Detection of Genomically Aberrant Cells within Circulating Tumor Microemboli (CTMs) Isolated from Early-Stage Breast Cancer Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Establishment of a Method for Accurate Estimation of Tumor Fraction (TF) in CTMs
2.1.1. Building of a Calibration Curve for Tumor Genome Fraction (TGF) Estimation
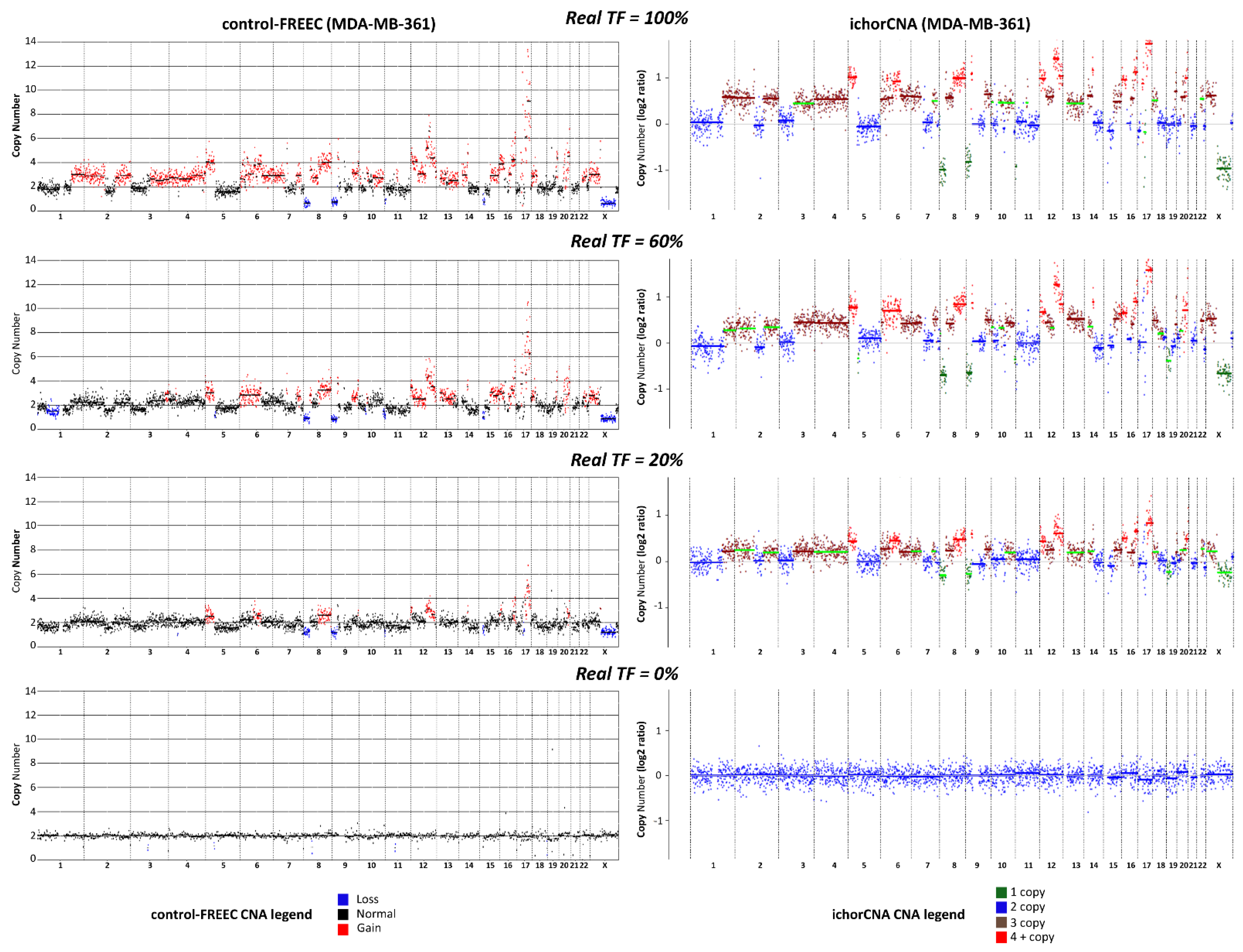
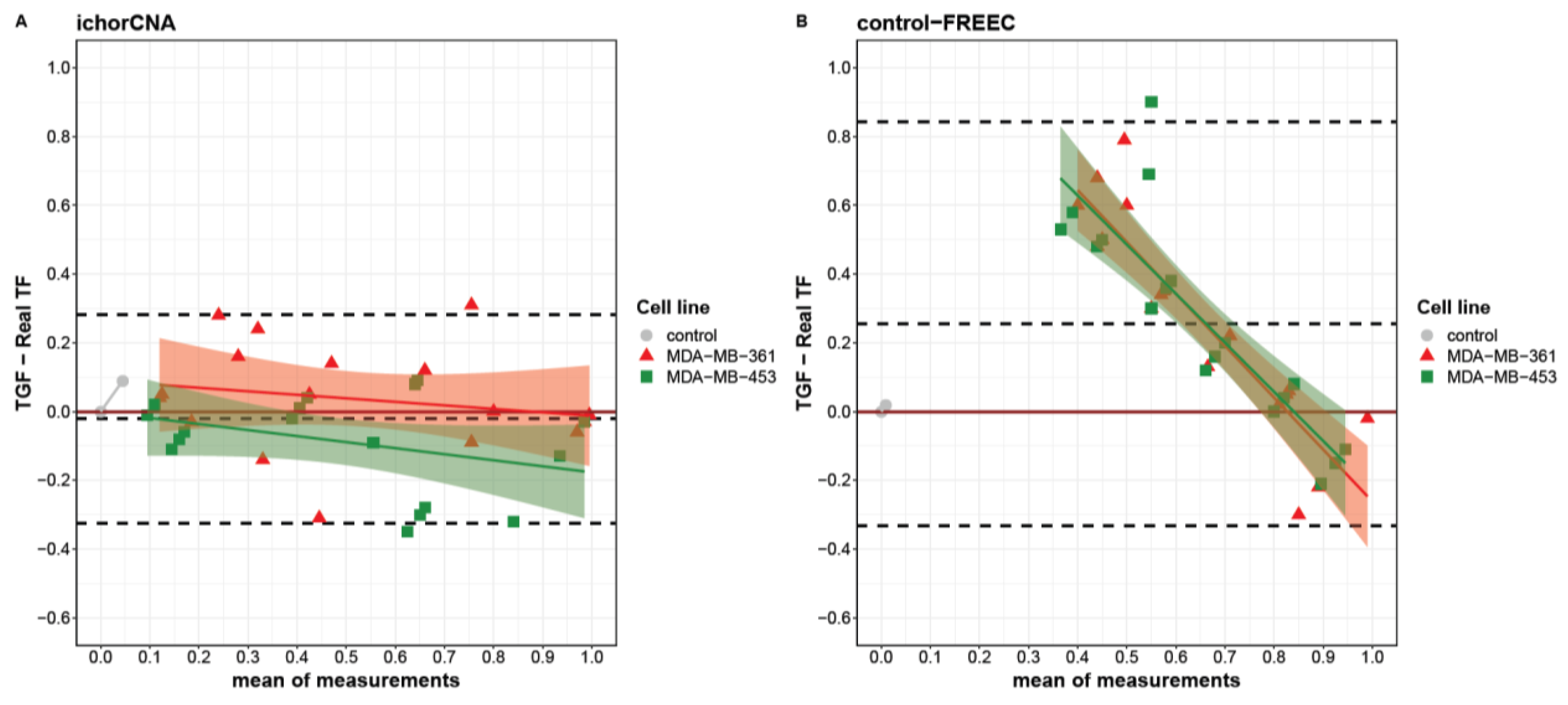
2.1.2. Comparison between Control-FREEC and ichorCNA for TGF Estimation
2.1.3. Validation of TGF in Stroma and Tumor Fraction from Microdissected Tissues
2.1.4. Building of a Statistical Model to Predict TF in Clinical Samples Starting from TGF Values
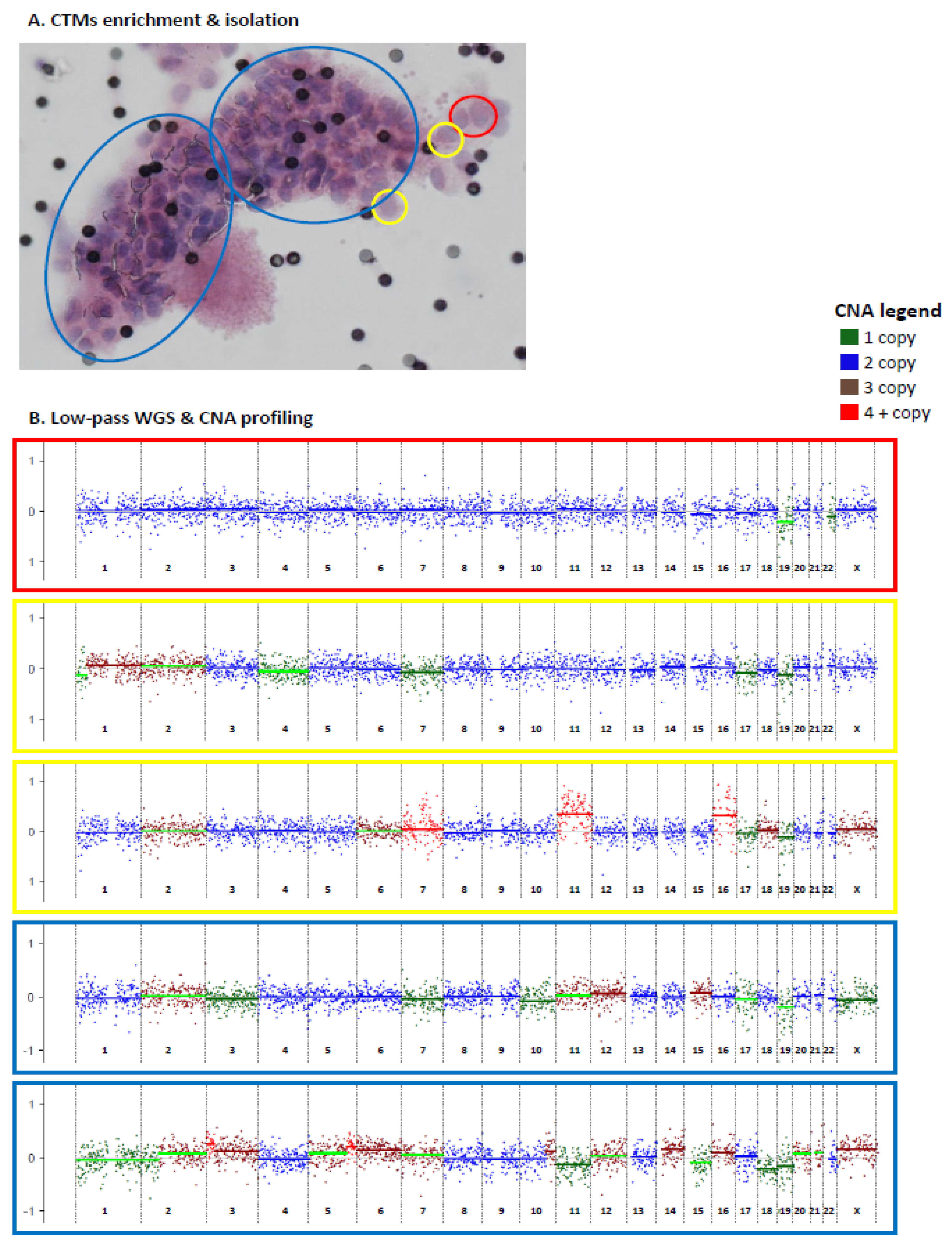
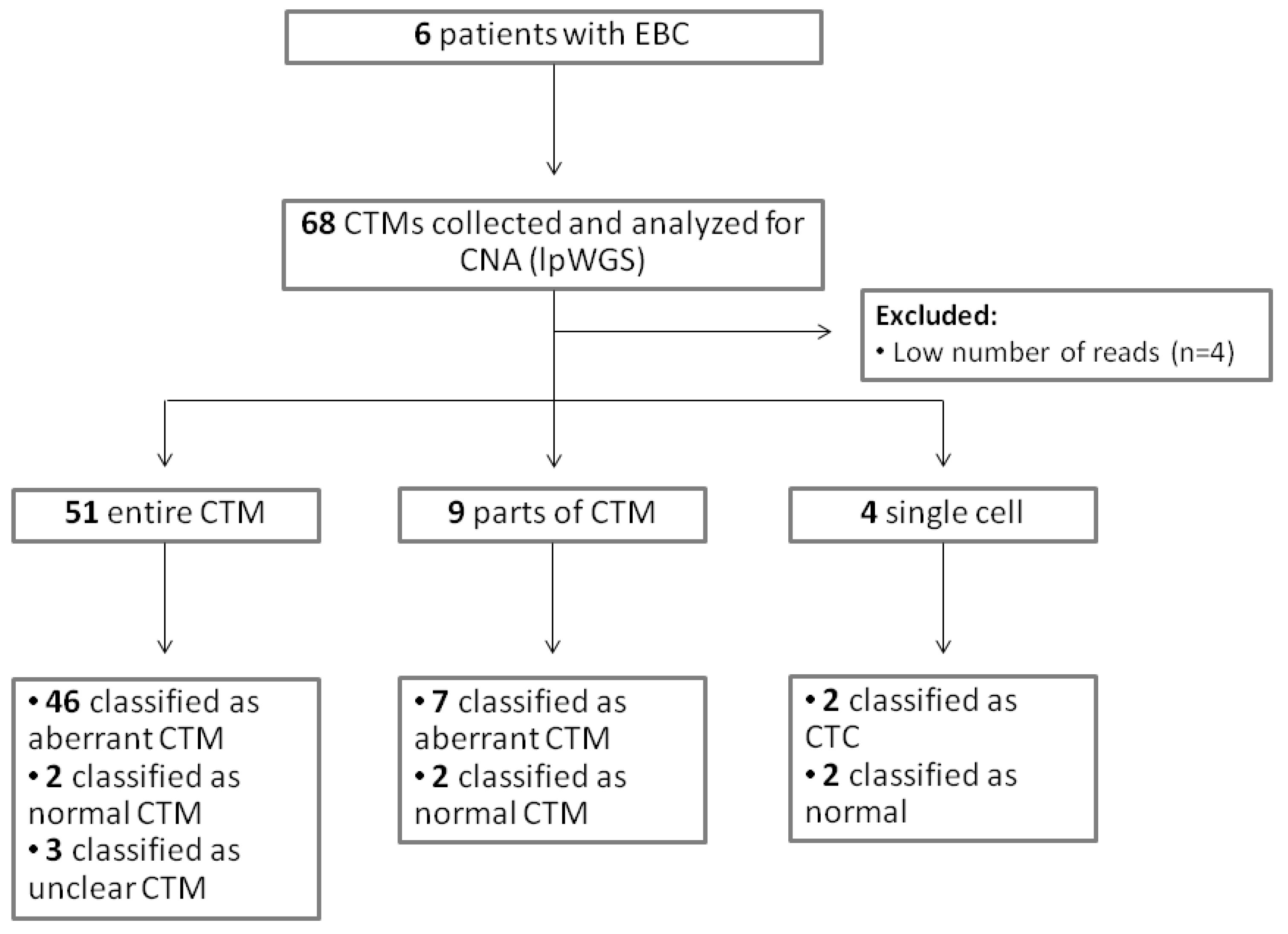
2.2. Genomic Analysis of CTMs Isolated from Blood of Women with EBC
2.2.1. Predicted TF in CTMs of EBC Patients
2.2.2. Comparison of Genomic Alterations of CTMs with Primary Tumor Tissue
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Generation of Cell Mixtures
4.2. Case Series
4.3. Blood Sample Collection from Breast Cancer Patients
4.4. CTM Enrichment by a Size-Based Approach
4.5. Cell Isolation, Ampli1TM Whole Genome Amplification, DNA Library Construction and Whole Genome Sequencing
4.6. Isolation of Cancer Cell Populations from Human Formalin-Fixed, Paraffin-Embedded Tissue Sections
4.7. Sequencing Data Analysis
- control-FREEC. coefficientofVariation = 0.05, mateOrientation = 0, normal control = TRUE, window = 1 Mb, ploidy = 2.65 (MDA-MB-361) or 4.2 (MDA-MB-453);
- ichorCNA. Window = 1 Mb, ploidy = 2.65 (MDA-MB-361) or 4.2 (MDA-MB-453), estimatePloidy = FALSE, estimateNormal = TRUE, normalPanel = TRUE, normal state = c(0.1, 0.2, 0.3, 0.4, 0.5, 0.6, 0.7, 0.8, 0.9).
- Normal profile but only 1 genomic region with amplification/deletion lower than 125 Mb;
- Normal profile but sum of amplification/deletion of different genomic regions lower than 375 Mb.
4.8. Statistical Analysis
- Both tumor samples of the same patients (when possible);
- Both tumor samples and at least one CTM of the same patient;
- At least two CTMs of the same patients.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fidler, I.J.; Kripke, M.L. The challenge of targeting metastasis. Cancer Metastasis Rev. 2015, 34, 635–641. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Mph, K.D.M.; Sauer, A.G.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Pantel, K.; Alix-Panabières, C. Circulating tumour cells in cancer patients: Challenges and perspectives. Trends Mol. Med. 2010, 16, 398–406. [Google Scholar] [CrossRef] [PubMed]
- Piñeiro, R.; Martínez-Pena, I.; López-López, R. Relevance of CTC Clusters in Breast Cancer Metastasis. Adv. Exp. Med. Biol. 2020, 1220, 93–115. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, C.; Li, Y.; Muñiz, M.C.; Kidwell, K.M.; Aung, K.; Thomas, D.G.; Brown, M.E.; Abramson, V.G.; Irvin, W.J., Jr.; Lin, N.U.; et al. Significance of Circulating Tumor Cells in Metastatic Triple-Negative Breast Cancer Patients within a Randomized, Phase II Trial: TBCRC 019. Clin. Cancer Res. 2015, 21, 2771–2779. [Google Scholar] [CrossRef] [PubMed]
- Jansson, S.; Bendahl, P.-O.; Larsson, A.-M.; Aaltonen, K.E.; Rydén, L. Prognostic impact of circulating tumor cell apoptosis and clusters in serial blood samples from patients with metastatic breast cancer in a prospective observational cohort. BMC Cancer 2016, 16, 1–15. [Google Scholar] [CrossRef]
- Wang, C.; Mu, Z.; Chervoneva, I.; Austin, L.; Ye, Z.; Rossi, G.; Palazzo, J.P.; Sun, C.; Abu-Khalaf, M.; Myers, R.E.; et al. Longitudinally collected CTCs and CTC-clusters and clinical outcomes of metastatic breast cancer. Breast Cancer Res. Treat. 2017, 161, 83–94. [Google Scholar] [CrossRef]
- Paoletti, C.; Miao, J.; Dolce, E.M.; Darga, E.P.; Repollet, M.I.; Doyle, G.V.; Gralow, J.R.; Hortobagyi, G.N.; Smerage, J.B.; Barlow, W.E.; et al. Circulating Tumor Cell Clusters in Patients with Metastatic Breast Cancer: A SWOG S0500 Translational Medicine Study. Clin. Cancer Res. 2019, 25, 6089–6097. [Google Scholar] [CrossRef]
- Costa, C.; Muinelo-Romay, L.; Cebey-López, V.; Pereira-Veiga, T.; Martínez-Pena, I.; Abreu, M.; Abalo, A.; Lago-Lestón, R.M.; Abuín, C.; Palacios, P.; et al. Analysis of a Real-World Cohort of Metastatic Breast Cancer Patients Shows Circulating Tumor Cell Clusters (CTC-clusters) as Predictors of Patient Outcomes. Cancers 2020, 12, 1111. [Google Scholar] [CrossRef]
- Mu, Z.; Wang, C.; Ye, Z.; Austin, L.; Civan, J.; Hyslop, T.; Palazzo, J.P.; Jaslow, R.; Li, B.; Myers, R.E.; et al. Prospective assessment of the prognostic value of circulating tumor cells and their clusters in patients with advanced-stage breast cancer. Breast Cancer Res. Treat. 2015, 154, 563–571. [Google Scholar] [CrossRef]
- Larsson, A.-M.; Jansson, S.; Bendahl, P.-O.; Jörgensen, C.L.T.; Loman, N.; Graffman, C.; Lundgren, L.; Aaltonen, K.; Rydén, L. Longitudinal enumeration and cluster evaluation of circulating tumor cells improve prognostication for patients with newly diagnosed metastatic breast cancer in a prospective observational trial. Breast Cancer Res. 2018, 20, 48. [Google Scholar] [CrossRef]
- Fina, E.; Reduzzi, C.; Motta, R.; Di Cosimo, S.; Bianchi, G.V.; Martinetti, A.; Wechsler, J.; Cappelletti, V.; Daidone, M.G. Did Circulating Tumor Cells Tell us all they Could? The Missed Circulating Tumor Cell Message in Breast Cancer. Int. J. Biol. Markers 2015, 30, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Reduzzi, C.; Di Cosimo, S.; Gerratana, L.; Motta, R.; Martinetti, A.; Vingiani, A.; D’Amico, P.; Zhang, Y.; Vismara, M.; Depretto, C.; et al. Dissemination of circulating tumor cell clusters occurs early in non metastatic breast cancer patients. under revision.
- Hüsemann, Y.; Geigl, J.B.; Schubert, F.; Musiani, P.; Meyer, M.; Burghart, E.; Forni, G.; Eils, R.; Fehm, T.; Riethmüller, G.; et al. Systemic Spread Is an Early Step in Breast Cancer. Cancer Cell 2008, 13, 58–68. [Google Scholar] [CrossRef] [PubMed]
- Hosseini, H.; Obradović, M.M.S.; Hoffmann, M.; Harper, K.L.; Sosa, M.S.; Werner-Klein, M.; Nanduri, S.L.K.; Werno, C.; Ehrl, C.; Maneck, M.; et al. Early dissemination seeds metastasis in breast cancer. Nature 2016, 540, 552–558. [Google Scholar] [CrossRef]
- Aceto, N.; Bardia, A.; Miyamoto, D.T.; Donaldson, M.C.; Wittner, B.S.; Spencer, J.A.; Yu, M.; Pely, A.; Engstrom, A.; Zhu, H.; et al. Circulating Tumor Cell Clusters Are Oligoclonal Precursors of Breast Cancer Metastasis. Cell 2014, 158, 1110–1122. [Google Scholar] [CrossRef] [PubMed]
- Aceto, N. Bring along your friends: Homotypic and heterotypic circulating tumor cell clustering to accelerate metastasis. Biomed. J. 2020, 43, 18–23. [Google Scholar] [CrossRef] [PubMed]
- Heeke, S.; Mograbi, B.; Alix-Panabières, C.; Hofman, P. Never Travel Alone: The Crosstalk of Circulating Tumor Cells and the Blood Microenvironment. Cells 2019, 8, 714. [Google Scholar] [CrossRef]
- Szczerba, B.M.; Castro-Giner, F.; Vetter, M.; Krol, I.; Gkountela, S.; Landin, J.; Scheidmann, M.C.; Donato, C.; Scherrer, R.; Singer, J.; et al. Neutrophils escort circulating tumour cells to enable cell cycle progression. Nature 2019, 566, 553–557. [Google Scholar] [CrossRef]
- Guo, B.; Oliver, T.G. Partners in Crime: Neutrophil–CTC Collusion in Metastasis. Trends Immunol. 2019, 40, 556–559. [Google Scholar] [CrossRef] [PubMed]
- Sprouse, M.L.; Welte, T.; Boral, D.; Liu, H.N.; Yin, W.; Vishnoi, M.; Goswami-Sewell, D.; Li, L.; Pei, G.; Jia, P.; et al. PMN-MDSCs Enhance CTC Metastatic Properties through Reciprocal Interactions via ROS/Notch/Nodal Signaling. Int. J. Mol. Sci. 2019, 20, 1916. [Google Scholar] [CrossRef]
- Liu, Q.; Liao, Q.; Zhao, Y. Myeloid-derived suppressor cells (MDSC) facilitate distant metastasis of malignancies by shielding circulating tumor cells (CTC) from immune surveillance. Med. Hypotheses 2016, 87, 34–39. [Google Scholar] [CrossRef]
- Bolognesi, C.; Forcato, C.; Buson, G.; Fontana, F.; Mangano, C.; Doffini, A.; Sero, V.; Lanzellotto, R.; Signorini, G.; Calanca, A.; et al. Digital Sorting of Pure Cell Populations Enables Unambiguous Genetic Analysis of Heterogeneous Formalin-Fixed Paraffin-Embedded Tumors by Next Generation Sequencing. Sci. Rep. 2016, 6, 20944. [Google Scholar] [CrossRef] [PubMed]
- Norton, M.E.; Jacobsson, B.; Swamy, G.K.; Laurent, L.C.; Ranzini, A.C.; Brar, H.; Tomlinson, M.W.; Pereira, L.; Spitz, J.L.; Hollemon, D.; et al. Cell-free DNA Analysis for Noninvasive Examination of Trisomy. N. Engl. J. Med. 2015, 372, 1589–1597. [Google Scholar] [CrossRef] [PubMed]
- Amant, F.; Verheecke, M.; Wlodarska, I.; Dehaspe, L.; Brady, P.; Brison, N.; Bogaert, K.V.D.; Dierickx, D.; Vandecaveye, V.; Tousseyn, T.; et al. Presymptomatic Identification of Cancers in Pregnant Women During Noninvasive Prenatal Testing. JAMA Oncol. 2015, 1, 814–819. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.W.; Chudova, D.; Sehnert, A.J.; Bhatt, S.; Murray, K.; Prosen, T.L.; Garber, J.E.; Wilkins-Haug, L.; Vora, N.L.; Warsof, S.; et al. Noninvasive Prenatal Testing and Incidental Detection of Occult Maternal Malignancies. JAMA 2015, 314, 162–169. [Google Scholar] [CrossRef]
- Hovelson, D.H.; Liu, C.-J.; Wang, Y.; Kang, Q.; Henderson, J.; Gursky, A.; Brockman, S.; Ramnath, N.; Krauss, J.C.; Talpaz, M.; et al. Rapid, ultra low coverage copy number profiling of cell-free DNA as a precision oncology screening strategy. Oncotarget 2017, 8, 89848–89866. [Google Scholar] [CrossRef]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable whole-exome sequencing of cell-free DNA reveals high concordance with metastatic tumors. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Stover, D.G.; Parsons, H.A.; Ha, G.; Freeman, S.S.; Barry, W.T.; Guo, H.; Choudhury, A.D.; Gydush, G.; Reed, S.C.; Rhoades, J.; et al. Association of Cell-Free DNA Tumor Fraction and Somatic Copy Number Alterations with Survival in Metastatic Triple-Negative Breast Cancer. J. Clin. Oncol. 2018, 36, 543–553. [Google Scholar] [CrossRef]
- Choudhury, A.D.; Werner, L.; Francini, E.; Wei, X.X.; Ha, G.; Freeman, S.S.; Rhoades, J.; Reed, S.C.; Gydush, G.; Rotem, D.; et al. Tumor fraction in cell-free DNA as a biomarker in prostate cancer. JCI Insight 2018, 3. [Google Scholar] [CrossRef]
- Mohan, S.; Foy, V.; Ayub, M.; Leong, H.S.; Schofield, P.; Sahoo, S.; Descamps, T.; Kilerci, B.; Smith, N.K.; Carter, M.; et al. Profiling of Circulating Free DNA Using Targeted and Genome-wide Sequencing in Patients with SCLC. J. Thorac. Oncol. 2020, 15, 216–230. [Google Scholar] [CrossRef]
- Boeva, V.; Popova, T.; Bleakley, K.; Chiche, P.; Cappo, J.; Schleiermacher, G.; Janoueix-Lerosey, I.; Delattre, O.; Barillot, E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics 2011, 28, 423–425. [Google Scholar] [CrossRef]
- Zhang, L.; Feizi, N.; Chi, C.; Hu, P. Association Analysis of Somatic Copy Number Alteration Burden with Breast Cancer Survival. Front. Genet. 2018, 9, 421. [Google Scholar] [CrossRef]
- Reinholz, M.M.; Bruzek, A.K.; Visscher, D.W.; Lingle, W.L.; Schroeder, M.J.; Perez, A.E.; Jenkins, R.B. Breast cancer and aneusomy 17: Implications for carcinogenesis and therapeutic response. Lancet Oncol. 2009, 10, 267–277. [Google Scholar] [CrossRef]
- Hodgson, J.G.; Chin, K.; Collins, C.; Gray, J.W. Genome amplification of chromosome 20 in breast cancer. Breast Cancer Res. Treat. 2003, 78, 337–345. [Google Scholar] [CrossRef]
- Tanner, M.M.; Tirkkonen, M.; Kallioniemi, A.; Isola, J.; Kuukasjärvi, T.; Collins, C.; Kowbel, D.; Guan, X.Y.; Trent, J.; Gray, J.W.; et al. Independent amplification and frequent co-amplification of three nonsyntenic regions on the long arm of chromosome 20 in human breast cancer. Cancer Res. 1996, 56, 3441–3445. [Google Scholar]
- Ciriello, G.; Miller, M.L.; Aksoy, B.A.; Senbabaoglu, Y.; Schultz, N.; Sander, C. Emerging landscape of oncogenic signatures across human cancers. Nat. Genet. 2013, 45, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Cailleau, R.; Olivé, M.; Cruciger, Q.V.J. Long-term human breast carcinoma cell lines of metastatic origin: Preliminary characterization. Vitr. Cell. Dev. Biol. Anim. 1978, 14, 911–915. [Google Scholar] [CrossRef] [PubMed]
- Dowsett, M.; Nielsen, T.O.; A’Hern, R.; Bartlett, J.; Coombes, R.C.; Cuzick, J.; Ellis, M.; Henry, N.L.; Hugh, J.C.; Lively, T.; et al. Assessment of Ki67 in Breast Cancer: Recommendations from the International Ki67 in Breast Cancer Working Group. J. Natl. Cancer Inst. 2011, 103, 1656–1664. [Google Scholar] [CrossRef] [PubMed]
- Weidele, K.; Stojanović, N.; Feliciello, G.; Markiewicz, A.; Scheitler, S.; Alberter, B.; Renner, P.; Haferkamp, S.; Klein, C.A.; Polzer, B. Microfluidic enrichment, isolation and characterization of disseminated melanoma cells from lymph node samples. Int. J. Cancer 2019, 145, 232–241. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.A.; Schmidt-Kittler, O.; Schardt, J.A.; Pantel, K.; Speicher, M.R.; Riethmüller, G. Comparative genomic hybridization, loss of heterozygosity, and DNA sequence analysis of single cells. Proc. Natl. Acad. Sci. USA 1999, 96, 4494–4499. [Google Scholar] [CrossRef]
- Hammond, M.E.H.; Hayes, D.F.; Dowsett, M.; Allred, D.C.; Hagerty, K.L.; Badve, S.; Fitzgibbons, P.L.; Francis, G.; Goldstein, N.S.; Hayes, M.; et al. American Society of Clinical Oncology/College of American Pathologists Guideline Recommendations for Immunohistochemical Testing of Estrogen and Progesterone Receptors in Breast Cancer. J. Clin. Oncol. 2010, 28, 2784–2795. [Google Scholar] [CrossRef]
- Wolff, A.C.; Hammond, M.E.H.; Hicks, D.G.; Dowsett, M.; McShane, L.M.; Allison, K.H.; Allred, D.C.; Bartlett, J.M.; Bilous, M.; Fitzgibbons, P.; et al. Recommendations for Human Epidermal Growth Factor Receptor 2 Testing in Breast Cancer: American Society of Clinical Oncology/College of American Pathologists Clinical Practice Guideline Update. J. Clin. Oncol. 2013, 31, 3997–4013. [Google Scholar] [CrossRef]
- Fina, E.; Necchi, A.; Bottelli, S.; Reduzzi, C.; Pizzamiglio, S.; Iacona, C.; Daidone, M.G.; Verderio, P.; Cappelletti, V. Detection of Circulating Tumour Cells in Urothelial Cancers and Clinical Correlations: Comparison of Two Methods. Dis. Markers 2017, 2017, 3414910. [Google Scholar] [CrossRef] [PubMed]
- Klein, C.A. Single cell amplification methods for the study of cancer and cellular ageing. Mech. Ageing Dev. 2005, 126, 147–151. [Google Scholar] [CrossRef]
- Polzer, B.; Medoro, G.; Pasch, S.; Fontana, F.; Zorzino, L.; Pestka, A.; Andergassen, U.; Meier-Stiegen, F.; Czyz, Z.T.; Alberter, B.; et al. Molecular profiling of single circulating tumor cells with diagnostic intention. EMBO Mol. Med. 2014, 6, 1371–1386. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Conesa, A.; García-Alcalde, F. Qualimap 2: Advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics 2016, 32, 292–294. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Wang, Q.; Wang, Q.; Jia, P.; Zhao, Z. Computational tools for copy number variation (CNV) detection using next-generation sequencing data: Features and perspectives. BMC Bioinform. 2013, 14. [Google Scholar] [CrossRef] [PubMed]
- Reduzzi, C.; Vismara, M.; Silvestri, M.; Celio, L.; Niger, M.; Peverelli, G.; De Braud, F.; Daidone, M.G.; Cappelletti, V. A novel circulating tumor cell subpopulation for treatment monitoring and molecular characterization in biliary tract cancer. Int. J. Cancer 2020, 146, 3495–3503. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.I.-K.A. Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255. [Google Scholar] [CrossRef]
- Bland, J.M.; Altman, D. Statistical Methods for Assessing Agreement Between Two Methods of Clinical Measurement. Lancet 1986, 327, 307–310. [Google Scholar] [CrossRef]
- Lindstrom, M.J.; Bates, D.M. Newton—Raphson and EM Algorithms for Linear Mixed-Effects Models for Repeated-Measures Data. J. Am. Stat. Assoc. 1988, 83, 1014–1022. [Google Scholar] [CrossRef]
- Graybill, F.A.; Iyer, H.K. Regression Analysis: Concepts and Applications; Duxbury Press: Belmont, CA, USA, 1994. [Google Scholar]
- Maimon, O.; Rokach, L. Data Mining and Knowledge Discovery Handbook; Springer International Publishing: Berlin/Heidelberg, Germany, 2005; pp. 321–352. [Google Scholar]
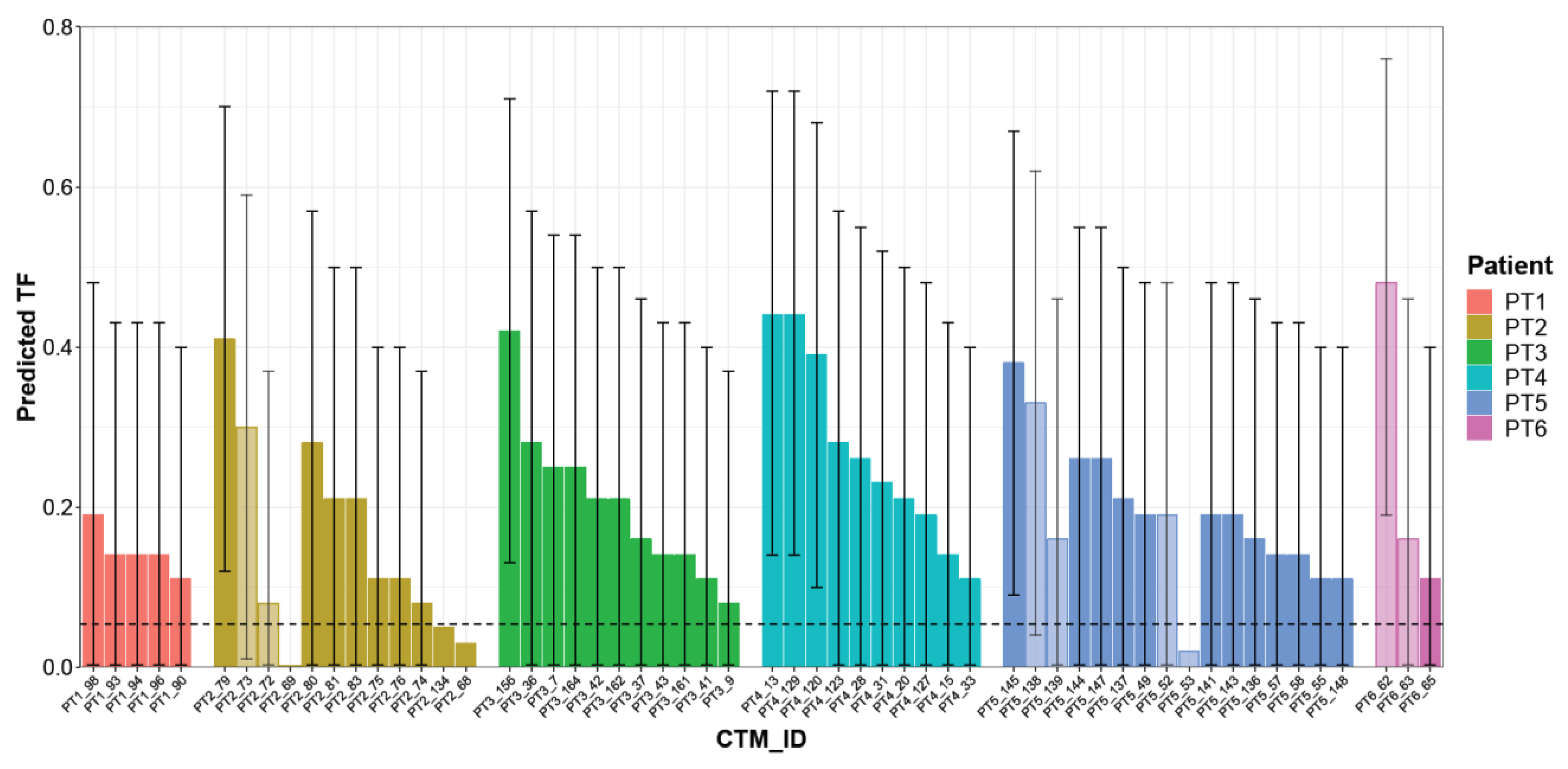
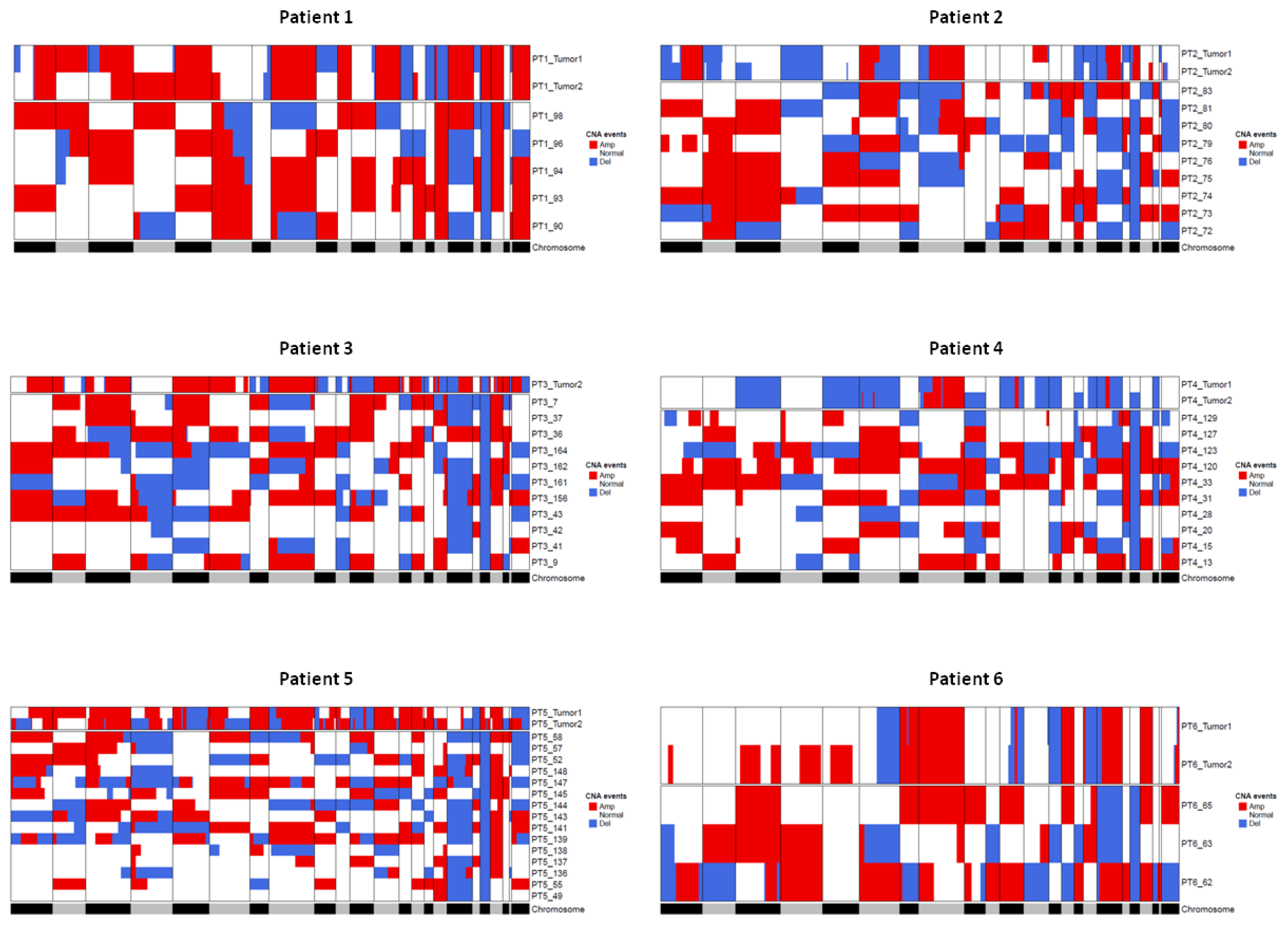
| PT1 | PT2 | PT3 | PT4 | PT5 | PT6 | |
|---|---|---|---|---|---|---|
| pT | 1.5 | 2.2 | 2.5 | 1.2 | / | 1.7 |
| Histotype | IDC | IDC | IDC | IDC muc | IC | IDC |
| Histological grade | G2 | G3 | G3 | G2 | G3 | G2 |
| pN | N+ | N+ | N+ | N0 | / | N+ |
| ER | Pos | Neg | Pos | Pos | / | Pos |
| PgR | Pos | Pos | Pos | Pos | / | Pos |
| HER2 | 2+ | 3+ | 1+ | 0 | / | 1+ |
| Ki67 | 35% | 70% | 75% | 30% | / | 22% |
| Patient | Stromal | Tumor 1 * | Tumor 2 * |
|---|---|---|---|
| PT1 | 0.01 | 0.25 | 0.53 |
| PT 2 | 0.004 | 0.24 | 0.72 |
| PT 3 | 0.01 | NA | 0.35 |
| PT 4 | 0.006 | 0.66 | 0.64 |
| PT 5 | 0.009 | 0.31 | 0.53 |
| PT 6 | 0.01 | 0.85 | 0.67 |
| Patient | CNA Private (Tumor) | CNA Private (CTM) | Sum of CNA Shared |
| PT1 | 0.08 | 0.52 | 0.44 |
| PT2 | 0.04 | 0.62 | 0.39 |
| PT3 | 0.05 | 0.3 | 0.71 |
| PT4 | 0.12 | 0.63 | 0.37 |
| PT5 | 0.01 | 0.44 | 0.56 |
| PT6 | 0.04 | 0.62 | 0.31 |
| Legend | |||
| CNA Private (Tumor) | Number of CNA exclusive of the tumors/total of genome | ||
| CNA Private (CTM) | Number of CNA exclusive of the CTMs/total of genome | ||
| Sum of CNA shared | Sum of CNA shared between both tumors and at least 1 CTMs/total of genome | ||
| TGF | + | CNA Profile | = | Final Output |
|---|---|---|---|---|
| 0 ≤ TGF ≤ 0.05 | + | Normal/Aberrant/Unclear | = | Normal CTM |
| 0.05 < TGF ≤ 1 | + | Aberrant | = | Aberrant CTM |
| 0.05 < TGF ≤ 1 | + | Unclear | = | Unclear CTM |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silvestri, M.; Reduzzi, C.; Feliciello, G.; Vismara, M.; Schamberger, T.; Köstler, C.; Motta, R.; Calza, S.; Ferraris, C.; Vingiani, A.; et al. Detection of Genomically Aberrant Cells within Circulating Tumor Microemboli (CTMs) Isolated from Early-Stage Breast Cancer Patients. Cancers 2021, 13, 1409. https://doi.org/10.3390/cancers13061409
Silvestri M, Reduzzi C, Feliciello G, Vismara M, Schamberger T, Köstler C, Motta R, Calza S, Ferraris C, Vingiani A, et al. Detection of Genomically Aberrant Cells within Circulating Tumor Microemboli (CTMs) Isolated from Early-Stage Breast Cancer Patients. Cancers. 2021; 13(6):1409. https://doi.org/10.3390/cancers13061409
Chicago/Turabian StyleSilvestri, Marco, Carolina Reduzzi, Giancarlo Feliciello, Marta Vismara, Thomas Schamberger, Cäcilia Köstler, Rosita Motta, Stefano Calza, Cristina Ferraris, Andrea Vingiani, and et al. 2021. "Detection of Genomically Aberrant Cells within Circulating Tumor Microemboli (CTMs) Isolated from Early-Stage Breast Cancer Patients" Cancers 13, no. 6: 1409. https://doi.org/10.3390/cancers13061409
APA StyleSilvestri, M., Reduzzi, C., Feliciello, G., Vismara, M., Schamberger, T., Köstler, C., Motta, R., Calza, S., Ferraris, C., Vingiani, A., Pruneri, G., Daidone, M. G., Klein, C. A., Polzer, B., & Cappelletti, V. (2021). Detection of Genomically Aberrant Cells within Circulating Tumor Microemboli (CTMs) Isolated from Early-Stage Breast Cancer Patients. Cancers, 13(6), 1409. https://doi.org/10.3390/cancers13061409







