer cell proliferation, migration and tumorigenesis. Overexpression of DPP9 in NSCLC is independently associated with poor 5-year overall survival [7]. Similarly, in colorectal cancer, greater DPP9 expression is associated with poor prognosis [8]. In contrast, in patients with oral squamous cell carcinoma, lower DPP9 expression correlates with poor survival [9]. These data suggest that DPP9 has different roles in various types of cancers.
Liver is rich in DPP9 enzyme activity [2], and DPP9 mRNA is upregulated in inflamed and fibrotic mouse liver [10]. Downregulating DPP9 or blocking its enzyme activity reduces cell adhesion and migration in the Huh7 human hepatoma cell line [11]. The role of DPP9 in human liver cancer has not been studied, particularly in liver hepatocellular carcinoma (HCC). HCC is the most common primary liver cancer [12]. The major risk for HCC is chronic liver injury, with causes including non-alcoholic fatty liver disease (NAFLD), obesity, smoking tobacco, chronic hepatitis B or C, and chronic excessive alcohol consumption [13,14,15]. NAFLD, obesity and associated metabolic diseases are becoming the leading cause of HCC [16].
The DPP4 enzyme family is the S9b subfamily of the prolyl oligopeptidase (S9) family and share 27–60% protein sequence identity with each other [17,18]. The proteases in the DPP4 enzyme family are able to hydrolyze peptides after a proline residue and can remove a dipeptide from the N-terminus of substrates [18,19]. Very few proteases have this capability. DPP4 and FAP are expressed on the cell surface, whereas DPP8 and DPP9 are intracellular [1]. DPP8 and DPP9 share the greatest similarity in protein sequence and crystal structure in this gene family [20]. There are two isoforms of DPP9 with distinct subcellular localizations: the long form (892 amino acids) is in both cytosol and nucleus because it contains a nuclear localization sequence while the short form (863 amino acids) is only cytosolic [21]. The S9b proteases are active as dimers [22], with each monomer having two domains, the α/β-hydrolase domain and the β-propeller domain [20]. The β−propeller domain binds to protein ligands, and DPP9 has been found to bind to SUMO-1 [23] and filamin A [6] and associate with H-Ras [3], which are probably involved in roles of DPP9 in cell adhesion and migration. Selective inhibitors for DPP9 versus DPP8 are lacking, so individual functions of DPP9 are poorly resolved and derive primarily from genetic manipulations of mice and cell lines [1,24,25].
Large-scale human genetic databases have become increasingly powerful resources to study the impacts of somatic mutation in diseases, especially in cancer. Human gene sequence variants that are predicted to cause loss of function (LoF) of an encoded protein provide valuable data on studying human gene inactivation [26,27]. Genome-wide association studies (GWAS) have found several risk loci mapping to the DPP9 gene on chromosome 19 (19p13.3) that contribute to human diseases, including alcoholism [28], interstitial lung abnormalities [29], pulmonary fibrosis [29,30] and adolescent idiopathic scoliosis [31]. The intronic single nucleotide polymorphism (SNP) rs113683471 in DPP9 is associated with alcoholism [28]. Intronic SNPs rs12610495 and rs2109069 in DPP9 are genome-wide significant loci for severe COVID-19, idiopathic pulmonary fibrosis and idiopathic interstitial pneumonia [29,30,32,33]. However, there are limited information on exonic SNPs in DPP9 and DPP9 LoF variants, or their associations with diseases.
This study identifies DPP9 exonic LoF variants in human genomic databases and their associations with cancers. This study also reports the first comprehensive data mining of associations between DPP9 mRNA expression, survival and gene signature in human HCC.
2. Materials and Methods
2.1. Protein Sequences
The amino acid sequences of the short (863 amino acids) and long forms (892 amino acids) of DPP9 were downloaded from the UniProt Consortium (https://www.uniprot.org/uniprot/Q86TI2, accessed on 7 April 2020). The long form of DPP9 and its amino acid numbering was used in this study. Protein crystal structure of the four enzymatic members of the DPP4 enzyme family were depicted using the PyMOL Molecular Graphics System Version 2.4.2, and a multiple-sequence alignment was made using a Clustal Omega bioinformatics tools for multiple sequence alignment (https://www.ebi.ac.uk/Tools/msa/, accessed on 6 March 2021) (Supplementary Figure S1).
2.2. Public Databases for Accessing Loss of Function Variants
Public databases assessed in this study are listed in Table 1.

Table 1.
Public databases sourced for loss of function data in this study.
2.3. Loss of Function Variants
The exonic variants that are predicted to cause LoF of DPP9 were exported from gnomAD, TCGA and COSMIC. Only the LoF variants that are annotated as either stop gained or frameshift in the databases were studied. The numbering of the amino acid sequence of DPP9 in GnomAD is of the short form of DPP9. Therefore, to use the DPP9 long form, 29 was added to all protein sequence numbers for the gnomAD DPP9 LoF analyses. DPP9 variants from TCGA and COSMIC were found to be of the long form. Variants with a sequence number mismatch with the long form of DPP9 were excluded. In addition, if frameshift variants exported from gnomAD and TCGA occurred where there is no corresponding structural residue, these frameshift variants were considered unlikely to be DPP9 exonic LoF variants, and thus were excluded from this study.
2.4. Intronic Variants
The two DPP9 intronic variants rs12610495 and rs2109069 have the smallest p values that have been associated with critical illness caused by COVID-19 [33]. More genetic information about these two intronic variants was exported from PhenoScanner V2 [40].
2.5. Genetic Variant Tolerance Parameters
The observed to expected ratio (o:e) of LoF, the probability of LoF intolerance (pLI) and haploinsufficiency score were retrieved from gnomAD and DECIPHER [38]. Tolerance percentile and GDI values were retrieved from GeneCards human gene database.
2.6. Gene Correlation Analysis
UALCAN is a comprehensive web database for investigating complete genetic or molecular data of cancers and allows in silico validation of genes of interest [41]. It is available online at http://ualcan.path.uab.edu/index.html, accessed on 16 October 2020. Both positively and negatively correlated genes of DPPs and FAP were accessed using liver hepatocellular carcinoma (LIHC; HCC) dataset of TCGA. The expression level of each gene was displayed as transcripts per million (TPM).
2.7. Survival Analysis
OncoLnc database contains TCGA survival data to mRNA expression level for 8647 patients from 21 cancer projects [42]. Survival data for 360 patients with HCC and 540 patients with UCEC were exported from OncoLnc. Normal patients’ data (n = 50) were exported from UALCAN [41]. HCC patient survival was also analyzed based on patients’ BMI (Table 2). High and low levels of gene expression were stratified based on either median expression, or the upper or lower quartile, based on the biological questions we were addressing.

Table 2.
Categories of BMI.
SurvExpress is an online database for cancer gene expression data using survival analysis, which is able to access the influence of combined high expression of multiple genes on survival in a specific cancer type [43]. The genes that correlate in-common amongst the DPP4 gene family were imported into SurvExpress and analyzed for their associations with HCC patient survival (June 2016 dataset). Patient survival time and status were exported from SurvExpress and Kaplan-Meier curves were plotted.
2.8. Reactome Pathways Enrichment Analysis
Reactome enrichment pathways of positively or negatively correlated genes in-common between DPP9 and DPP8, and of the positively correlated genes in-common amongst DPP9, DPP8 and DPP4 were explored using ConcensusPathDB (CPDB) [44]. Genes were imported into ConcensusPathDB and identified as gene symbols. The analysis was performed under gene set analysis and over-representation analysis. In ConsensusPathDB, each p value was calculated in a hypergeometric test based on the number of entities present in both the predefined set and a user defined list of entities, and the p values were corrected for multiple testing using false discovery rate method.
2.9. Data and Statistical Analyses
Figures and Kaplan-Meier curves were generated in GraphPad Prism (version 8.4.2, San Diego, CA, USA). Data statistics were analyzed in GraphPad Prism. Survival analysis was performed in Prism using logrank (Mantel-Cox) to assess the significance between the groups. The Mann-Whitney U test was used to compare differences between two independent groups. The Kruskal-Wallis one-way analysis of variance with Tukey’s post-hoc test was used to compare differences amongst multiple groups. Significance was assigned to p values < 0.05, <0.01 or <0.0001.
3. Results
3.1. DPP9 Loss of Function (LoF) Variants and Disease Associations
Large-scale human genetic databases provide useful tools to study protein LoF variants and their associations with diseases. DPP9 LoF has not been explored previously. Therefore, we assessed data from several human genomic databases to identify DPP9 LoF and potentially associated diseases. Gaining a premature termination codon (PTC), or stop codon, anywhere in a gene of the DPP4 gene family is expected to ablate enzyme activity [45]. We have shown previously that mature termination mutants of DPP8 are inactive, even if lacking only the C-terminal alpha-helix [45]. Similarly, any frameshift variant is almost certain to lack activity due to nonsense sequence causing a PTC. Therefore, the exonic gene variants considered to be LoF are very likely true LoF.
In gnomAD, there were 18 exonic variants of DPP9 that were tagged as LoF. However, examining the sequence of long-form DPP9 in the UniProt Consortium database revealed that some variants were mismatched with UniProt, and thus were excluded. Only exonic LoF variants that were annotated as either stop-gained or frameshift variants were included in this study. We identified three frameshift variants and seven stop gained variants that were exonic in gnomAD (Table 3). There was no record of associated diseases for each mutation variant. In TCGA, we identified eight DPP9 LoF variants (Table 4), and none of these variants were the same as found in gnomAD. Four DPP9 variants were designated as stop gained, with three of them associated with primary uterine corpus endometrial carcinoma (UCEC). Another four DPP9 variants were designated as frameshift mutation, with half of them found in patients diagnosed with stomach adenocarcinoma (Table 4). None of the DPP9 variants in TCGA were found to be associated with HCC. There were many more DPP8 than DPP9 LoF gene variants in TCGA, found in diverse types of cancer, mostly in UCEC and one in HCC (Supplementary Table S1). In a separate database, COSMIC, eleven DPP9 variants had been tagged as nonsense mutation and were substitution mutations resulting in PTC codons (Table 5). These variants were associated with different types of carcinoma, most often lung carcinoma. All variants that were likely to cause LoF for DPP8 or DPP9 have a PTC. All exonic DPP9 variants found in each of the three databases were mapped to the DPP9 protein structure (Supplementary Figure S2).

Table 3.
DPP9 LoF variants from gnomAD. ins = insertion. * Premature termination codon (PTC).

Table 4.
DPP9 LoF variants in TCGA. Simple somatic mutation (SSM) affected frequency was calculated as the number of cases affected by a specific mutation in a TCGA disease project divided by the number of cases tested for SSM in that disease project in TCGA. ins = insertion. del = deletion. * Premature termination codon (PTC).

Table 5.
DPP9 LoF variants in COSMIC. Nonsense mutation that is a substitution mutation resulting in a premature termination codon (*). CDS = coding sequence; AA = amino acid; SSM = simple somatic mutation.
There were 22 DPP8 nonsense mutations in COSMIC, most often in colon adenocarcinoma and one in HCC (Supplementary Table S2). DPP9 variants Glutamic acid (Glu) 154 to PTC and Arginine (Arg) 303 to PTC were both reported in both TCGA and COSMIC, but with different nucleotide substitutions. Another anomaly was that in COSMIC, both endometrioid carcinoma and colon adenocarcinoma were associated with the Arg303* variant, but in TCGA only endometrioid carcinoma was recorded.
3.2. Intronic SNPs in DPP9
Intronic SNPs rs12610495 and rs2109069 in DPP9 are genome-wide significant loci for severe COVID-19, idiopathic pulmonary fibrosis and idiopathic interstitial pneumonia [29,30,32,33]. These SNPs are located 5′ to the open reading frame, at nucleotides 4717672 and 4719443 of chromosome 19. They are important DPP9 SNPs because they are relatively common, with allelic frequencies of 0.706 to 0.828 for rs12610495 and 0.186 to 0.321 for rs2109069 (Supplementary Table S3). These SNPs have been associated with lower DPP9 expression levels in human lung [33]. We found that these SNPs are not associated with liver fibrosis or cancer in the PhenoScanner database.
3.3. Genetic Variation Tolerance in DPP9 and the DPP4 Gene Family
Some databases incorporate scoring systems that determine tolerance towards genetic variations. We analyzed genetic variation tolerance in the DPP4 gene family using five scores: observed to expected ratio (o:e), probability of LoF intolerance (pLI), haploinsufficiency score, tolerance percentile, and gene damage index (GDI) scores. When o:e is approximately 0.1, the gene is haploinsufficient and heterozygous LoF is very probably not tolerated; when o:e is between 0.1 and 0.5, the heterozygous LoF may be tolerated; when o:e is close to 1, LoF is tolerated [46]. DPP8 and DPP9 had low o:e, little more than 0.1, DPP4 had a moderate o:e less than 0.5 and FAP has a high o:e (Table 6). The probability of LoF intolerance (pLI) is the probability of a given gene being haploinsufficient; genes with pLI ≥ 0.9 are extremely intolerant to LoF variation and genes with pLI ≤ 0.1 are LoF tolerant [46]. The pLI scores for DPP9, DPP8, DPP4 and FAP (Table 6) suggested that DPP8 and DPP9 LoF variants are poorly tolerated. Haploinsufficient genes only contain a single copy of a diploid genome that changes the organism phenotype from a normal to disease state, and many of the LoF-associated haploinsufficient genes contribute to metabolic disorders and tumorigenesis [47]. The haploinsufficiency score was greater for DPP9 than for DPP8, DPP4 or FAP, and, like the o:e and pLI, indicated that DPP9 is likely to exhibit haploinsufficiency whereas DPP4 and FAP are unlikely to exhibit haploinsufficiency. The gene damage index (GDI) scores for these four enzymes were low (Table 6), suggesting that LoF variants are likely to be disease-causing [48]. Genes in the 25th tolerance percentile and below are considered relatively intolerant to variation. Unlike the other haploinsufficiency indications, GDI and tolerance percentile did not discriminate amongst the four genes (Table 6).

Table 6.
Genetic variation tolerance in the DPP4 gene family. The o:e ratio, pLI and haploinsufficiency score were retrieved from gnomAD and DECIPHER. Tolerance percentile and GDI values were retrieved from the GeneCards human gene database. o:e, the observed to expected ratio. pLI, the probability of LoF intolerance. GDI, gene damage index score.
3.4. Association of DPP9 Expression with Patient Survival in HCC and UCEC
Previous studies reported associations of DPP9 expression with patient survival in non-small-cell lung cancer [7], colorectal cancer [8] and oral squamous cell carcinoma [9]. Many DPP8 and DPP9 variants were identified in patients with UCEC and HCC in our study (Table 4 and Supplementary Tables S1 and S2). Therefore, we analyzed whether DPPs expression is associated with patient survival in HCC and UCEC. Clinical parameters and mRNA expression of DPPs in patients with HCC and UCEC were exported from TCGA. Patient demographics are presented in Supplementary Table S4. Patients were stratified by upper or lower quartile expression of DPPs. Kaplan-Meier analysis for DPP9, DPP8, DPP4 and FAP mRNA suggested that expression of none of these four genes was associated with HCC overall survival (OS) (Figure 1A–D). In contrast, patients with low DPP9 mRNA expression had significantly shorter survival time for UCEC (Figure 1E). Less than 15% of UCEC patients with low DPP9 expression had survived for 10 years after diagnosis. Unlike DPP9, DPP8, DPP4 and FAP expression did not associate with OS in UCEC (Figure 1F–H). This data indicates that DPP9 might have an important role in UCEC.
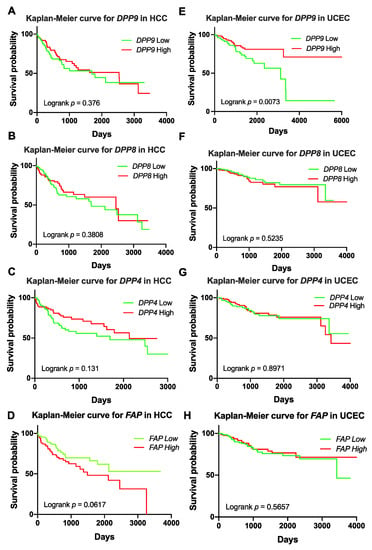
Figure 1.
Kaplan-Meier curves for DPP9, DPP8, DPP4 and FAP in liver tumors (hepatocellular carcinoma, HCC; (A–D)) (n = 360) and uterine corpus endometrial carcinoma (UCEC; (E–H)) (n = 540). High or low was defined as the mRNA expression values in the upper or lower quartile, respectively. p values were calculated by logrank (Mantel-Cox) test.
3.5. Association of DPP9 Expression with Clinical Parameters and Survival Outcome in HCC
We next explored closely the association of DPP9 with clinical parameters relevant to HCC, including gender, age, BMI and cancer stages. In HCC patients, all four DPP genes were significantly upregulated in tumors compared to normal tissue (Figure 2A–D). Epidemiology studies have shown that globally, the numbers of male HCC patients are two to four times greater than female HCC patients [15,49]. Our results showed that significantly greater expression of DPP9 was detected in liver tumors from females than males (Figure 2E). It may be expected that HCC patients with greater age and advance stage of cancer will have more expression of DPP9 and therefore poorer prognosis. However, DPP9 expression did not differ significantly amongst six age groups (Figure 2F), or amongst different cancer stages (Figure 2G). In addition, we used Cox proportional hazards model with gender as a covariate to re-evaluate the associations between DPP9 expression and survival. DPP9 expression was not significantly associated with poor prognosis based on patients’ gender (Supplementary Table S5).
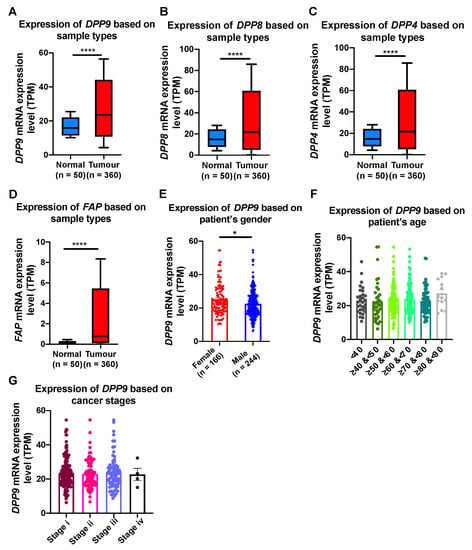
Figure 2.
The relative mRNA expression level of DPP9, DPP8, DPP4 and FAP in liver tumors (A–D) and association of DPP9 expression with gender (E), age (F) and cancer stages (G) in HCC patients. p values were calculated by Mann-Whitney U test (A–E) and Kruskal-Wallis one-way analysis of variance with Tukey’s post-hoc test (F,G). * p < 0.05, **** p < 0.0001.
Obesity is a well-known risk factor for HCC [16], so we examined DPP9 expression with HCC patient survival based on body mass index (BMI; BMI categories in Table 2). We stratified HCC patients based on BMI and found that DPP9 mRNA was significantly upregulated in the obese and extreme obese patients compared to patients with normal body weight (Figure 3A). Survival analysis suggested that DPP9 was not significantly associated with patient survival when patients are overweight (Figure 3B), or obese/extremely obese (Figure 3C). We then used a Cox proportional hazards model with BMI as a covariate to evaluate associations between DPP9 expression and survival. DPP9 expression was not significantly associated with poor prognosis based on patients’ BMI (Supplementary Table S6). High DPP9 expression in the overweight group had a hazard ratio of 1.54, but it lacked statistical significance.
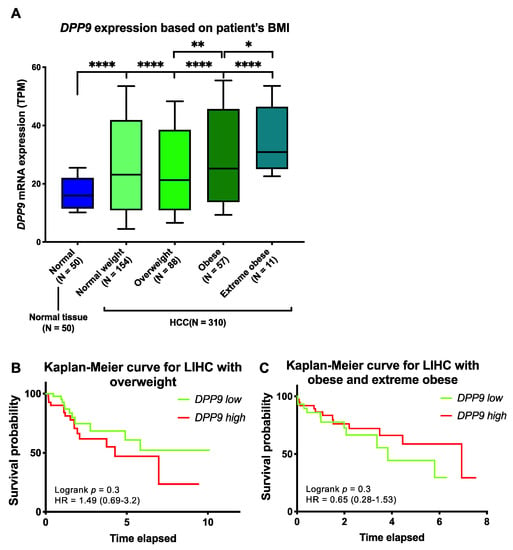
Figure 3.
The relative mRNA expression and survival analysis of DPP9 in HCC based on patient BMI. (A) The mRNA expression of DPP9 based on HCC patient (n = 360) BMI compared to non-diseased subjects. p values were calculated by Kruskal-Wallis one-way analysis of variance with Tukey’s post-hoc test, with * p < 0.05, ** p < 0.01, **** p < 0.0001. (B) Kaplan-Meier curve for DPP9 expression in overweight HCC patients. (C) Kaplan-Meier curve for DPP9 expression in obese and extreme obese HCC patients. The high (red) and low (green) mRNA expression levels of genes in liver tumors were stratified based on median expression value. p values and hazard ratio (HR) with confidence interval were calculated by logrank (Mantel-Cox) test.
3.6. Association of Expression of DPPs with Poor Survival of Patients with HCC
To further investigate DPP4 gene family expression in liver cancer prognosis, we stratified DPP9, DPP8 and DPP4 based on median expression in HCC. High co-expression of DPP9, DPP8 and DPP4 was not associated with OS (Figure 4A). However, high co-expression of DPP9, DPP8, DPP4 and FAP in the liver tumor was significantly associated with poor survival in HCC patients (Figure 4B). This data strengthens evidence that FAP might contribute to poor outcomes, possibly due to its role in liver cirrhosis and steatosis, which is strongly associated with poor liver function and liver cancer [50,51].
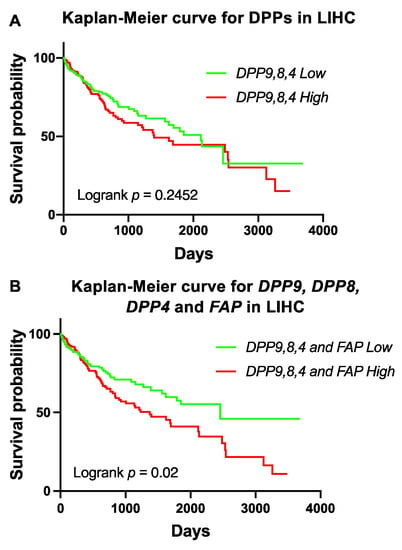
Figure 4.
Survival analysis of high co-expression of DPP9, DPP8, DPP4 and FAP in HCC patients. (A) Kaplan-Meier curve for high co-expression of DPP9, DPP8 and DPP4 in HCC patients (n = 360). (B) Kaplan-Meier curve for high co-expression of DPP9, DPP8, DPP4 and FAP in HCC patients. The high (red) and low (green) mRNA expression levels of genes in liver tumors were stratified based on median expression value. p values were calculated by logrank (Mantel-Cox) test.
3.7. Genes Correlated with DPPs Significantly Associated with Poor Prognosis in HCC and Multiple Oncogenic and Epigenetic Pathways
Having shown that DPP9, DPP8, DPP4 and FAP were overexpressed in liver tumors (Figure 2A–D), and that high co-expression of the DPP4 gene family members was significantly associated with shorter survival times in HCC (Figure 4B), we next assessed the gene expression signature correlated with the DPP4 gene family in HCC patients to gain mechanistic insights. Firstly, we analyzed the two homologs DPP8 and DPP9 and found that 661 genes were positively correlated in-common between DPP8 and DPP9 (Figure 5A). Reactome pathway enrichment analysis revealed that these genes were associated with tumor suppressor TP53 regulation and with chromatin remodeling pathways in HCC (Figure 5B). In contrast, only 10 negatively correlated genes in-common between DPP8 and DPP9 were identified and they were associated with metabolism related pathways (Supplementary Figure S3B).
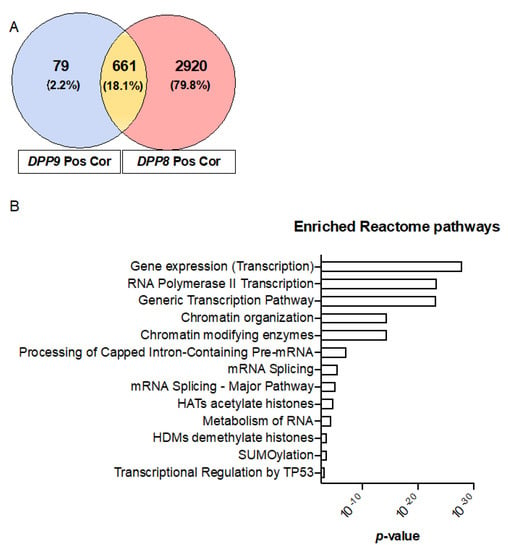
Figure 5.
Enriched reactome pathways of genes that were positively correlated with DPP9 and DPP8. (A) Venn diagram of the genes that were positively correlated with DPP9 and DPP8, with Pearson correlation coefficient greater or equal 0.5. “Pos Cor” refers to positive correlation. (B) Enriched reactome pathways associated with positively correlated genes in-common between DPP9 and DPP8.
A similar approach was undertaken to study the gene expression signature correlated with all three genes, DPP9, DPP8 and DPP4. We found that 268 genes were positively correlated and in-common amongst DPP9, DPP8 and DPP4 in HCC patients (Figure 6A), and stratified the expression of these genes based on median expression. Most importantly, high expression of these positively correlated gene signatures was significantly associated with poor prognosis in HCC patients (Figure 6B, logrank p < 0.0001).
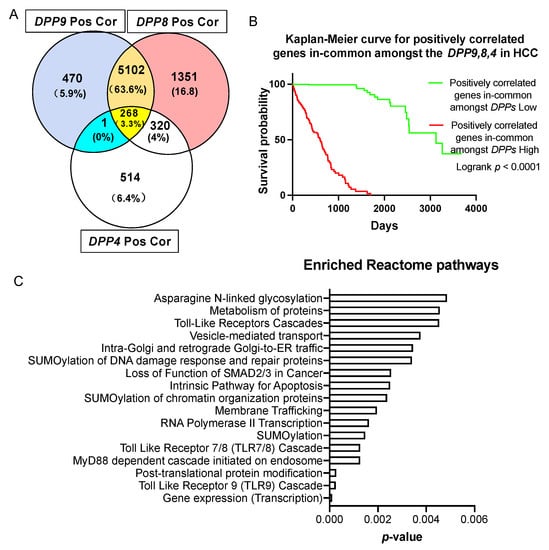
Figure 6.
Survival analysis and enriched reactome pathways analysis on genes that were positively correlated with DPP9, DPP8 and DPP4. (A) Venn diagram of the gens that were positively correlated with DPP9, DPP8 and DPP4. The gene in the blue section is TPRA1. “Pos Cor” refers to positive correlation. (B) Kaplan-Meier curve for the 268 genes that were positively correlated and in-common amongst DPP9, DPP8 and DPP4 in HCC patients (n = 360). The high (red) and low (green) mRNA expression levels of genes in liver tumors were stratified based on median expression value. p values were calculated by logrank (Mantel-Cox) test. (C) Enriched reactome pathways of genes that were positively correlated in-common amongst DPP9, DPP8 and DPP4.
To gain a better understanding of how DPPs may influence survival, we analyzed the biological pathways that were enriched in genes that were positively correlated with DPPs. There were 46 significantly enriched pathways according to reactome annotations for those 268 genes (Supplementary Table S5). Amongst the 15 top-ranked pathways (Figure 6C), the Toll-like receptors (TLRs) cascade, especially the TLR9 and TLR7/8 cascade, MyD88 dependent cascade initiated on endosome, N-glycosylation and SUMOylation pathways were dominant. When FAP was added to this analysis, only seven in-common genes emerged, most of which are the zinc finger gene regulators LIMS1, PHF20L1, ZEB1 and ZNFX1 (Supplementary Figure S4).
4. Discussion
This is the first set of analysis of human genomic databases to examine DPP9 exonic LoF variants and the of association of DPP9 expression with cancers. There were 27 unique DPP9 LoF variants found from our analyses of gnomAD, TCGA and COSMIC. Notably, patients with DPP9 LoF variants were most commonly diagnosed with cancers. The two important DPP9 intronic variants (rs12610495 and rs2109069) were found to be associated with pulmonary fibrosis and severe COVID, as shown previously [29,30,32,33], but not with liver cancer. Uterine corpus endometrial carcinoma (UCEC) and lung carcinoma were the most common diagnosed cancers in patients with DPP9 LoF variants. It is likely that DPP9 LoF variants will result in lowered abundance of DPP9 enzyme activity. Our datamining evidence that DPP9 LoF was associated with lung carcinoma perhaps does not align with a previous study that linked high DPP9 mRNA expression with poor survival in human lung cancer [7]. Perhaps the role of DPP9 that is dominant differs in different cancer types and differs depending upon whether high or low DPP9 expression occurred in the tumor or was life-long. Due to the limited case numbers of DPP9 LoF variants, we were unable to associate these variants with patient survival.
We observed that DPP8 and DPP9 LoF variants were more commonly diagnosed with a gynecological cancer, such as UCEC, cervical cancer or ovarian cancer. Together with the observation that low DPP9 expression was associated with poor survival in UCEC (Figure 1E), it might suggest that DPP9 is a suppressor of gynecological cancer, which would align with the recently discovered role of DPP9 in BRCA2 homeostasis and DNA repair [52]. However, in both TCGA and COSMIC, there was limited clinical data concerning patient outcomes, especially in COSMIC, where many cases were filed with unknown gender, age and diseases. This is a common limitation of in silico investigations.
Our investigation found that DPP9 and DPP8 are LoF intolerant in humans. Genetic variation tolerance parameters universally suggested that DPP9 was LoF intolerant (Table 6). LoF intolerance has been associated with successful targets of medicinal therapies for cancer [26]. The necessity of DPP9 is conserved in mice, where constitutive homozygous ablation of DPP9 enzyme activity is neonate lethal [25,53,54,55]. There were fewer than expected numbers of LoF variants observed in the three large-scale databases that we interrogated. The reason could be that the association with gynecological cancers, mentioned above, also associates with poor reproductive fitness. In addition, people who are homozygous or biallelic for a DPP9 LoF variant may not survive to be included in the searched databases, or survivors possibly possess a modification in a molecular pathway that compensates for DPP9 deficiency. An example is that deletion of NLRP1 and its downstream signaling molecules rescues DPP9 deficient mice from lethality [56]. Another consideration is that the LoF DPP9 variants in genomic databases are expected to be heterozygous and would therefore have lifelong low DPP9 expression throughout the body. Taken together, these data show that DPP9 is LoF intolerant but suggest that DPP9 LoF variants do not necessarily cause early death.
The databases contained some data mismatches about DPP9 sequences, presumably due to automation of data importation. There were more LoF variants in gnomAD than in COSMIC, but some of the listed LoF locations were not found in either long form or short form DPP9. Moreover, in both TCGA and COSMIC, variants of Arg303* and Glu154* were reported, but the mutations of nucleotides differed between the two databases. Also, in patients with the Arg303* variant, COSMIC reported both endometrial carcinoma and large intestinal adenocarcinoma, while TCGA only reported endometrial carcinoma. Hence, using a single database can be misleading and unmatched information is inevitable when using multiple databases. Therefore, database-derived information needs to be carefully and critically assessed.
Our TCGA datamining on the DPP4 gene family showed that in HCC patients, DPP9, DPP8, DPP4 and FAP were all overexpressed in liver tumors. Most notably, high expression of all four genes showed poorer survival in HCC patients (Figure 4B). Therefore, DPP targeted therapy might need to target all four enzymes together. Supporting this concept, VbP (Talabostat; PT-100; BXCL-701), which exerts potent inhibition of DPP9 and DPP8 and moderate inhibition of DPP4 and FAP, has been shown to lessen tumor burden in lung cancer and sarcoma models [57,58,59].
We have examined the associations between DPP9 mRNA expression and several potential prognostic factors, including gender, age, cancer stage and BMI. The impacts of these factors on HCC prognosis remain controversial. Our data analysis showed that DPP9 expression is greater in females than males. Even though age is known not to be an independent risk factor associated with cancer survival, the cumulative 10-year survival rate was significantly higher in younger patients [60]. Here, we saw no significant difference in DPP9 expression among different age groups. Moreover, patients diagnosed with advanced cancer stages would generally have shorter life spans, but our results showed that DPP9 expression was not associated with cancer stage. Taken together, the difference in survival probability amongst age groups and patients at different cancer stages may not be attributed to the expression of DPP9. Obesity is an independent risk factor for developing HCC. Our data showed that DPP9 expression was significantly upregulated in HCC patients who were obese/extremely obese, but significantly downregulated in overweight HCC patients. However, survival analysis on BMI showed no significant association with DPP9 expression, despite a hazard ratio greater than 1. As liver cancer is a multifactorial disease, further investigations may combine several risk factors together and investigate multiple comparisons between these parameters.
Enriched biochemical pathways analysis provided insights into differentially expressed genes correlated with DPPs in human HCC. Positively correlated genes in-common between DPP8 and DPP9 were associated with oncogenic and chromatin remodeling pathways in HCC, whilst 10 negatively correlated genes were associated with metabolism related pathways (Supplementary Figure S3A). Mutation of the tumor suppressor TP53 is associated with poor outcome for HCC patients [61]. The top ranking TP53 regulation pathway in our analyses indicates that DPP8 and DPP9 overexpression could be oncogenic in human HCC. The downregulation of metabolism genes with DPP8 and DPP9 upregulation concords with previous data showing that DPP9 deficiency dysregulates metabolic pathways in neonatal liver and gut [54], and that DPP9 hydrolyses metabolism related substrates including adenylate kinase 2 [62,63] and nucleobindin1 [64] and regulates preadipocyte differentiation [65]. Moreover, people with obesity are more likely to develop HCC [66,67]. DPP9 expression was significantly upregulated in HCC patients who were obese and extremely obese compared to those with normal weight. Taken together, this information suggests that DPP9 has an important role in liver manifestations of the metabolic syndrome, which drives progression to NASH, fibrosis and HCC.
The high expression of positively correlated gene signatures in-common amongst DPPs were associated with considerably shorter survival time in HCC patients. Amongst the identified reactome pathways, the TLR cascade, glycosylation and SUMOylation were the most frequent pathways enriched with increased DPP9 mRNA expression. No association between DPP9 and glycosylation is known. SUMOylation is mostly associated with SUMOylation of chromatin organization proteins, and SUMOylation of DNA damage response and repair proteins, which could have roles in cancer development. SUMO1 is a DPP9 interacting protein, acting as an allosteric regulator of DPP9 and thereby influencing its enzymatic activity [23]. TLR7/8 and TLR9 signaling pathways have not been linked to DPP9 previously. The expression of TLRs is low in healthy liver [68]. TLRs are upregulated in HCC tissues [69], and inhibition of TLR7 and TLR9 eliminates the proliferation of liver cancer cells [69,70]. Therefore, overexpression of DPPs might activate these TLR cascades, thereby inducing overactivation of the innate immune system and pathological changes.
5. Conclusions
This study reports the first comprehensive data mining of associations between DPP9 expression, survival and gene expression signatures in human HCC. DPP9 was found to be very LoF-intolerant and its LoF exonic variants were mostly associated with cancers. Two intronic variants that lower DPP9 expression and have been linked to lung fibrosis and COVID-19 were not associated with HCC or liver fibrosis. In HCC patients, DPP9 and its closely related genes were significantly upregulated in liver tumors. High co-expression of genes that were positively correlated and in-common amongst DPP4, DPP8 and DPP9 were associated with poor survival of HCC patients, possibly via affecting the following enriched pathways: Toll-like receptors (TLRs), glycosylation and SUMOylation. These findings strongly implicate the DPP4 gene family, especially DPP9, in the pathogenesis of human HCC and therefore requires future functional studies.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cancers13071637/s1, Table S1: DPP8 LoF variants in TCGA, Table S2: DPP8 LoF variants in COSMIC, Table S3: Genome-wide significant loci for severe COVID-19: Intronic DPP9 variants rs12610495 and rs2109069, Table S4: The LIHC/HCC (n = 360) and UCEC (n = 540) patient demographics, Table S5: Cox proportional hazards model with gender as the covariate to evaluate associations between DPP9 expression and survival, Table S6: Cox proportional hazards model with BMI as the covariate to evaluate associations between DPP9 expression and survival. Figure S1: Crystal structure and protein sequence of the four enzymatic members in the DPP4 gene family, Figure S2: DPP9 variants mapping to the DPP9 protein structure, Figure S3: Enriched reactome pathways of genes that were negatively correlated and in-common between DPP9 and DPP8, Figure S4: Survival analysis on genes that were positively correlated and in-common amongst DPP9, DPP8, DPP4 and FAP in HCC.
Author Contributions
Conceptualization, J.C.H., M.D.G. and H.E.Z.; methodology, J.C.H. and A.A.E.; formal analysis, J.C.H., A.A.E. and J.M.E.; investigation, J.C.H., A.A.E., M.D.G. and H.E.Z.; data curation, J.C.H., A.A.E. and J.M.E.; writing—original draft preparation, J.C.H., M.D.G. and H.E.Z.; writing—review and editing, J.C.H., A.A.E., J.M.E., G.W.M., M.D.G. and H.E.Z.; supervision, M.D.G. and H.E.Z.; funding acquisition, M.D.G., H.E.Z. and G.W.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Australian National Health and Medical Research Council Grant 1113842, a Perpetual Trustees IMPACT Philanthropy grant IPAP2019/1295, and a Kickstart grant from The University of Sydney.
Institutional Review Board Statement
All research activities were conducted in accordance with the Helsinki Declaration of 1975 by the TCGA Research Network [71]. Local ethical review and approval were waived for this study because all data were obtained from public access databases and individuals were not identifiable.
Informed Consent Statement
Written informed consent of all participants was obtained by the TCGA Research Network [71].
Data Availability Statement
TCGA data are available through Oncolnc and UALCAN (R).
Acknowledgments
We thank Ulf Schmitz and Richard D. Bagnall from Centenary Institute for kindly providing expert guidance. We thank the many people involved in making the TCGA, COSMIC and LIHC (HCC) data available for study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Zhang, H.; Chen, Y.; Keane, F.M.; Gorrell, M.D. Advances in understanding the expression and function of dipeptidyl peptidase 8 and 9. Mol. Cancer Res. 2013, 11, 1487–1496. [Google Scholar] [CrossRef]
- Yu, D.M.; Ajami, K.; Gall, M.G.; Park, J.; Lee, C.S.; Evans, K.A.; McLaughlin, E.A.; Pitman, M.R.; Abbott, C.A.; McCaughan, G.W.; et al. The in vivo expression of dipeptidyl peptidases 8 and 9. J. Histochem. Cytochem. 2009, 57, 1025–1040. [Google Scholar] [CrossRef]
- Yao, T.W.; Kim, W.S.; Yu, D.M.; Sharbeen, G.; McCaughan, G.W.; Choi, K.Y.; Xia, P.; Gorrell, M.D. A novel role of dipeptidyl peptidase 9 in epidermal growth factor signaling. Mol. Cancer Res. 2011, 9, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Okondo, M.C.; Rao, S.D.; Taabazuing, C.Y.; Chui, A.J.; Poplawski, S.E.; Johnson, D.C.; Bachovchin, D.A. Inhibition of Dpp8/9 activates the Nlrp1b inflammasome. Cell Chem. Biol. 2018, 25, 262–267.e265. [Google Scholar] [CrossRef] [PubMed]
- Okondo, M.C.; Johnson, D.C.; Sridharan, R.; Go, E.B.; Chui, A.J.; Wang, M.S.; Poplawski, S.E.; Wu, W.; Liu, Y.; Lai, J.H.; et al. DPP8 and DPP9 inhibition induces pro-caspase-1-dependent monocyte and macrophage pyroptosis. Nat. Chem. Biol. 2017, 13, 46–53. [Google Scholar] [CrossRef]
- Justa-Schuch, D.; Silva-Garcia, M.; Pilla, E.; Engelke, M.; Kilisch, M.; Lenz, C.; Möller, U.; Nakamura, F.; Urlaub, H.; Geiss-Friedlander, R. DPP9 is a novel component of the N-end rule pathway targeting the tyrosine kinase Syk. eLife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, J.; Shen, Q.; Feng, J.; Liu, H.; Wang, W.; Xu, L.; Shi, G.; Ye, X.; Ge, M.; et al. Contribution of upregulated dipeptidyl peptidase 9 (DPP9) in promoting tumoregenicity, metastasis and the prediction of poor prognosis in non-small cell lung cancer (NSCLC). Int. J. Cancer 2017, 140, 1620–1632. [Google Scholar] [CrossRef]
- Yokobori, T. Editorial Comment on: DPP9 Increases Chemoresistance and is an Indicator of Poor Prognosis in Colorectal Cancer. Ann. Surg. Oncol. 2020, 27, 4084–4085. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.Q.; Zhao, M.; Huang, G.Z.; Zheng, Z.N.; Chen, Y.; Zeng, W.S.; Lv, X.Z. Fibroblast Activation Protein (FAP) Overexpression Induces Epithelial-Mesenchymal Transition (EMT) in Oral Squamous Cell Carcinoma by Down-Regulating Dipeptidyl Peptidase 9 (DPP9). Onco Targets Ther. 2020, 13, 2599–2611. [Google Scholar] [CrossRef]
- Chowdhury, S.; Chen, Y.; Yao, T.W.; Ajami, K.; Wang, X.M.; Popov, Y.; Schuppan, D.; Bertolino, P.; McCaughan, G.W.; Yu, D.M.; et al. Regulation of dipeptidyl peptidase 8 and 9 expression in activated lymphocytes and injured liver. World J. Gastroenterol. 2013, 19, 2883–2893. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, Y.; Wadham, C.; McCaughan, G.W.; Keane, F.M.; Gorrell, M.D. Dipeptidyl peptidase 9 subcellular localization and a role in cell adhesion involving focal adhesion kinase and paxillin. Biochim. Biophys. Acta 2015, 1853, 470–480. [Google Scholar] [CrossRef]
- Badvie, S. Hepatocellular carcinoma. Postgrad Med. J. 2000, 76, 4–11. [Google Scholar] [CrossRef]
- Forner, A.; Reig, M.; Bruix, J. Hepatocellular carcinoma. Lancet 2018, 391, 1301–1314. [Google Scholar] [CrossRef]
- El-Serag, H.B. Hepatocellular carcinoma: An epidemiologic view. J. Clin. Gastroenterol. 2002, 35, S72–S78. [Google Scholar] [CrossRef]
- El–Serag, H.B.; Rudolph, K.L. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef] [PubMed]
- Ray, K. NAFLD-the next global epidemic. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 621. [Google Scholar] [CrossRef] [PubMed]
- Ajami, K.; Abbott, C.A.; McCaughan, G.W.; Gorrell, M.D. Dipeptidyl peptidase 9 has two forms, a broad tissue distribution, cytoplasmic localization and DPIV-like peptidase activity. Biochim. Biophys. Acta 2004, 1679, 18–28. [Google Scholar] [CrossRef] [PubMed]
- Yu, D.M.; Yao, T.W.; Chowdhury, S.; Nadvi, N.A.; Osborne, B.; Church, W.B.; McCaughan, G.W.; Gorrell, M.D. The dipeptidyl peptidase IV family in cancer and cell biology. FEBS J. 2010, 277, 1126–1144. [Google Scholar] [CrossRef]
- Dunaevsky, Y.E.; Tereshchenkova, V.F.; Oppert, B.; Belozersky, M.A.; Filippova, I.Y.; Elpidina, E.N. Human proline specific peptidases: A comprehensive analysis. Biochim. Biophys. Acta. Gen. Subj. 2020, 1864, 129636. [Google Scholar] [CrossRef]
- Ross, B.; Krapp, S.; Augustin, M.; Kierfersauer, R.; Arciniega, M.; Geiss-Friedlander, R.; Huber, R. Structures and mechanism of dipeptidyl peptidases 8 and 9, important players in cellular homeostasis and cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E1437–E1445. [Google Scholar] [CrossRef]
- Justa-Schuch, D.; Möller, U.; Geiss-Friedlander, R. The amino terminus extension in the long dipeptidyl peptidase 9 isoform contains a nuclear localization signal targeting the active peptidase to the nucleus. Cell Mol. Life Sci. 2014, 71, 3611–3626. [Google Scholar] [CrossRef]
- Lee, H.J.; Chen, Y.S.; Chou, C.Y.; Chien, C.H.; Lin, C.H.; Chang, G.G.; Chen, X. Investigation of the dimer interface and substrate specificity of prolyl dipeptidase DPP8. J. Biol. Chem. 2006, 281, 38653–38662. [Google Scholar] [CrossRef]
- Pilla, E.; Möller, U.; Sauer, G.; Mattiroli, F.; Melchior, F.; Geiss-Friedlander, R. A novel SUMO1-specific interacting motif in dipeptidyl peptidase 9 (DPP9) that is important for enzymatic regulation. J. Biol. Chem. 2012, 287, 44320–44329. [Google Scholar] [CrossRef]
- Gall, M.G.; Gorrell, M.D. The multifunctional post-proline dipeptidyl peptidase, DPP9, in mice, cell biology and immunity. In Pathophysiological Aspects of Proteases; Chakraborti, S., Dhalla, N.S., Eds.; Springer: Singapore, 2017; Volume 1, pp. 23–46. [Google Scholar]
- Zhang, H.E.; Gorrell, M.D. DPP9. In Encyclopedia of Signaling Molecules, 2nd ed.; Choi, S., Ed.; Springer International Publishing: New York, NY, USA, 2018; pp. 1418–1422. [Google Scholar] [CrossRef]
- Minikel, E.V.; Karczewski, K.J.; Martin, H.C.; Cummings, B.B.; Whiffin, N.; Rhodes, D.; Alföldi, J.; Trembath, R.C.; van Heel, D.A.; Daly, M.J.; et al. Evaluating drug targets through human loss-of-function genetic variation. Nature 2020, 581, 459–464. [Google Scholar] [CrossRef]
- Plenge, R.M.; Scolnick, E.M.; Altshuler, D. Validating therapeutic targets through human genetics. Nat. Rev. Drug Discov. 2013, 12, 581–594. [Google Scholar] [CrossRef] [PubMed]
- Gelernter, J.; Kranzler, H.R.; Sherva, R.; Almasy, L.; Koesterer, R.; Smith, A.H.; Anton, R.; Preuss, U.W.; Ridinger, M.; Rujescu, D.; et al. Genome-wide association study of alcohol dependence:significant findings in African- and European-Americans including novel risk loci. Mol. Psychiatry 2014, 19, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, B.D.; Putman, R.K.; Araki, T.; Nishino, M.; Gudmundsson, G.; Gudnason, V.; Eiriksdottir, G.; Zilhao Nogueira, N.R.; Dupuis, J.; Xu, H.; et al. Overlap of Genetic Risk between Interstitial Lung Abnormalities and Idiopathic Pulmonary Fibrosis. Am. J. Respir Crit Care Med. 2019, 200, 1402–1413. [Google Scholar] [CrossRef] [PubMed]
- Fingerlin, T.E.; Murphy, E.; Zhang, W.; Peljto, A.L.; Brown, K.K.; Steele, M.P.; Loyd, J.E.; Cosgrove, G.P.; Lynch, D.; Groshong, S.; et al. Genome-wide association study identifies multiple susceptibility loci for pulmonary fibrosis. Nat. Genet. 2013, 45, 613–620. [Google Scholar] [CrossRef]
- Chan, V.; Fong, G.C.; Luk, K.D.; Yip, B.; Lee, M.K.; Wong, M.S.; Lu, D.D.; Chan, T.K. A genetic locus for adolescent idiopathic scoliosis linked to chromosome 19p13.3. Am. J. Hum. Genet. 2002, 71, 401–406. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic mechanisms of critical illness in Covid-19. Nature 2020. [Google Scholar] [CrossRef]
- Wang, L.; Balmat, T.J.; Antonia, A.L.; Constantine, F.J.; Henao, R.; Burke, T.W.; Ingham, A.; McClain, M.T.; Tsalik, E.L.; Ko, E.R.; et al. An atlas connecting shared genetic architecture of human diseases and molecular phenotypes provides insight into COVID-19 susceptibility. medRxiv 2020. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef] [PubMed]
- Grossman, R.L.; Heath, A.P.; Ferretti, V.; Varmus, H.E.; Lowy, D.R.; Kibbe, W.A.; Staudt, L.M. Toward a Shared Vision for Cancer Genomic Data. N. Engl. J. Med. 2016, 375, 1109–1112. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [CrossRef]
- Firth, H.V.; Richards, S.M.; Bevan, A.P.; Clayton, S.; Corpas, M.; Rajan, D.; Van Vooren, S.; Moreau, Y.; Pettett, R.M.; Carter, N.P. DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am. J. Hum. Genet. 2009, 84, 524–533. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The GeneCards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1.30.1–1.30.33. [Google Scholar] [CrossRef]
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A database of human genotype–phenotype associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Anaya, J. OncoRank: A pan-cancer method of combining survival correlations and its application to mRNAs, miRNAs, and lncRNAs. PeerJ Prepr. 2016, 4. [Google Scholar] [CrossRef]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martínez-Ledesma, E.; Martínez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Peña, J.G.; Treviño, V. SurvExpress: An Online Biomarker Validation Tool and Database for Cancer Gene Expression Data Using Survival Analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef]
- Herwig, R.; Hardt, C.; Lienhard, M.; Kamburov, A. Analyzing and interpreting genome data at the network level with ConsensusPathDB. Nat. Protoc. 2016, 11, 1889–1907. [Google Scholar] [CrossRef] [PubMed]
- Ajami, K.; Abbott, C.A.; Obradovic, M.; Gysbers, V.; Kähne, T.; McCaughan, G.W.; Gorrell, M.D. Structural requirements for catalysis, expression, and dimerization in the CD26/DPIV gene family. Biochemistry 2003, 42, 694–701. [Google Scholar] [CrossRef] [PubMed]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. Variation across 141,456 human exomes and genomes reveals the spectrum of loss-of-function intolerance across human protein-coding genes. bioRxiv 2019. [Google Scholar] [CrossRef]
- Dang, V.T.; Kassahn, K.S.; Marcos, A.E.; Ragan, M.A. Identification of human haploinsufficient genes and their genomic proximity to segmental duplications. Eur. J. Hum. Genet. 2008, 16, 1350–1357. [Google Scholar] [CrossRef]
- Itan, Y.; Shang, L.; Boisson, B.; Patin, E.; Bolze, A.; Moncada-Vélez, M.; Scott, E.; Ciancanelli, M.J.; Lafaille, F.G.; Markle, J.G.; et al. The human gene damage index as a gene-level approach to prioritizing exome variants. Proc. Natl. Acad. Sci. USA 2015, 112, 13615–13620. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer incidence and mortality worldwide: Sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef]
- Lay, A.J.; Zhang, H.E.; McCaughan, G.W.; Gorrell, M.D. Fibroblast activation protein in liver fibrosis. Front. Biosci. 2019, 24, 1–17. [Google Scholar]
- Cho, J.M.; Yang, E.H.; Quan, W.; Nam, E.H.; Cheon, H.G. Discovery of a novel fibroblast activation protein (FAP) inhibitor, BR103354, with anti-diabetic and anti-steatotic effects. Sci. Rep. 2020, 10, 21280. [Google Scholar] [CrossRef]
- Silva-Garcia, M.; Bolgi, O.; Ross, B.; Pilla, E.; Kari, V.; Killisch, M.; Stark, N.; Lenz, C.; Spitzner, M.; Gorrell, M.D.; et al. Dipeptidyl peptidase 9 triggers BRCA2 degradation by the N-degron pathway to promote DNA-damage repair. bioRxiv 2020. [Google Scholar] [CrossRef]
- Gall, M.G.; Chen, Y.; Vieira de Ribeiro, A.J.; Zhang, H.; Bailey, C.G.; Spielman, D.S.; Yu, D.M.; Gorrell, M.D. Targeted inactivation of dipeptidyl peptidase 9 enzymatic activity causes mouse neonate lethality. PLoS ONE 2013, 8, e78378. [Google Scholar] [CrossRef]
- Chen, Y.; Gall, M.G.; Zhang, H.; Keane, F.M.; McCaughan, G.W.; Yu, D.M.; Gorrell, M.D. Dipeptidyl peptidase 9 enzymatic activity influences the expression of neonatal metabolic genes. Exp. Cell Res. 2016, 342, 72–82. [Google Scholar] [CrossRef]
- Kim, M.; Minoux, M.; Piaia, A.; Kueng, B.; Gapp, B.; Weber, D.; Haller, C.; Barbieri, S.; Namoto, K.; Lorenz, T.; et al. DPP9 enzyme activity controls survival of mouse migratory tongue muscle progenitors and its absence leads to neonatal lethality due to suckling defect. Dev. Biol. 2017, 431, 297–308. [Google Scholar] [CrossRef]
- Harapas, C.R.; Robinson, K.; Lay, K.; Wong, J.; Raas-Rothschild, A.; Laohamonthonkul, P.; Boisson, B.; Drutman, S.B.; Gorrell, M.; Bonner, D.; et al. DPP9 deficiency: An inflammasomopathy which can be rescued by lowering NLRP1/IL-1 signaling. medRxiv 2021. [Google Scholar] [CrossRef]
- Duncan, B.B.; Highfill, S.L.; Qin, H.; Bouchkouj, N.; Larabee, S.; Zhao, P.; Woznica, I.; Liu, Y.; Li, Y.; Wu, W.; et al. A pan-inhibitor of DASH family enzymes induces immune-mediated regression of murine sarcoma and is a potent adjuvant to dendritic cell vaccination and adoptive T-cell therapy. J. Immunother. 2013, 36, 400–411. [Google Scholar] [CrossRef] [PubMed]
- Donahue, R.N.; Duncan, B.B.; Fry, T.J.; Jones, B.; Bachovchin, W.W.; Kiritsy, C.P.; Lai, J.H.; Wu, W.; Zhao, P.; Liu, Y.; et al. A pan inhibitor of DASH family enzymes induces immunogenic modulation and sensitizes murine and human carcinoma cells to antigen-specific cytotoxic T lymphocyte killing: Implications for combination therapy with cancer vaccines. Vaccine 2014, 32, 3223–3231. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Miller, G.T.; Jesson, M.I.; Watanabe, T.; Jones, B.; Wallner, B.P. PT-100, a small molecule dipeptidyl peptidase inhibitor, has potent antitumor effects and augments antibody-mediated cytotoxicity via a novel immune mechanism. Cancer Res. 2004, 64, 5471–5480. [Google Scholar] [CrossRef] [PubMed]
- Su, C.-W.; Lei, H.-J.; Chau, G.-Y.; Hung, H.-H.; Wu, J.-C.; Hsia, C.-Y.; Lui, W.-Y.; Su, Y.-H.; Wu, C.-W.; Lee, S.-D. The Effect of Age on the Long-term Prognosis of Patients with Hepatocellular Carcinoma After Resection Surgery: A Propensity Score Matching Analysis. Arch. Surg. 2012, 147, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Zhan, P.; Ji, Y.N.; Yu, L.K. TP53 mutation is associated with a poor outcome for patients with hepatocellular carcinoma: Evidence from a meta-analysis. Hepatobiliary Surg. Nutr. 2013, 2, 260–265. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.H.; Indarto, D.; Doucet, A.; Pogson, L.D.; Pitman, M.R.; McNicholas, K.; Menz, R.I.; Overall, C.M.; Abbott, C.A. Identifying natural substrates for dipeptidyl peptidases 8 and 9 using terminal amine isotopic labeling of substrates (TAILS) reveals in vivo roles in cellular homeostasis and energy metabolism. J. Biol. Chem. 2013, 288, 13936–13949. [Google Scholar] [CrossRef]
- Finger, Y.; Habich, M.; Gerlich, S.; Urbanczyk, S.; van de Logt, E.; Koch, J.; Schu, L.; Lapacz, K.J.; Ali, M.; Petrungaro, C.; et al. Proteasomal degradation induced by DPP9-mediated processing competes with mitochondrial protein import. EMBO J. 2020, 39, e103889. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Maqsudi, S.; Rainczuk, A.; Duffield, N.; Lawrence, J.; Keane, F.M.; Justa-Schuch, D.; Geiss-Friedlander, R.; Gorrell, M.D.; Stephens, A.N. Identification of novel dipeptidyl peptidase 9 substrates by two-dimensional differential in-gel electrophoresis. FEBS J. 2015, 282, 3737–3757. [Google Scholar] [CrossRef] [PubMed]
- Han, R.; Wang, X.; Bachovchin, W.; Zukowska, Z.; Osborn, J.W. Inhibition of dipeptidyl peptidase 8/9 impairs preadipocyte differentiation. Sci. Rep. 2015, 5, 12348. [Google Scholar] [CrossRef] [PubMed]
- Wolk, A.; Gridley, G.; Svensson, M.; Nyrén, O.; McLaughlin, J.K.; Fraumeni, J.F.; Adami, H.-O. A prospective study of obesity and cancer risk (Sweden). Cancer Causes Control 2001, 12, 13–21. [Google Scholar] [CrossRef]
- Calle, E.E.; Rodriguez, C.; Walker-Thurmond, K.; Thun, M.J. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003, 348, 1625–1638. [Google Scholar] [CrossRef]
- Sun, L.; Dai, J.J.; Hu, W.F.; Wang, J. Expression of toll-like receptors in hepatic cirrhosis and hepatocellular carcinoma. Genet. Mol. Res. 2016, 15. [Google Scholar] [CrossRef]
- Mohamed, F.E.; Al-Jehani, R.M.; Minogue, S.S.; Andreola, F.; Winstanley, A.; Olde Damink, S.W.; Habtesion, A.; Malagó, M.; Davies, N.; Luong, T.V.; et al. Effect of toll-like receptor 7 and 9 targeted therapy to prevent the development of hepatocellular carcinoma. Liver Int. 2015, 35, 1063–1076. [Google Scholar] [CrossRef]
- Seki, E.; Park, E.; Fujimoto, J. Toll-like receptor signaling in liver regeneration, fibrosis and carcinogenesis. Hepatol. Res. 2011, 41, 597–610. [Google Scholar] [CrossRef]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).