CD19 Chimeric Antigen Receptor-Exosome Targets CD19 Positive B-lineage Acute Lymphocytic Leukemia and Induces Cytotoxicity
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
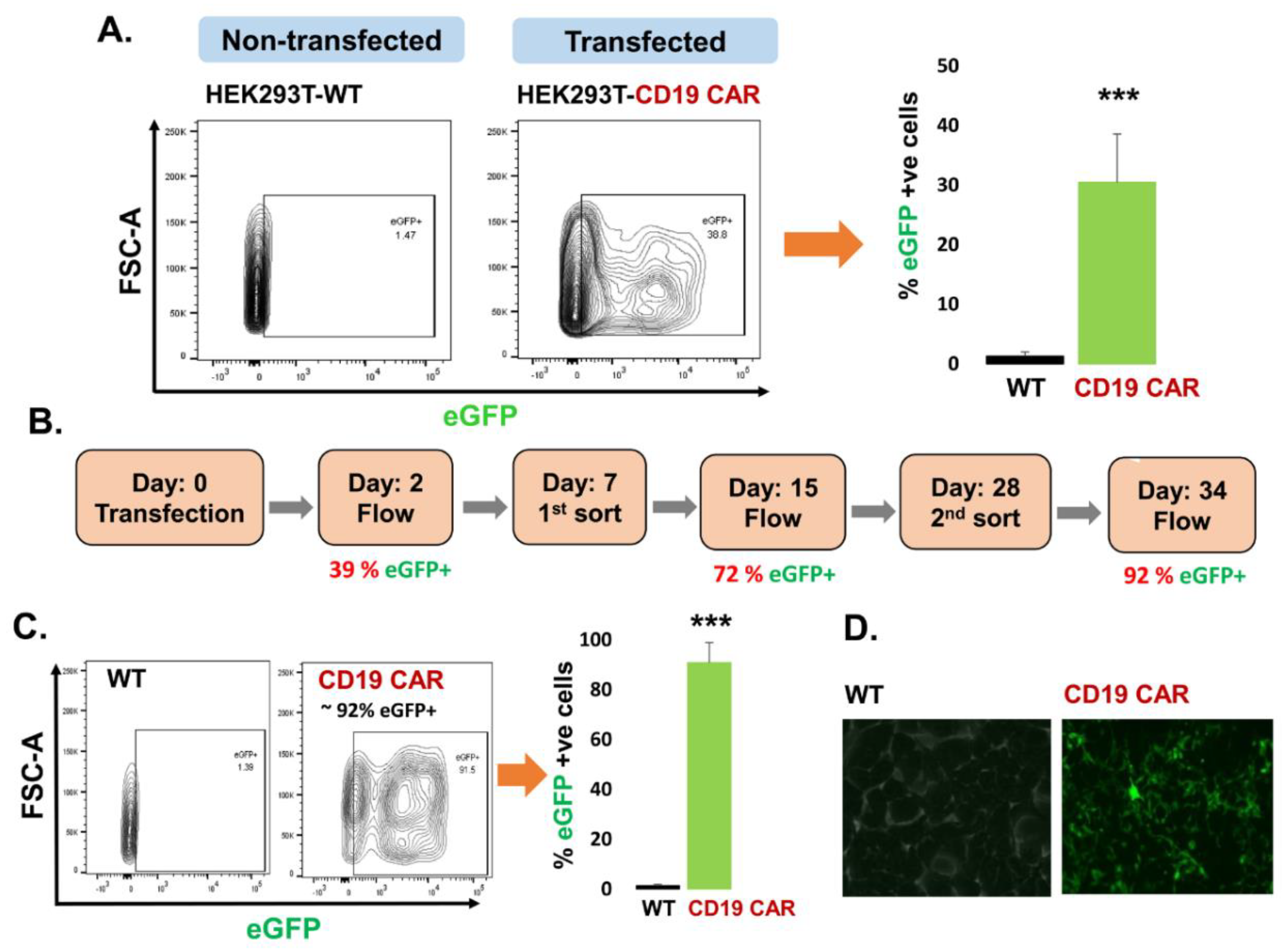
2.1. Transfection of CD19 CAR Plasmids into HEK293T Producer Cells
2.1.1. Characterization of CD19 CAR Plasmid
2.1.2. Transfection of Producer Cell Line
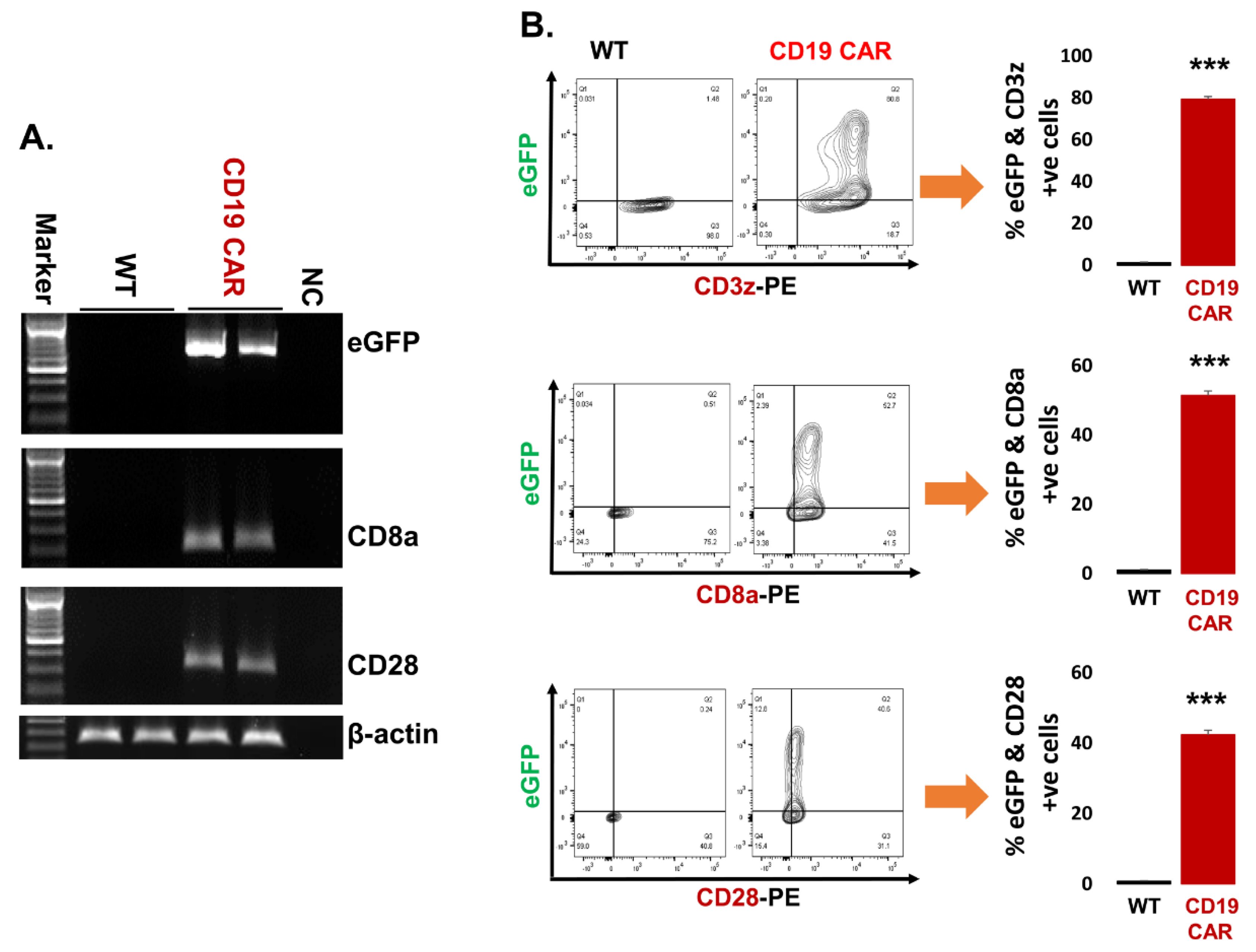
2.2. Confirmation of CD19 CAR Plasmid into the Transfected Producer Cell Line
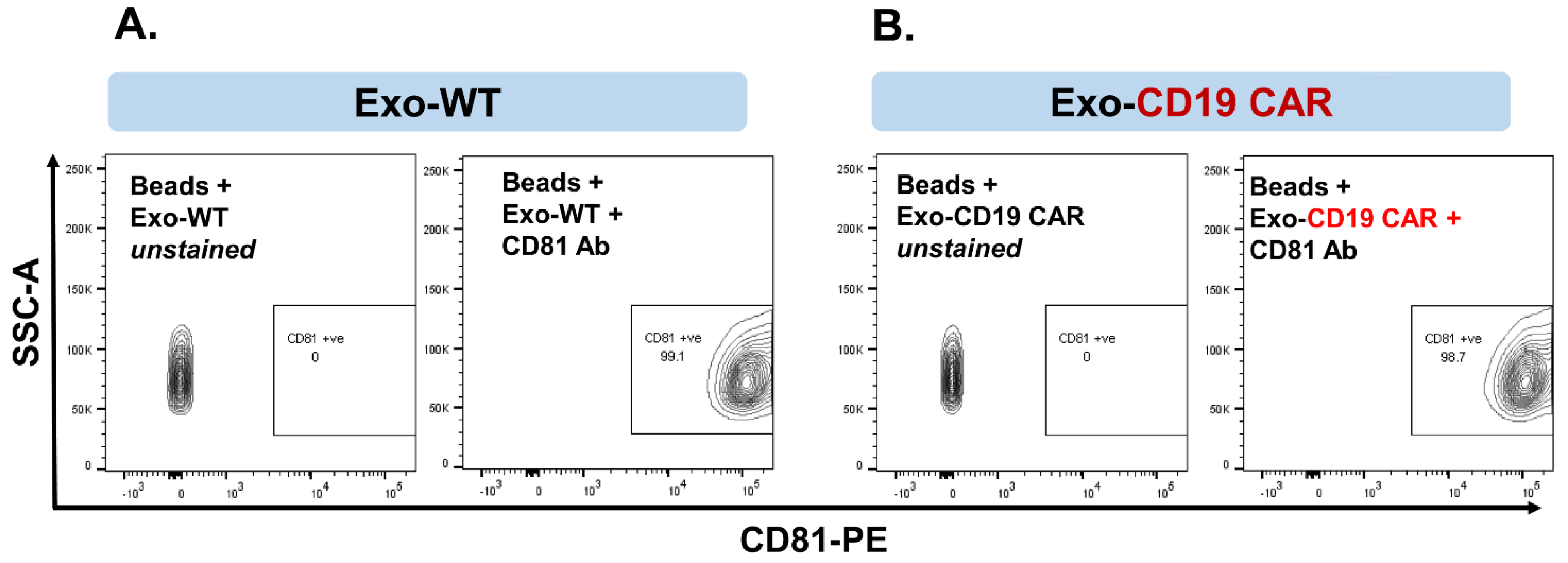
2.3. Characterization of Purified Exosomes
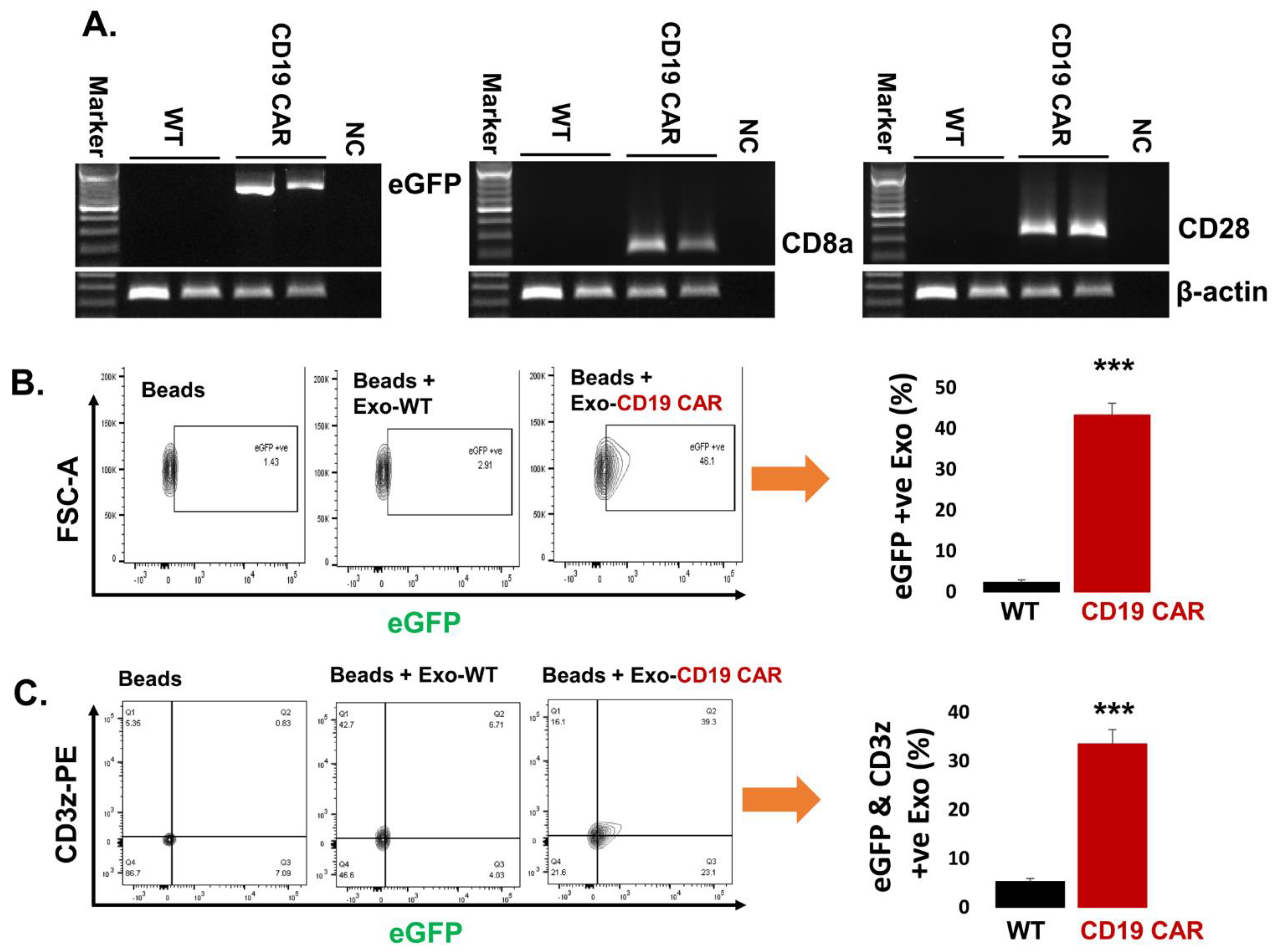
2.4. Confirmation of CD19 CAR Molecules Expression in Harvested Exosomes
2.5. Delivery of Exo-CD19 CAR Molecules into the Target Cells
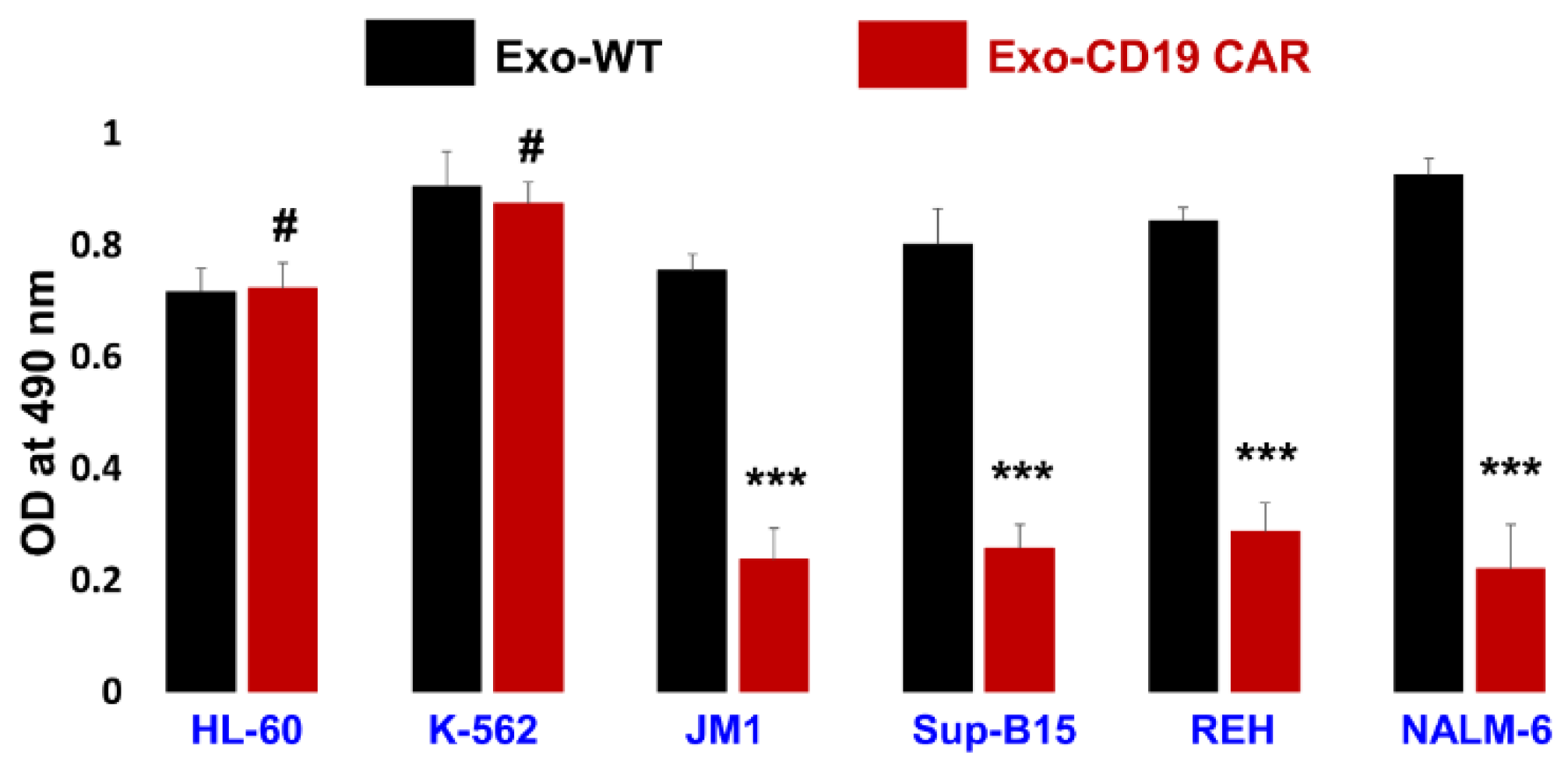
2.6. Exo-CD19 CAR Effect on CD19-Negative and CD19-Positive Cells
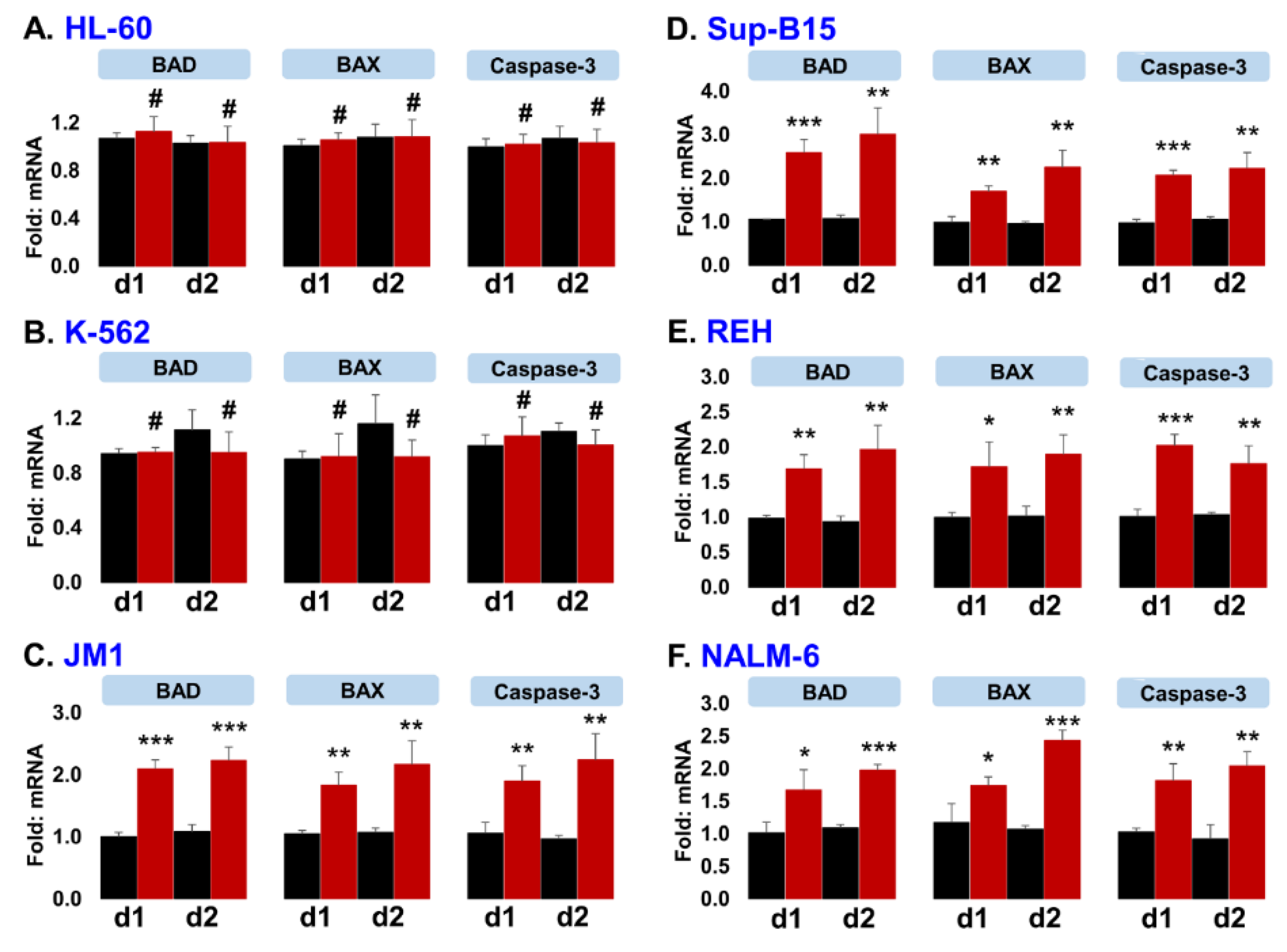
2.7. Exo-CD19 CAR Exposure Induces Pro-Apoptotic Genes in CD19-Positive B-Cell Leukemia
3. Discussion
4. Materials and Methods
4.1. Cell Lines
4.2. CD19 CAR Plasmid Isolation
4.2.1. Bacterial (Escherichia coli) Culture and Plasmid Isolation
4.2.2. CD19 CAR Plasmid Verification by Semi-Quantitative PCR
4.3. Production of CAR-Exosomes in HEK293T Parent Cells
4.3.1. Plasmid Transfection, Enrichment of Transfected Cells by FACS Cell Sorting, and Fluorescence Microscopy
4.3.2. Cell Staining to Analyze Transfection Efficiency by Flow Cytometry
4.4. Exosomes (Exo-WT and Exo-CD19 CAR) Production and Isolation
4.4.1. Exosomes Production
4.4.2. Preparation of Exo-Free FBS
4.4.3. Exosomes Isolation
4.4.4. Exosomal Markers CD63 and CD81 Expression Analysis by Flow Cytometry
4.4.5. Expression of CD19 CAR Markers Proteins on Exosomes by Flow Cytometry
4.4.6. Exosomal RNA Extraction
4.5. Exo-CD19 CAR Induced Cytotoxicity (Contact-Dependent) in CD19-Positive B-Leukemia Cells
4.5.1. Gene Expression by qPCR
4.5.2. Cytotoxicity Measurement by MTS Assay
4.6. Statistical Analysis
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Otero, D.C.; Rickert, R.C. CD19 Function in Early and Late B Cell Development. II. CD19 Facilitates the Pro-B/Pre-B Transition. J. Immunol. 2003, 171, 5921–5930. [Google Scholar] [CrossRef] [PubMed]
- Diamant, E.; Keren, Z.; Melamed, D. CD19 regulates positive selection and maturation in B lymphopoiesis: Lack of CD19 imposes developmental arrest of immature B cells and consequential stimulation of receptor editing. Blood 2005, 105, 3247–3254. [Google Scholar] [CrossRef] [PubMed]
- Scheuermann, R.H.; Racila, E. CD19 Antigen in Leukemia and Lymphoma Diagnosis and Immunotherapy. Leuk. Lymphoma 1995, 18, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Mills, D.M.; Stolpa, J.C.; Cambier, J.C. Modulation of MHC Class II Signal Transduction by CD19. Tissue Eng. 2007, 596, 139–148. [Google Scholar] [CrossRef]
- Pui, C.-H. Recent Research Advances in Childhood Acute Lymphoblastic Leukemia. J. Formos. Med. Assoc. 2010, 109, 777–787. [Google Scholar] [CrossRef]
- Grupp, S.A. Advances in T-cell therapy for ALL. Best Pract. Res. Clin. Haematol. 2014, 27, 222–228. [Google Scholar] [CrossRef]
- Maude, S.L.; Frey, N.; Shaw, P.A.; Aplenc, R.; Barrett, D.M.; Bunin, N.J.; Chew, A.; Gonzalez, V.E.; Zheng, Z.; Lacey, S.F.; et al. Chimeric antigen receptor T cells for sustained remissions in leukemia. N. Engl. J. Med. 2014, 371, 1507–1517. [Google Scholar] [CrossRef]
- Teachey, D.T.; Bishop, M.R.; Maloney, D.G.; Grupp, S.A. Toxicity management after chimeric antigen receptor T cell therapy: One size does not fit ’ALL’. Nat. Rev. Clin. Oncol. 2018, 15, 218. [Google Scholar] [CrossRef]
- Boyiadzis, M.M.; Dhodapkar, M.V.; Brentjens, R.J.; Kochenderfer, J.N.; Neelapu, S.S.; Maus, M.V.; Porter, D.L.; Maloney, D.G.; Grupp, S.A.; Mackall, C.L.; et al. Chimeric antigen receptor (CAR) T therapies for the treatment of hematologic malignancies: Clinical perspective and significance. J. Immunother. Cancer 2018, 6, 137. [Google Scholar] [CrossRef]
- Singh, N.; Frey, N.V.; Grupp, S.A.; Maude, S.L. CAR T Cell Therapy in Acute Lymphoblastic Leukemia and Potential for Chronic Lymphocytic Leukemia. Curr. Treat. Options Oncol. 2016, 17, 1–11. [Google Scholar] [CrossRef]
- Neelapu, S.S.; Tummala, S.; Kebriaei, P.; Wierda, W.; Gutierrez, C.; Locke, F.L.; Komanduri, K.V.; Lin, Y.; Jain, N.; Daver, N.; et al. Chimeric antigen receptor T-cell therapy—Assessment and management of toxicities. Nat. Rev. Clin. Oncol. 2018, 15, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Sommermeyer, D.; Hill, T.; Shamah, S.M.; Salter, A.; Chen, Y.; Mohler, K.M.; Riddell, S.R. Fully human CD19-specific chimeric antigen receptors for T-cell therapy. Leukemia 2017, 31, 2191–2199. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Li, D.; Ma, L.; He, T.; Qi, F.; Shen, J.; Lu, X.-A. The novel anti-CD19 chimeric antigen receptors with humanized scFv (single-chain variable fragment) trigger leukemia cell killing. Cell. Immunol. 2016, 304–305, 49–54. [Google Scholar] [CrossRef]
- Maude, S.L.; Teachey, D.T.; Porter, D.L.; Grupp, S.A. CD19-targeted chimeric antigen receptor T-cell therapy for acute lymphoblastic leukemia. Blood 2015, 125, 4017–4023. [Google Scholar] [CrossRef]
- Jena, B.; Maiti, S.; Huls, H.; Singh, H.; Lee, D.A.; Champlin, R.E.; Cooper, L.J.N. Chimeric Antigen Receptor (CAR)-Specific Monoclonal Antibody to Detect CD19-Specific T Cells in Clinical Trials. PLoS ONE 2013, 8, e57838. [Google Scholar] [CrossRef]
- Lee, D.W.; Gardner, R.; Porter, D.L.; Louis, C.U.; Ahmed, N.; Jensen, M.C.; Grupp, S.A.; Mackall, C.L. Current concepts in the diagnosis and management of cytokine release syndrome. Blood 2014, 124, 188–195. [Google Scholar] [CrossRef] [PubMed]
- Brudno, J.N.; Kochenderfer, J.N. Toxicities of chimeric antigen receptor T cells: Recognition and management. Blood 2016, 127, 3321–3330. [Google Scholar] [CrossRef]
- Nagamune, T. Biomolecular engineering for nanobio/bionanotechnology. Nano Converg. 2017, 4, 1–56. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef]
- Théry, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Vaiselbuh, S. Exosomes in Cancer Research. Cancer Res. Front. 2015, 1, 11–24. [Google Scholar] [CrossRef]
- Haque, S.; Vaiselbuh, S.R. Silencing of Exosomal miR-181a Reverses Pediatric Acute Lymphocytic Leukemia Cell Proliferation. Pharmaceuticals 2020, 13, 241. [Google Scholar] [CrossRef] [PubMed]
- Peters, P.J.; Geuze, H.J.; Der Van Donk, H.A.; Slot, J.W.; Griffith, J.M.; Stam, N.J.; Clevers, H.C.; Borst, J. Molecules relevant for T cell-target cell interaction are present in cytolytic granules of human T lymphocytes. Eur. J. Immunol. 1989, 19, 1469–1475. [Google Scholar] [CrossRef]
- Wu, C.-H.; Li, J.; Li, L.; Sun, J.; Fabbri, M.; Wayne, A.S.; Seeger, R.C.; Jong, A.Y. Extracellular vesicles derived from natural killer cells use multiple cytotoxic proteins and killing mechanisms to target cancer cells. J. Extracell. Vesicles 2019, 8, 1588538. [Google Scholar] [CrossRef] [PubMed]
- Peters, P.J.; Borst, J.; Oorschot, V.; Fukuda, M.; Krähenbühl, O.; Tschopp, J.; Slot, J.W.; Geuze, H.J. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J. Exp. Med. 1991, 173, 1099–1109. [Google Scholar] [CrossRef]
- Fu, W.; Lei, C.; Liu, S.; Cui, Y.; Wang, C.; Qian, K.; Li, T.; Shen, Y.; Fan, X.; Lin, F.; et al. CAR exosomes derived from effector CAR-T cells have potent antitumour effects and low toxicity. Nat. Commun. 2019, 10, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Haque, S.; Vaiselbuh, S.R. Exosomes molecular diagnostics: Direct conversion of exosomes into the cDNA for gene amplification by two-step polymerase chain reaction. J. Biol. Methods 2018, 5, e96. [Google Scholar] [CrossRef]
- Grupp, S.A.; Kalos, M.; Barrett, D.; Aplenc, R.; Porter, D.L.; Rheingold, S.R.; Teachey, D.T.; Chew, A.; Hauck, B.; Wright, J.F.; et al. Chimeric Antigen Receptor–Modified T Cells for Acute Lymphoid Leukemia. N. Engl. J. Med. 2013, 368, 1509–1518. [Google Scholar] [CrossRef]
- Grupp, S.A.; June, C.H. Adoptive Cellular Therapy. Curr. Top. Microbiol. Immunol. 2010, 344, 149–172. [Google Scholar] [CrossRef]
- Li, H.; Huang, Y.; Jiang, D.-Q.; Cui, L.-Z.; He, Z.; Wang, C.; Zhang, Z.-W.; Zhu, H.-L.; Ding, Y.-M.; Li, L.-F.; et al. Antitumor activity of EGFR-specific CAR T cells against non-small-cell lung cancer cells in vitro and in mice. Cell Death Dis. 2018, 9, 1–11. [Google Scholar] [CrossRef]
- Velasquez, M.P.; Torres, D.; Iwahori, K.; Kakarla, S.; Arber, C.; Rodriguez-Cruz, T.; Szoor, A.; Bonifant, C.L.; Gerken, C.; Cooper, L.J.N.; et al. T cells expressing CD19-specific Engager Molecules for the Immunotherapy of CD19-positive Malignancies. Sci. Rep. 2016, 6, 27130. [Google Scholar] [CrossRef]
- Morita, D.; Nishio, N.; Saito, S.; Tanaka, M.; Kawashima, N.; Okuno, Y.; Suzuki, S.; Matsuda, K.; Maeda, Y.; Wilson, M.H.; et al. Enhanced Expression of Anti-CD19 Chimeric Antigen Receptor in piggyBac Transposon-Engineered T Cells. Mol. Ther. Methods Clin. Dev. 2018, 8, 131–140. [Google Scholar] [CrossRef]
- Tsukahara, T.; Ohmine, K.; Yamamoto, C.; Uchibori, R.; Ido, H.; Teruya, T.; Urabe, M.; Mizukami, H.; Kume, A.; Nakamura, M.; et al. CD19 target-engineered T-cells accumulate at tumor lesions in human B-cell lymphoma xenograft mouse models. Biochem. Biophys. Res. Commun. 2013, 438, 84–89. [Google Scholar] [CrossRef] [PubMed]
- An, N.; Tao, Z.; Li, S.; Xing, H.; Tang, K.; Tian, Z.; Rao, Q.; Wang, M.; Wang, J. Construction of a new anti-CD19 chimeric antigen receptor and the anti-leukemia function study of the transduced T cells. Oncotarget 2016, 7, 10638–10649. [Google Scholar] [CrossRef]
- Nakajima, M.; Sakoda, Y.; Adachi, K.; Nagano, H.; Tamada, K. Improved survival of chimeric antigen receptor-engineered T (CAR -T) and tumor-specific T cells caused by anti-programmed cell death protein 1 single-chain variable fragment-producing CAR-T cells. Cancer Sci. 2019, 110, 3079–3088. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Fan, M.H.; Miao, C.H.; Liao, Y.J.; Yuan, R.-H.; Liu, C.L. Engineering Chimeric Antigen Receptor T Cells against Immune Checkpoint Inhibitors PD-1/PD-L1 for Treating Pancreatic Cancer. Mol. Ther. Oncolytics 2020, 17, 571–585. [Google Scholar] [CrossRef]
- Fitzgerald, J.C.; Weiss, S.L.; Maude, S.L.; Barrett, D.M.; Lacey, S.F.; Melenhorst, J.J.; Shaw, P.; Berg, R.A.; June, C.H.; Porter, D.L.; et al. Cytokine Release Syndrome After Chimeric Antigen Receptor T Cell Therapy for Acute Lymphoblastic Leukemia. Crit. Care Med. 2017, 45, e124–e131. [Google Scholar] [CrossRef] [PubMed]
- Ha, D.; Yang, N.; Nadithe, V. Exosomes as therapeutic drug carriers and delivery vehicles across biological membranes: Current perspectives and future challenges. Acta Pharm. Sin. B 2016, 6, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Viaud, S.; Terme, M.; Flament, C.; Taieb, J.; André, F.; Novault, S.; Escudier, B.; Robert, C.; Caillat-Zucman, S.; Tursz, T.; et al. Dendritic Cell-Derived Exosomes Promote Natural Killer Cell Activation and Proliferation: A Role for NKG2D Ligands and IL-15Rα. PLoS ONE 2009, 4, e4942. [Google Scholar] [CrossRef] [PubMed]
- Ruella, M.; Xu, J.; Barrett, D.M.; Fraietta, J.A.; Reich, T.J.; Ambrose, D.E.; Klichinsky, M.; Shestova, O.; Patel, P.R.; Kulikovskaya, I.; et al. Induction of resistance to chimeric antigen receptor T cell therapy by transduction of a single leukemic B cell. Nat. Med. 2018, 24, 1499–1503. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J.A. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Sverdlov, E.D. Amedeo Avogadro’s cry: What is 1 µg of exosomes? BioEssays 2012, 34, 873–875. [Google Scholar] [CrossRef] [PubMed]
- Morrissey, M.; Williamson, A.P.; Steinbach, A.M.; Roberts, E.W.; Kern, N.; Headley, M.B.; Vale, R.D. Chimeric antigen receptors that trigger phagocytosis. eLife 2018, 7, e36688. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Haque, S.; Nieto, J.; Trott, J.; Inman, J.K.; McCormick, S.; Chiorazzi, N.; Mongini, P.K.A. A p53 Axis Regulates B Cell Receptor-Triggered, Innate Immune System-Driven B Cell Clonal Expansion. J. Immunol. 2012, 188, 6093–6108. [Google Scholar] [CrossRef]
- Haque, S.; Patil, G.; Mishra, A.; Lan, X.; Popik, W.; Malhotra, A.; Skorecki, K.; Singhal, P.C. Effect of APOL1 disease risk variants on APOL1 gene product. Biosci. Rep. 2017, 37. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Curr. Protoc. Cell Biol. 2006, 30, 3.22.1–3.22.29. [Google Scholar] [CrossRef]

| SN | Gene Name | Gene ID | Primers |
|---|---|---|---|
| 1 | eGFP | JQ064507.1 | For 5′-CTGGTCGAGCTGGACGGCGACG-3′ |
| Rev 5′-CACGAACTCCAGCAGGACCATG-3′ | |||
| 2 | CD8a | NM_001768.6 | For 5′-CACGACGCCAGCGCCGCGACCACC-3′ |
| Rev 5′-GGGTGATAACCAGTGACAGGAGAA-3′ | |||
| 3 | CD28 | AJ937363.1 | For 5′-GGAGGGGGGACCAAGCTGGAGA-3′ |
| Rev 5′-TGCAGACTGTTCATTTTTAAG-3′ | |||
| 4 | β-actin | NM_001101.4 | For 5′-GTCCTCTCCCAAGTCCACACA-3′ |
| Rev 5′-CTGGTCTCAAGTCAGTGTACAGGTAA-3′ |
| SN | Gene Name | Gene ID | Primers | UPL # |
|---|---|---|---|---|
| 1 | BAD | AF031523.1 | For: 5′-CGAGTTTGTGGACTCCTTTAAGA-3′ | 78 |
| Rev: 5′-CACCAGGACTGGAAGACTCG-3′ | ||||
| 2 | BAX | U19599.1 | For: 5′-CAAGACCAGGGTGGTTGG-3′ | 55 |
| Rev: 5′-CACTCCCGCCACAAAGAT-3′ | ||||
| 2 | Caspase-3 | DD346274.1 | For: 5′-AATGGACCAGGACGATGAAG-3′ | 35 |
| Rev: 5′-CATCTCATCACCCACTGCTC-3′ | ||||
| 4 | GAPDH | NM_002046.3 | For: 5′-AGCCACATCGCTCAGACAC-3′ | 60 |
| Rev: 5′-GCCCAATACGACCAAATCC-3′ | ||||
| 5 | IFN-g | X13274.1 | For: 5′-GGCATTTTGAAGAATTGGAAAG-3′ | 21 |
| Rev: 5′-TTTGGATGCTCTGGTCATCTT-3′ | ||||
| 6 | TNF-a | X02910.1 | For: 5′-GTCCAGGCTTGTCCTGCTAC-3′ | 7 |
| Rev: 5′-AGTCCTGAGGCCTGTGTTTG-3′ | ||||
| 7 | IL-2 | S77834.1 | For: 5′-AAGTTTTACATGCCCAAGAAGG-3′ | 65 |
| Rev:5′-AAGTGAAAGTTTTTGCTTTGAGCTA-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haque, S.; Vaiselbuh, S.R. CD19 Chimeric Antigen Receptor-Exosome Targets CD19 Positive B-lineage Acute Lymphocytic Leukemia and Induces Cytotoxicity. Cancers 2021, 13, 1401. https://doi.org/10.3390/cancers13061401
Haque S, Vaiselbuh SR. CD19 Chimeric Antigen Receptor-Exosome Targets CD19 Positive B-lineage Acute Lymphocytic Leukemia and Induces Cytotoxicity. Cancers. 2021; 13(6):1401. https://doi.org/10.3390/cancers13061401
Chicago/Turabian StyleHaque, Shabirul, and Sarah R. Vaiselbuh. 2021. "CD19 Chimeric Antigen Receptor-Exosome Targets CD19 Positive B-lineage Acute Lymphocytic Leukemia and Induces Cytotoxicity" Cancers 13, no. 6: 1401. https://doi.org/10.3390/cancers13061401
APA StyleHaque, S., & Vaiselbuh, S. R. (2021). CD19 Chimeric Antigen Receptor-Exosome Targets CD19 Positive B-lineage Acute Lymphocytic Leukemia and Induces Cytotoxicity. Cancers, 13(6), 1401. https://doi.org/10.3390/cancers13061401






