The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells—A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
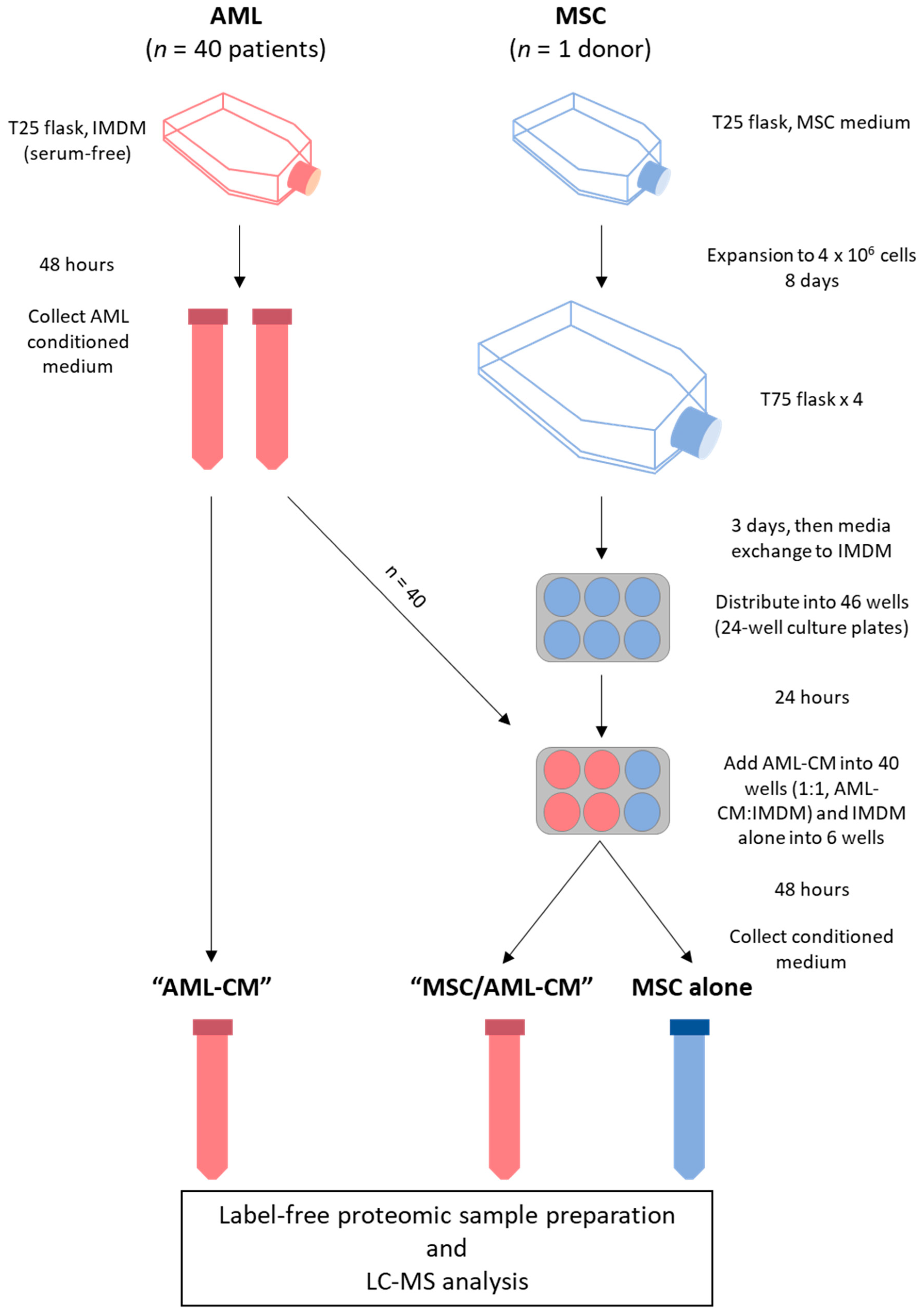
2.1. AML Patients and Preparation of Enriched AML Cells
2.2. Cell Culture
2.3. Proteomics Sample Preparation and Liquid Chromatography (LC)–Tandem Mass Spectrometry (MS/MS) Analysis
2.4. Statistical and Bioinformatical Analyses
3. Results
3.1. Constitutive Extracellular Protein Release by AML; Characterization of Patient Heterogeneity and the Overlap with MSC Release
3.2. Primary Human AML Cells Release of Proteins Derived from Different Cellular Compartments
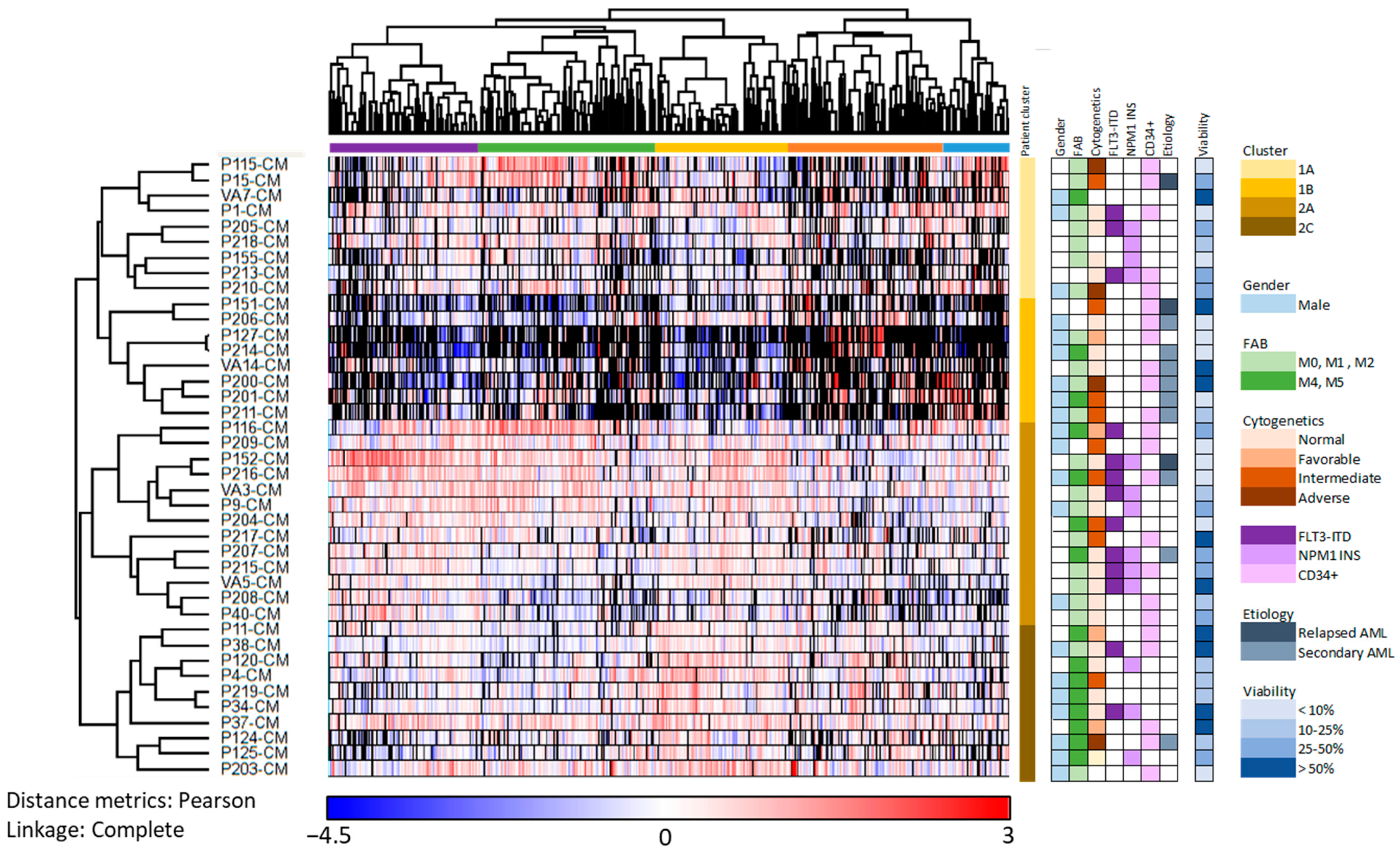
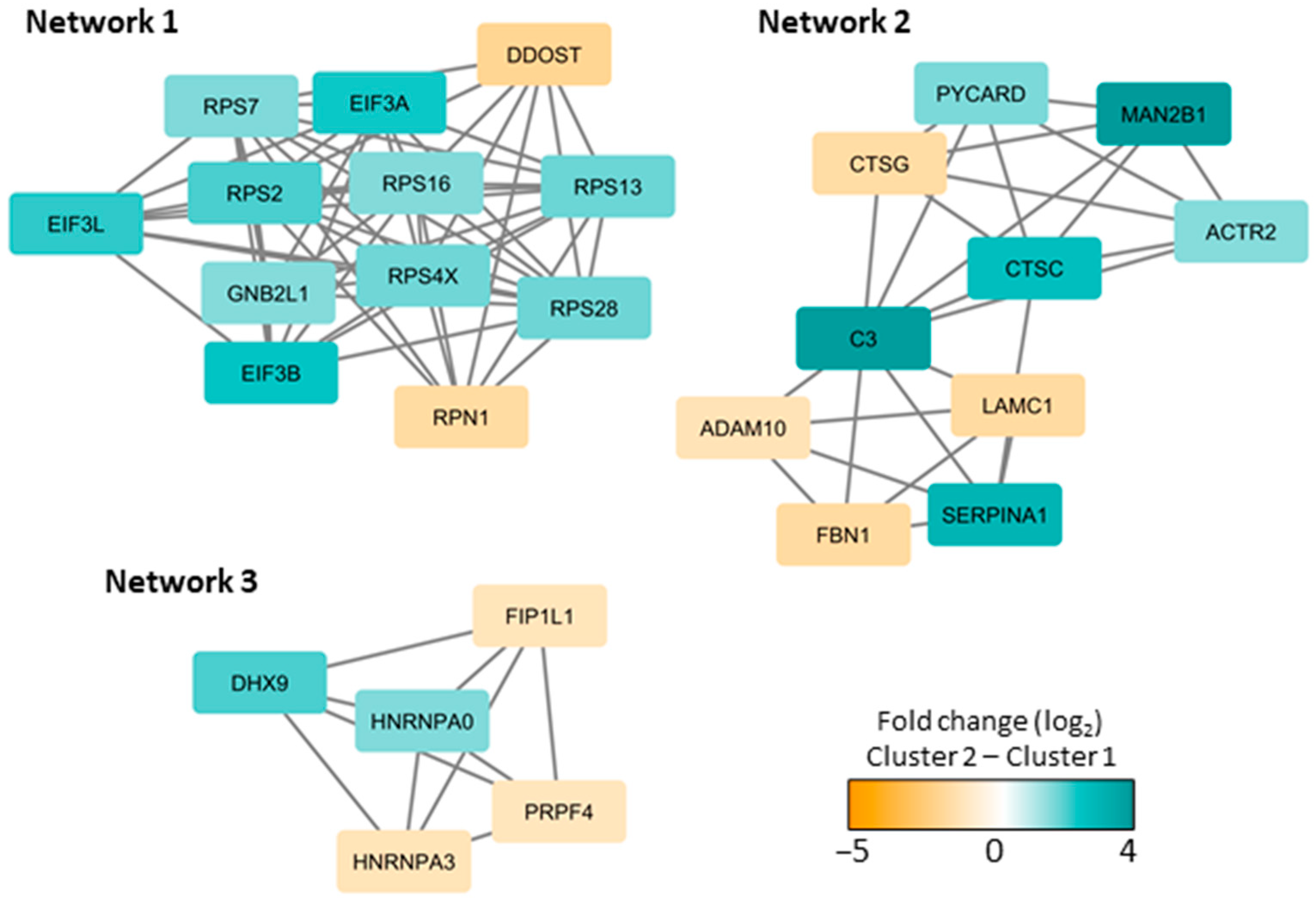
3.3. Subclassification of AML Patients Based on the Constitutive Protein Release Profile of Their In Vitro Cultured Leukemic Cells
3.4. The AML-Associated Heterogeneity of the Extracellular Protein Profile Is Largely Maintained also in the Presence of MSCs
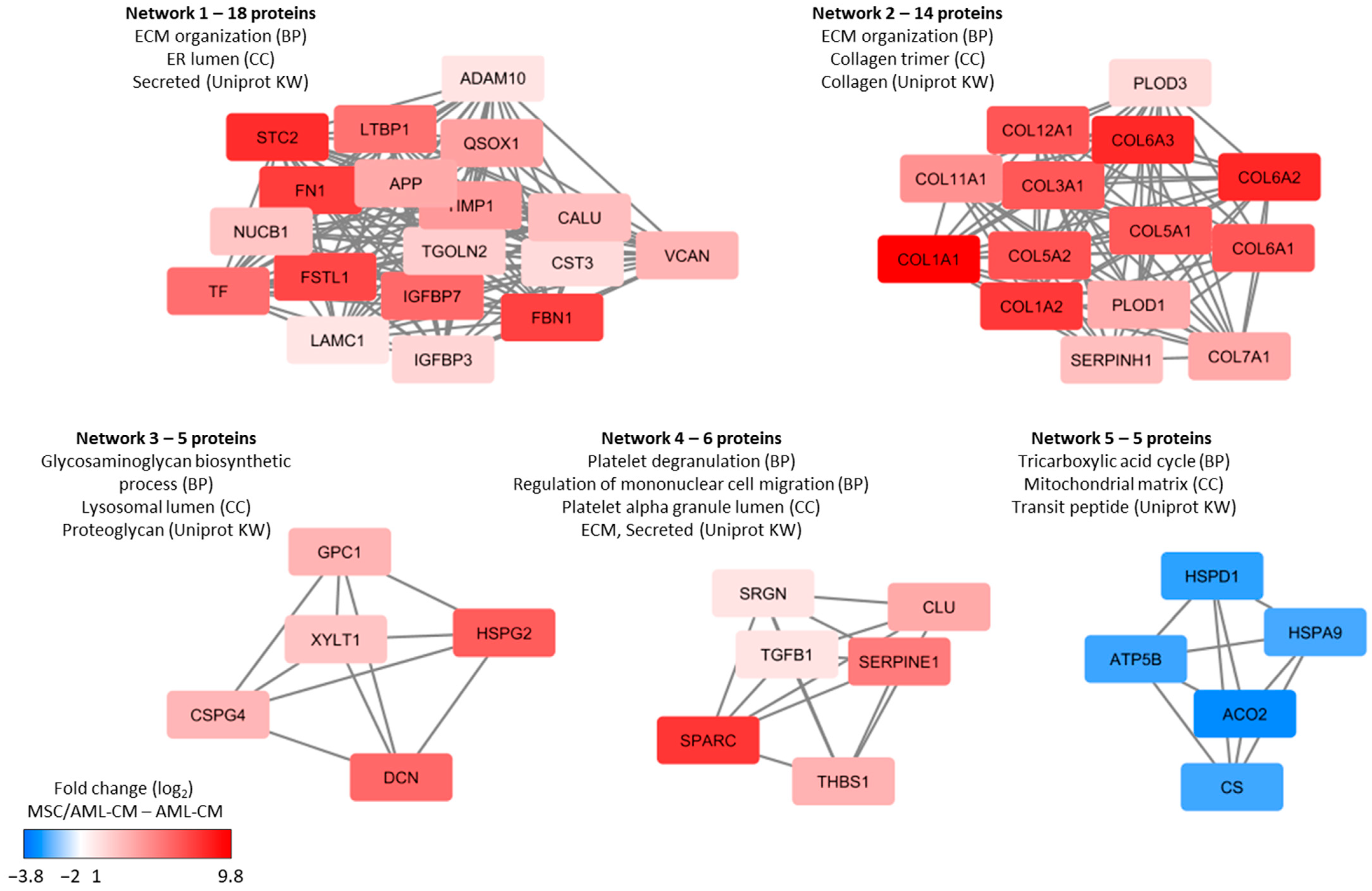
3.5. Reduction in Patient Heterogeneity by MSCs; a Small Subset of Proteins Show Heterogeneous AML Cell Release but Are Released at High Levels by MSCs
3.6. The Effect of MSCs on the AML-Associated Heterogeneity of of Their Common Extracellular Protein Profile; Relatively Few of the Quantified Proteins Are Significantly Altered by the Presence of MSCs
3.7. Identification of AML Patient Subsets by a Clustering Analysis Based on the Protein Profile of Supernatants Derived from MSC Cultures Supplemented with AML-Conditioned Media
3.8. Both MSCs and AML Cells Show Extracellular Release of a Wide Range of Exosomal Proteins
3.9. Culture of MSCs in the Presence of AML-CM; the Supernatant Levels of MSC-Specific Proteins Are Decreased in the Presence of AML-CM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Döhner, H.; Estey, E.; Grimwade, D.; Amadori, S.; Appelbaum, F.R.; Büchner, T.; Dombret, H.; Ebert, B.L.; Fenaux, P.; Larson, R.A.; et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood 2017, 129, 424–447. [Google Scholar] [CrossRef] [PubMed]
- Sanz, M.A.; Fenaux, P.; Tallman, M.S.; Estey, E.H.; Löwenberg, B.; Naoe, T.; Lengfelder, E.; Döhner, H.; Burnett, A.K.; Chen, S.J.; et al. Management of acute promyelocytic leukemia: Updated recommendations from an expert panel of the European LeukemiaNet. Blood 2019, 133, 1630–1643. [Google Scholar] [CrossRef]
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Geyh, S.; Rodríguez-Paredes, M.; Jäger, P.; Khandanpour, C.; Cadeddu, R.P.; Gutekunst, J.; Wilk, C.M.; Fenk, R.; Zilkens, C.; Hermsen, D.; et al. Functional inhibition of mesenchymal stromal cells in acute myeloid leukemia. Leukemia 2016, 30, 683–691. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Aasebø, E.; Hernandez-Valladares, M.; Tsykunova, G.; Reikvam, H. Therapeutic targeting of leukemic stem cells in acute myeloid leukemia–the biological background for possible strategies. Expert Opin. Drug Discov. 2017, 12, 1053–1065. [Google Scholar] [CrossRef] [PubMed]
- Brenner, A.K.; Nepstad, I.; Bruserud, Ø. Mesenchymal Stem Cells Support Survival and Proliferation of Primary Human Acute Myeloid Leukemia Cells through Heterogeneous Molecular Mechanisms. Front. Immunol. 2017, 8, 106. [Google Scholar] [CrossRef]
- Ehninger, A.; Trumpp, A. The bone marrow stem cell niche grows up: Mesenchymal stem cells and macrophages move in. J. Exp. Med. 2011, 208, 421–428. [Google Scholar] [CrossRef]
- Brenner, A.K.; Tvedt, T.H.; Nepstad, I.; Rye, K.P.; Hagen, K.M.; Reikvam, H.; Bruserud, Ø. Patients with acute myeloid leukemia can be subclassified based on the constitutive cytokine release of the leukemic cells; the possible clinical relevance and the importance of cellular iron metabolism. Expert Opin. Ther. Targets 2017, 21, 357–369. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Gjertsen, B.T.; Foss, B.; Huang, T.S. New strategies in the treatment of acute myelogenous leukemia (AML): In vitro culture of aml cells--the present use in experimental studies and the possible importance for future therapeutic approaches. Stem Cells 2001, 19, 1–11. [Google Scholar] [CrossRef]
- Eltoukhy, H.S.; Sinha, G.; Moore, C.A.; Gergues, M.; Rameshwar, P. Secretome within the bone marrow microenvironment: A basis for mesenchymal stem cell treatment and role in cancer dormancy. Biochimie 2018, 155, 92–103. [Google Scholar] [CrossRef]
- Kaushansky, K.; Zhan, H. The regulation of normal and neoplastic hematopoiesis is dependent on microenvironmental cells. Adv. Biol. Regul. 2018, 69, 11–15. [Google Scholar] [CrossRef]
- Aasebø, E.; Birkeland, E.; Selheim, F.; Berven, F.; Brenner, A.K.; Bruserud, Ø. The Extracellular Bone Marrow Microenvironment-A Proteomic Comparison of Constitutive Protein Release by In Vitro Cultured Osteoblasts and Mesenchymal Stem Cells. Cancers 2020, 13, 62. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Cox, J.; Matic, I.; Hilger, M.; Nagaraj, N.; Selbach, M.; Olsen, J.V.; Mann, M. A practical guide to the MaxQuant computational platform for SILAC-based quantitative proteomics. Nat. Protoc. 2009, 4, 698–705. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]
- Scholz, C.; Lyon, D.; Refsgaard, J.C.; Jensen, L.J.; Choudhary, C.; Weinert, B.T. Avoiding abundance bias in the functional annotation of post-translationally modified proteins. Nat. Methods 2015, 12, 1003–1004. [Google Scholar] [CrossRef] [PubMed]
- Arntzen, M.Ø.; Koehler, C.J.; Barsnes, H.; Berven, F.S.; Treumann, A.; Thiede, B. IsobariQ: Software for isobaric quantitative proteomics using IPTL, iTRAQ, and TMT. J. Proteome Res. 2011, 10, 913–920. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: Quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45, D362–D368. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Bader, G.D.; Hogue, C.W. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform. 2003, 4, 2. [Google Scholar] [CrossRef]
- Brenner, A.K.; Aasebø, E.; Hernandez-Valladares, M.; Selheim, F.; Berven, F.; Grønningsæter, I.S.; Bartaula-Brevik, S.; Bruserud, Ø. The Capacity of Long-Term in Vitro Proliferation of Acute Myeloid Leukemia Cells Supported Only by Exogenous Cytokines Is Associated with a Patient Subset with Adverse Outcome. Cancers 2019, 11, 73. [Google Scholar] [CrossRef]
- Ryningen, A.; Ersvaer, E.; Øyan, A.M.; Kalland, K.H.; Vintermyr, O.K.; Gjertsen, B.T.; Bruserud, Ø. Stress-induced in vitro apoptosis of native human acute myelogenous leukemia (AML) cells shows a wide variation between patients and is associated with low BCL-2:Bax ratio and low levels of heat shock protein 70 and 90. Leuk. Res. 2006, 30, 1531–1540. [Google Scholar] [CrossRef]
- Reikvam, H.; Aasebø, E.; Brenner, A.K.; Bartaula-Brevik, S.; Grønningsæter, I.S.; Forthun, R.B.; Hovland, R.; Bruserud, Ø. High Constitutive Cytokine Release by Primary Human Acute Myeloid Leukemia Cells Is Associated with a Specific Intercellular Communication Phenotype. J. Clin. Med. 2019, 8, 970. [Google Scholar] [CrossRef] [PubMed]
- Honnemyr, M.; Bruserud, Ø.; Brenner, A.K. The constitutive protease release by primary human acute myeloid leukemia cells. J. Cancer Res. Clin. Oncol. 2017, 143, 1985–1998. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø. Effects of endogenous interleukin 1 on blast cells derived from acute myelogenous leukemia patients. Leuk. Res. 1996, 20, 65–73. [Google Scholar] [CrossRef]
- Bruserud, Ø.; Ryningen, A.; Olsnes, A.M.; Stordrange, L.; Øyan, A.M.; Kalland, K.H.; Gjertsen, B.T. Subclassification of patients with acute myelogenous leukemia based on chemokine responsiveness and constitutive chemokine release by their leukemic cells. Haematologica 2007, 92, 332–341. [Google Scholar] [CrossRef]
- Domingues, M.J.; Cao, H.; Heazlewood, S.Y.; Cao, B.; Nilsson, S.K. Niche Extracellular Matrix Components and Their Influence on HSC. J. Cell Biochem. 2017, 118, 1984–1993. [Google Scholar] [CrossRef]
- Sangaletti, S.; Chiodoni, C.; Tripodo, C.; Colombo, M.P. Common extracellular matrix regulation of myeloid cell activity in the bone marrow and tumor microenvironments. Cancer Immunol. Immunother. 2017, 66, 1059–1067. [Google Scholar] [CrossRef]
- Klamer, S.; Voermans, C. The role of novel and known extracellular matrix and adhesion molecules in the homeostatic and regenerative bone marrow microenvironment. Cell Adh. Migr. 2014, 8, 563–577. [Google Scholar] [CrossRef]
- Vedi, A.; Santoro, A.; Dunant, C.F.; Dick, J.E.; Laurenti, E. Molecular landscapes of human hematopoietic stem cells in health and leukemia. Ann. N. Y. Acad. Sci. 2016, 1370, 5–14. [Google Scholar] [CrossRef]
- Yu, V.W.; Scadden, D.T. Hematopoietic Stem Cell and Its Bone Marrow Niche. Curr. Top. Dev. Biol. 2016, 118, 21–44. [Google Scholar]
- Lee, D.; Kim, D.W.; Cho, J.Y. Role of growth factors in hematopoietic stem cell niche. Cell Biol. Toxicol. 2020, 36, 131–144. [Google Scholar] [CrossRef]
- Izzi, V.; Lakkala, J.; Devarajan, R.; Savolainen, E.R.; Koistinen, P.; Heljasvaara, R.; Pihlajaniemi, T. Vanin 1 (VNN1) levels predict poor outcome in acute myeloid leukemia. Am. J. Hematol. 2018, 93, E4–E7. [Google Scholar] [CrossRef]
- Izzi, V.; Lakkala, J.; Devarajan, R.; Savolainen, E.R.; Koistinen, P.; Heljasvaara, R.; Pihlajaniemi, T. Expression of a specific extracellular matrix signature is a favorable prognostic factor in acute myeloid leukemia. Leuk. Res. Rep. 2017, 9, 9–13. [Google Scholar] [CrossRef] [PubMed]
- Izzi, V.; Lakkala, J.; Devarajan, R.; Ruotsalainen, H.; Savolainen, E.R.; Koistinen, P.; Heljasvaara, R.; Pihlajaniemi, T. An extracellular matrix signature in leukemia precursor cells and acute myeloid leukemia. Haematologica 2017, 102, e245–e248. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Eto, H.; Tanabe, K.K. Involvement of CD44 in matrix metalloproteinase-2 regulation in human melanoma cells. Int. J. Cancer 1999, 80, 387–395. [Google Scholar] [CrossRef]
- Ponta, H.; Sherman, L.; Herrlich, P.A. CD44: From adhesion molecules to signalling regulators. Nat. Rev. Mol. Cell Biol. 2003, 4, 33–45. [Google Scholar] [CrossRef] [PubMed]
- Klingbeil, P.; Marhaba, R.; Jung, T.; Kirmse, R.; Ludwig, T.; Zöller, M. CD44 variant isoforms promote metastasis formation by a tumor cell-matrix cross-talk that supports adhesion and apoptosis resistance. Mol. Cancer Res. 2009, 7, 168–179. [Google Scholar] [CrossRef]
- Schuurhuis, G.J.; Meel, M.H.; Wouters, F.; Min, L.A.; Terwijn, M.; de Jonge, N.A.; Kelder, A.; Snel, A.N.; Zweegman, S.; Ossenkoppele, G.J.; et al. Normal hematopoietic stem cells within the AML bone marrow have a distinct and higher ALDH activity level than co-existing leukemic stem cells. PLoS ONE 2013, 8, e78897. [Google Scholar] [CrossRef]
- Ishii, S.; Ford, R.; Thomas, P.; Nachman, A.; Steele, G., Jr.; Jessup, J.M. CD44 participates in the adhesion of human colorectal carcinoma cells to laminin and type IV collagen. Surg. Oncol. 1993, 2, 255–264. [Google Scholar] [CrossRef]
- Spertini, C.; Baïsse, B.; Bellone, M.; Gikic, M.; Smirnova, T.; Spertini, O. Acute Myeloid and Lymphoblastic Leukemia Cell Interactions with Endothelial Selectins: Critical Role of PSGL-1, CD44 and CD43. Cancers 2019, 11, 1253. [Google Scholar] [CrossRef] [PubMed]
- Winkler, I.G.; Barbier, V.; Nowlan, B.; Jacobsen, R.N.; Forristal, C.E.; Patton, J.T.; Magnani, J.L.; Lévesque, J.P. Vascular niche E-selectin regulates hematopoietic stem cell dormancy, self renewal and chemoresistance. Nat. Med. 2012, 18, 1651–1657. [Google Scholar] [CrossRef] [PubMed]
- Yokota, A.; Ishii, G.; Sugaya, Y.; Nishimura, M.; Saito, Y.; Harigaya, K. Expression of exon v6-containing CD44 isoforms is related to poor prognosis of acute myelocytic leukemia. Hematol. Oncol. 1998, 16, 131–141. [Google Scholar] [CrossRef]
- Legras, S.; Günthert, U.; Stauder, R.; Curt, F.; Oliferenko, S.; Kluin-Nelemans, H.C.; Marie, J.P.; Proctor, S.; Jasmin, C.; Smadja-Joffe, F. A strong expression of CD44-6v correlates with shorter survival of patients with acute myeloid leukemia. Blood 1998, 91, 3401–3413. [Google Scholar] [CrossRef]
- Aasebø, E.; Berven, F.S.; Bartaula-Brevik, S.; Stokowy, T.; Hovland, R.; Vaudel, M.; Døskeland, S.O.; McCormack, E.; Batth, T.S.; Olsen, J.V.; et al. Proteome and Phosphoproteome Changes Associated with Prognosis in Acute Myeloid Leukemia. Cancers 2020, 12, 709. [Google Scholar] [CrossRef]
- Gjertsen, B.T.; Øyan, A.M.; Marzolf, B.; Hovland, R.; Gausdal, G.; Døskeland, S.O.; Dimitrov, K.; Golden, A.; Kalland, K.H.; Hood, L.; et al. Analysis of acute myelogenous leukemia: Preparation of samples for genomic and proteomic analyses. J. Hematother. Stem Cell Res. 2002, 11, 469–481. [Google Scholar] [CrossRef]
- Wheatley, K.; Burnett, A.K.; Goldstone, A.H.; Gray, R.G.; Hann, I.M.; Harrison, C.J.; Rees, J.K.; Stevens, R.F.; Walker, H.A. Simple, robust, validated and highly predictive index for the determination of risk-directed therapy in acute myeloid leukaemia derived from the MRC AML 10 trial. United Kingdom Medical Research Council’s Adult and Childhood Leukaemia Working Parties. Br. J. Haematol. 1999, 107, 69–79. [Google Scholar] [CrossRef]
- de Jonge, H.J.; Valk, P.J.; de Bont, E.S.; Schuringa, J.J.; Ossenkoppele, G.; Vellenga, E.; Huls, G. Prognostic impact of white blood cell count in intermediate risk acute myeloid leukemia: Relevance of mutated NPM1 and FLT3-ITD. Haematologica 2011, 96, 1310–1317. [Google Scholar] [CrossRef] [PubMed]
- Lin, P.; Chen, L.; Luthra, R.; Konoplev, S.N.; Wang, X.; Medeiros, L.J. Acute myeloid leukemia harboring t(8;21)(q22;q22): A heterogeneous disease with poor outcome in a subset of patients unrelated to secondary cytogenetic aberrations. Mod. Pathol. 2008, 21, 1029–1036. [Google Scholar] [CrossRef][Green Version]
- Feng, S.; Zhou, L.; Zhang, X.; Tang, B.; Zhu, X.; Liu, H.; Sun, Z.; Zheng, C. Impact Of ELN Risk Stratification, Induction Chemotherapy Regimens And Hematopoietic Stem Cell Transplantation On Outcomes In Hyperleukocytic Acute Myeloid Leukemia With Initial White Blood Cell Count More Than 100 × 109/L. Cancer Manag. Res. 2019, 11, 9495–9503. [Google Scholar] [CrossRef] [PubMed]
- How, J.; Sykes, J.; Gupta, V.; Yee, K.W.; Schimmer, A.D.; Schuh, A.C.; Minden, M.D.; Kamel-Reid, S.; Brandwein, J.M. Influence of FLT3-internal tandem duplication allele burden and white blood cell count on the outcome in patients with intermediate-risk karyotype acute myeloid leukemia. Cancer 2012, 118, 6110–6117. [Google Scholar] [CrossRef] [PubMed]
- Thomas, D.; Majeti, R. Biology and relevance of human acute myeloid leukemia stem cells. Blood 2017, 129, 1577–1585. [Google Scholar] [CrossRef]
- Hernandez-Valladares, M.; Bruserud, Ø.; Selheim, F. The Implementation of Mass Spectrometry-Based Proteomics Workflows in Clinical Routines of Acute Myeloid Leukemia: Applicability and Perspectives. Int. J. Mol. Sci. 2020, 21, 6830. [Google Scholar] [CrossRef]
- Aasebø, E.; Berven, F.S.; Hovland, R.; Døskeland, S.O.; Bruserud, Ø.; Selheim, F.; Hernandez-Valladares, M. The Progression of Acute Myeloid Leukemia from First Diagnosis to Chemoresistant Relapse: A Comparison of Proteomic and Phosphoproteomic Profiles. Cancers 2020, 12, 1466. [Google Scholar] [CrossRef]
- Hernandez-Valladares, M.; Aasebø, E.; Berven, F.; Selheim, F.; Bruserud, Ø. Biological characteristics of aging in human acute myeloid leukemia cells: The possible importance of aldehyde dehydrogenase, the cytoskeleton and altered transcriptional regulation. Aging 2020, 12, 24734–24777. [Google Scholar] [CrossRef]
- Mer, A.S.; Lindberg, J.; Nilsson, C.; Klevebring, D.; Wang, M.; Grönberg, H.; Lehmann, S.; Rantalainen, M. Expression levels of long non-coding RNAs are prognostic for AML outcome. J. Hematol. Oncol. 2018, 11, 52. [Google Scholar] [CrossRef]
- Stäubert, C.; Bhuiyan, H.; Lindahl, A.; Broom, O.J.; Zhu, Y.; Islam, S.; Linnarsson, S.; Lehtiö, J.; Nordström, A. Rewired metabolism in drug-resistant leukemia cells: A metabolic switch hallmarked by reduced dependence on exogenous glutamine. J. Biol. Chem. 2015, 290, 8348–8359. [Google Scholar] [CrossRef]
- Lazarevic, V.; Hörstedt, A.S.; Johansson, B.; Antunovic, P.; Billström, R.; Derolf, Å.; Lehmann, S.; Möllgård, L.; Peterson, S.; Stockelberg, D.; et al. Failure matters: Unsuccessful cytogenetics and unperformed cytogenetics are associated with a poor prognosis in a population-based series of acute myeloid leukaemia. Eur. J. Haematol. 2015, 94, 419–423. [Google Scholar] [CrossRef] [PubMed]
- Grønbæk, K.; Müller-Tidow, C.; Perini, G.; Lehmann, S.; Bach Treppendahl, M.; Mills, K.; Plass, C.; Schlegelberger, B.; European Genomics and Epigenomics Study on MDS and AML (EuGESMA), COST Action BM0801. A critical appraisal of tools available for monitoring epigenetic changes in clinical samples from patients with myeloid malignancies. Haematologica 2012, 97, 1380–1388. [Google Scholar] [CrossRef] [PubMed]
- Eppert, K.; Takenaka, K.; Lechman, E.R.; Waldron, L.; Nilsson, B.; van Galen, P.; Metzeler, K.H.; Poeppl, A.; Ling, V.; Beyene, J.; et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat. Med. 2011, 17, 1086–1093. [Google Scholar] [CrossRef] [PubMed]
- Malhotra, V.; Erlmann, P. The pathway of collagen secretion. Annu. Rev. Cell Dev. Biol. 2015, 31, 109–124. [Google Scholar] [CrossRef]
- Granfeldt Østgård, L.S.; Medeiros, B.C.; Sengeløv, H.; Nørgaard, M.; Andersen, M.K.; Dufva, I.H.; Friis, L.S.; Kjeldsen, E.; Marcher, C.W.; Preiss, B.; et al. Epidemiology and Clinical Significance of Secondary and Therapy-Related Acute Myeloid Leukemia: A National Population-Based Cohort Study. J. Clin. Oncol. 2015, 33, 3641–3649. [Google Scholar] [CrossRef] [PubMed]
- Hulegårdh, E.; Nilsson, C.; Lazarevic, V.; Garelius, H.; Antunovic, P.; Rangert Derolf, Å.; Möllgård, L.; Uggla, B.; Wennström, L.; Wahlin, A.; et al. Characterization and prognostic features of secondary acute myeloid leukemia in a population-based setting: A report from the Swedish Acute Leukemia Registry. Am. J. Hematol. 2015, 90, 208–214. [Google Scholar] [CrossRef] [PubMed]
- Hatfield, K.J.; Reikvam, H.; Bruserud, Ø. The crosstalk between the matrix metalloprotease system and the chemokine network in acute myeloid leukemia. Curr. Med. Chem. 2010, 17, 4448–4461. [Google Scholar] [CrossRef] [PubMed]
- Afonina, I.S.; Müller, C.; Martin, S.J.; Beyaert, R. Proteolytic Processing of Interleukin-1 Family Cytokines: Variations on a Common Theme. Immunity 2015, 42, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Bruserud, Ø.; Aasen, I.; Akselsen, P.E.; Bergheim, J.; Rasmussen, G.; Nesthus, I. Interleukin 1 receptor antagonist (IL1RA) in acute leukaemia: IL1RA is both secreted spontaneously by myelogenous leukaemia blasts and is a part of the acute phase reaction in patients with chemotherapy-induced leucopenia. Eur. J. Haematol. 1996, 57, 87–95. [Google Scholar] [CrossRef]
- Fukushima, T.; Uchiyama, S.; Tanaka, H.; Kataoka, H. Hepatocyte Growth Factor Activator: A Proteinase Linking Tissue Injury with Repair. Int. J. Mol. Sci. 2018, 19, 3435. [Google Scholar] [CrossRef]
- Brenner, A.K.; Bruserud, Ø. Functional Toll-Like Receptors (TLRs) Are Expressed by a Majority of Primary Human Acute Myeloid Leukemia Cells and Inducibility of the TLR Signaling Pathway Is Associated with a More Favorable Phenotype. Cancers 2019, 11, 973. [Google Scholar] [CrossRef] [PubMed]
- Wensink, A.C.; Hack, C.E.; Bovenschen, N. Granzymes regulate proinflammatory cytokine responses. J. Immunol. 2015, 194, 491–497. [Google Scholar] [CrossRef] [PubMed]
- Baschuk, N.; Wang, N.; Watt, S.V.; Halse, H.; House, C.; Bird, P.I.; Strugnell, R.; Trapani, J.A.; Smyth, M.J.; Andrews, D.M. NK cell intrinsic regulation of MIP-1α by granzyme M. Cell Death Dis. 2014, 5, e1115. [Google Scholar] [CrossRef]
- Lee, S.E.; Jeong, S.K.; Lee, S.H. Protease and protease-activated receptor-2 signaling in the pathogenesis of atopic dermatitis. Yonsei Med. J. 2010, 51, 808–822. [Google Scholar] [CrossRef]
- Reiss, K.; Saftig, P. The "a disintegrin and metalloprotease" (ADAM) family of sheddases: Physiological and cellular functions. Semin. Cell Dev. Biol. 2009, 20, 126–137. [Google Scholar] [CrossRef] [PubMed]
- Qureshi, N.; Morrison, D.C.; Reis, J. Proteasome protease mediated regulation of cytokine induction and inflammation. Biochim. Biophys. Acta 2012, 1823, 2087–2093. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Miller, J. A review on protein markers of exosome from different bio- resources and the antibodies used for characterization. J. Histotechnol. 2019, 42, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Zhang, J.; Li, S.; Li, L.; Li, M.; Guo, C.; Yao, J.; Mi, S. Exosome and exosomal microRNA: Trafficking, sorting, and function. Genom. Proteom. Bioinform. 2015, 13, 17–24. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Kreitz, J.; Schönfeld, C.; Seibert, M.; Stolp, V.; Alshamleh, I.; Oellerich, T.; Steffen, B.; Schwalbe, H.; Schnütgen, F.; Kurrle, N.; et al. Metabolic Plasticity of Acute Myeloid Leukemia. Cells 2019, 8, 805. [Google Scholar] [CrossRef]
- Castro, I.; Sampaio-Marques, B.; Ludovico, P. Targeting Metabolic Reprogramming in Acute Myeloid Leukemia. Cells 2019, 8, 967. [Google Scholar] [CrossRef] [PubMed]
- Jones, C.L.; Stevens, B.M.; D’Alessandro, A.; Reisz, J.A.; Culp-Hill, R.; Nemkov, T.; Pei, S.; Khan, N.; Adane, B.; Ye, H.; et al. Inhibition of Amino Acid Metabolism Selectively Targets Human Leukemia Stem Cells. Cancer Cell 2018, 34, 724–740.e4. [Google Scholar] [CrossRef]
- Chen, W.L.; Wang, J.H.; Zhao, A.H.; Xu, X.; Wang, Y.H.; Chen, T.L.; Li, J.M.; Mi, J.Q.; Zhu, Y.M.; Liu, Y.F.; et al. A distinct glucose metabolism signature of acute myeloid leukemia with prognostic value. Blood 2014, 124, 1645–1654. [Google Scholar] [CrossRef]
- Ersvaer, E.; Brenner, A.K.; Vetås, K.; Reikvam, H.; Bruserud, Ø. Effects of cytarabine on activation of human T cells-cytarabine has concentration-dependent effects that are modulated both by valproic acid and all-trans retinoic acid. BMC Pharmacol. Toxicol. 2015, 16, 12. [Google Scholar] [CrossRef] [PubMed]
- Kvestad, H.; Evensen, L.; Lorens, J.B.; Bruserud, O.; Hatfield, K.J. In Vitro Characterization of Valproic Acid, ATRA, and Cytarabine Used for Disease-Stabilization in Human Acute Myeloid Leukemia: Antiproliferative Effects of Drugs on Endothelial and Osteoblastic Cells and Altered Release of Angioregulatory Mediators by Endothelial Cells. Leuk. Res. Treatmen 2014, 2014, 143479. [Google Scholar]
- Zheng, H.; Bae, Y.; Kasimir-Bauer, S.; Tang, R.; Chen, J.; Ren, G.; Yuan, M.; Esposito, M.; Li, W.; Wei, Y.; et al. Therapeutic Antibody Targeting Tumor- and Osteoblastic Niche-Derived Jagged1 Sensitizes Bone Metastasis to Chemotherapy. Cancer Cell 2017, 32, 731–747.e6. [Google Scholar] [CrossRef] [PubMed]



| Characteristics (n = 40) | |||
|---|---|---|---|
| Sex and age | Karyotype/Karyotype Abnormalities | ||
| Males/females | 21/19 | Normal | 20 |
| Age median (range) in years | 71 (18–87) | Favorable | 4 |
| Intermediate | 9 | ||
| Predisposition/previous disease | Adverse | 4 | |
| Previous chronic myeloid neoplasia | 1 | Not tested | 3 |
| Myelodysplastic syndrome | 8 | ||
| Relapsed AML | 3 | Flt3 abnormalities | |
| Chemotherapy related | 0 | ITD | 13 |
| Wild type | 19 | ||
| Morphology/FAB classification | Not tested | 8 | |
| M0/M1 | 17 | ||
| M2 | 8 | NPM1 abnormalities | |
| M4/M5 | 15 | Insertion | 13 |
| M6/M7 | 0 | Insertion + Flt3-ITD | 8 |
| Wild type | 20 | ||
| CD34 positive | 21 | Not tested | 7 |
| Cellular Compartment | Number of Proteins | Proteins in the Background Dataset | Percent of Proteins | Fold Enrichment | p-Value (Hypergeometric Test) | Bonferroni Corrected |
|---|---|---|---|---|---|---|
| Cytoplasm | 1171 | 5684 | 68.4 | 1.8 | 2.2 × 10−151 | 1.7 × 10−148 |
| Nucleus | 946 | 5847 | 55.3 | 1.4 | 2.83 × 10−41 | 2.22 × 10−38 |
| Exosomes | 746 | 2043 | 43.6 | 3.1 | 5.8 × 10−232 | 4.5 × 10−229 |
| Lysosome | 561 | 1620 | 32.8 | 2.9 | 1.8 × 10−151 | 1.4 × 10−148 |
| Nucleolus | 443 | 1257 | 25.9 | 3.0 | 1.4 × 10−118 | 1.1 × 10−115 |
| Cytosol | 407 | 1178 | 23.8 | 2.9 | 1.5 × 10−104 | 1.1 × 10−101 |
| Mitochondrion | 348 | 1259 | 20.3 | 2.4 | 2.36 × 10−59 | 1.85 × 10−56 |
| Centrosome | 343 | 656 | 20.0 | 4.4 | 4.8 × 10−152 | 3.7 × 10−149 |
| Plasma membrane | 316 | 3479 | 18.5 | 0.8 | 1 | 1 |
| Extracellular | 262 | 1825 | 15.3 | 1.2 | 0.000182 | 0.14294 |
| Endoplasmic reticulum | 148 | 1104 | 8.6 | 1.1 | 0.044427 | 1 |
| Cytoskeleton | 137 | 427 | 8.0 | 2.7 | 7.52 × 10−30 | 5.89 × 10−27 |
| Golgi apparatus | 127 | 897 | 7.4 | 1.2 | 0.013653 | 1 |
| Nucleoplasm | 106 | 449 | 6.2 | 2.0 | 7.13 × 10−13 | 5.59 × 10−10 |
| Cluster and Corresponding Go Terms | p-Value (Uncorrected) | FDR |
|---|---|---|
| Left purple protein cluster | ||
| Mitochondrial part | 7.77 × 10−8 | 4.68 × 10−5 |
| Ribosomal subunit | 1.72 × 10−6 | 0.000345 |
| Organelle inner membrane | 1.41 × 10−5 | 0.00212 |
| Cytosolic part | 1.48 × 10−5 | 0.00212 |
| Mitochondrion | 1.86 × 10−5 | 0.00212 |
| Middle left green protein cluster | ||
| Nucleoplasm part | 7.69 × 10−7 | 0.000557 |
| Nucleoplasm | 0.000116 | 0.0168 |
| Spliceosomal complex | 0.000621 | 0.0749 |
| Nuclear chromosome part | 0.00125 | 0.113 |
| Nucleoplasm part | 7.69 × 10−7 | 0.000557 |
| Middle yellow protein cluster | ||
| Extracellular exosome | 4.72 × 10−17 | 2.97 × 10−14 |
| Cytoplasmic vesicle part | 1.68 × 10−9 | 1.76 × 10−7 |
| Cytosol | 1.80 × 10−9 | 1.76 × 10−7 |
| Extracellular region | 1.42 × 10−8 | 1.11 × 10−6 |
| Vesicle lumen | 1.42 × 10−7 | 7.43 × 10−6 |
| Middle right orange protein cluster | ||
| Extracellular matrix organization | 2.67 × 10−10 | 1.33 × 10−6 |
| Anatomical structure morphogenesis | 1.96 × 10−5 | 0.0325 |
| Positive regulation of developmental process | 0.000231 | 0.231 |
| Vesicle-mediated transport | 0.000237 | 0.231 |
| Multicellular organismal process | 0.000243 | 0.231 |
| Right blue protein cluster | ||
| No significant GO terms |
| All identified proteins (alphabetic order) | ABI3BP, B4GALT1, BGN, C1R, CD248, CDH2, CDH11, CDH13, CFH, COL10A1, COL16A1, COL18A1, COL4A1, COL8A1, CRIM1, CTGF, CTHRC1, CTSK, CYR61, DAG1, DKK3, ECM1, ENPP1, ENPP2, FBLN1, FBLN5, FKBP10, GAS6, GOLM1, GREM1, IGFBP4, ISLR, ITGBL1, LAMA4, LOX, LOXL1, LOXL2, MFAP2, MMP13, MMP14, MXRA8, NBL1, NRP2, OLFML2B, PAPPA, PLOD2, PLTP, PROCR, PRSS23, PTPRK, SDC1, SMOC1, SPON2, SRPX2, SSC5D, STC2, TAGLN, THY1, TNC, VASN |
| Extracellular matrix (ECM) molecules | ABI3BP, BGN, COL10A1, COL16A1, COL18A1, COL4A1, COL8A1, ECM1, FBLN1, FBLN5, ISLR, LAMA4, MFAP2, MXRA8, SMOC1, SPON2, SRPX2, STC2, TNC ECM modulators: LOX, LOXL1, LOXL2 |
| Cytokines, extracellular soluble mediators | CRIM1 (TGFβ interaction), CTGF, CYR61/CCN1, DKK3 (extracellular Wnt inhibitor), IGFBP4 (IGF binding), PLTP (lipid metabolism), |
| Cytokine receptors and signaling | ECM1, GAS6, GREM1, NBL1, NRP2, SDC1, SSC5D, VASN (TGF signaling) |
| Cell surface molecules | Ig superfamily: CD248, THY1 Adhesion: CDH2, CDH11, CDH13, DAG1, ITGBL1 Others: OLFML2B, PROCR, PTPRK, VASN (TGF signaling) |
| Enzymes | Proteases: C1R, CFH, CTSK, ECM1, MMP13, MMP14, PAPPA, PRSS23 Other enzymes: B4GALT1, ENPP1, ENPP2, LOX, LOXL1, LOXL2 |
| Golgi/endoplasmatic reticulum | B4GALT1, FKBP10 (chaperon), GOLM1 |
| Cytoskeleton | DAG1, TAGLN |
| Intracellular signaling | CTHRC1, PTPRK |
| GO Term Identity | Percent Associated Foreground | Percent Associated Background | Fold_Enrichment_Fore- Ground to Background | Foreground_Count | Foreground_n | Background_Count | Background_n | p-Value Uncorrected | FDR | Description |
|---|---|---|---|---|---|---|---|---|---|---|
| CELLULAR COMPARTMENT | ||||||||||
| GO:0005615 | 58.3 | 13.4 | 4.4 | 35 | 60 | 402 | 2998 | 1.29 × 10−15 | 2.67 × 10−13 | extracellular space |
| GO:0031012 | 45.0 | 8.0 | 5.6 | 27 | 60 | 240 | 2998 | 4.68 × 10−14 | 4.82 × 10−12 | extracellular matrix |
| GO:0005576 | 53.3 | 17.5 | 3.0 | 32 | 60 | 525 | 2998 | 6.05 × 10−10 | 2.49 × 10−8 | extracellular region |
| GO:0044420 | 20.0 | 1.8 | 10.9 | 12 | 60 | 55 | 2998 | 2.73 × 10−9 | 9.38 × 10−8 | extracellular matrix component |
| GO:0005788 | 26.6 | 4.5 | 5.9 | 16 | 60 | 136 | 2998 | 1.43 × 10−8 | 4.21 × 10−7 | endoplasmic reticulum lumen |
| GO:0005581 | 11.6 | 1.1 | 10.6 | 7 | 60 | 33 | 2998 | 8.81 × 10−6 | 0.000202 | collagen trimer |
| GO:0009986 | 18.3 | 4.9 | 3.8 | 11 | 60 | 146 | 2998 | 0.000177 | 0.00331 | cell surface |
| GO:0005796 | 8.3 | 0.9 | 9.6 | 5 | 60 | 26 | 2998 | 0.000282 | 0.00484 | Golgi lumen |
| GO:0031224 | 30.0 | 13.0 | 2.3 | 18 | 60 | 391 | 2998 | 0.000543 | 0.0086 | intrinsic component of membrane |
| GO:0016323 | 6.7 | 0.9 | 7.4 | 4 | 60 | 27 | 2998 | 0.00284 | 0.0325 | basolateral plasma membrane |
| MOLECULAR FUNCTION | ||||||||||
| GO:0005509 | 25.5 | 5.1 | 5.0 | 14 | 55 | 152 | 2968 | 7.97 × 10−7 | 0.000223 | calcium ion binding |
| GO:0050840 | 12.7 | 0.9 | 14.0 | 7 | 55 | 27 | 2968 | 1.64 × 10−6 | 0.00023 | extracellular matrix binding |
| GO:0019838 | 14.5 | 1.4 | 10.0 | 8 | 55 | 43 | 2968 | 2.48 × 10−6 | 0.000231 | growth factor binding |
| GO:0005044 | 7.3 | 0.4 | 18.0 | 4 | 55 | 12 | 2968 | 0.000152 | 0.00777 | scavenger receptor activity |
| GO:0005178 | 10.9 | 1.7 | 6.5 | 6 | 55 | 50 | 2968 | 0.000443 | 0.0138 | integrin binding |
| GO:0001968 | 7.3 | 0.6 | 11.4 | 4 | 55 | 19 | 2968 | 0.000672 | 0.0188 | fibronectin binding |
| GO:0016641 | 5.5 | 0.2 | 20.2 | 3 | 55 | 8 | 2968 | 0.000848 | 0.0198 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor |
| GO:0016015 | 3.6 | 0.1 | 54.0 | 2 | 55 | 2 | 2968 | 0.00191 | 0.0333 | morphogen activity |
| GO:0019955 | 7.3 | 0.9 | 7.7 | 4 | 55 | 28 | 2968 | 0.00242 | 0.0398 | cytokine binding |
| GO:0005201 | 7.3 | 1.0 | 7.4 | 4 | 55 | 29 | 2968 | 0.00271 | 0.042 | extracellular matrix structural constituent |
| GO:0005539 | 10.9 | 2.5 | 4.4 | 6 | 55 | 74 | 2968 | 0.00293 | 0.042 | glycosaminoglycan binding |
| GO:0004528 | 3.6 | 0.1 | 36.0 | 2 | 55 | 3 | 2968 | 0.00314 | 0.042 | phosphodiesterase I activity |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aasebø, E.; Brenner, A.K.; Birkeland, E.; Tvedt, T.H.A.; Selheim, F.; Berven, F.S.; Bruserud, Ø. The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells—A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells. Cancers 2021, 13, 1509. https://doi.org/10.3390/cancers13071509
Aasebø E, Brenner AK, Birkeland E, Tvedt THA, Selheim F, Berven FS, Bruserud Ø. The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells—A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells. Cancers. 2021; 13(7):1509. https://doi.org/10.3390/cancers13071509
Chicago/Turabian StyleAasebø, Elise, Annette K. Brenner, Even Birkeland, Tor Henrik Anderson Tvedt, Frode Selheim, Frode S. Berven, and Øystein Bruserud. 2021. "The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells—A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells" Cancers 13, no. 7: 1509. https://doi.org/10.3390/cancers13071509
APA StyleAasebø, E., Brenner, A. K., Birkeland, E., Tvedt, T. H. A., Selheim, F., Berven, F. S., & Bruserud, Ø. (2021). The Constitutive Extracellular Protein Release by Acute Myeloid Leukemia Cells—A Proteomic Study of Patient Heterogeneity and Its Modulation by Mesenchymal Stromal Cells. Cancers, 13(7), 1509. https://doi.org/10.3390/cancers13071509







