Autocrine TGFβ1 Opposes Exogenous TGFβ1-Induced Cell Migration and Growth Arrest through Sustainment of a Feed-Forward Loop Involving MEK-ERK Signaling
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
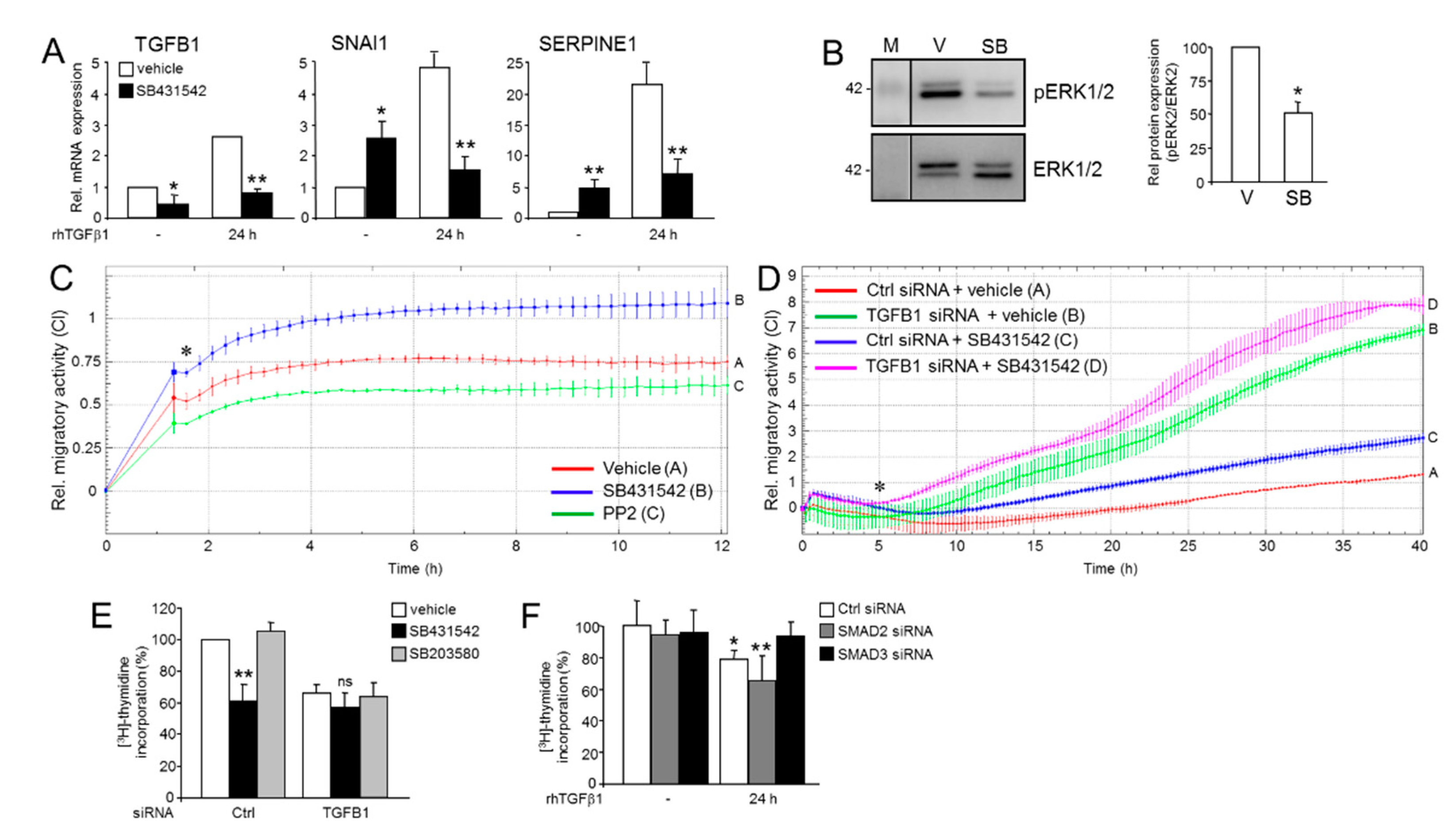
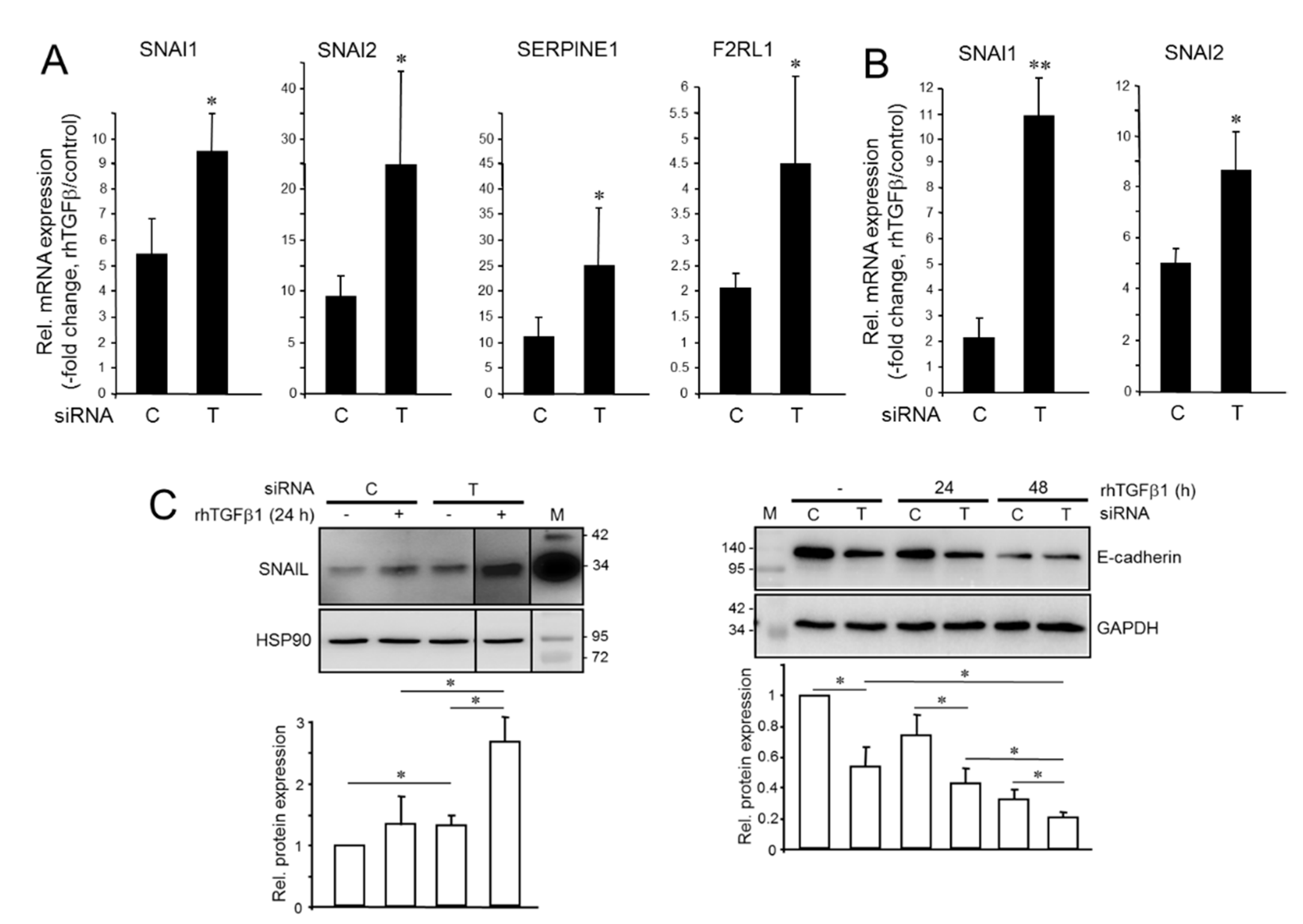
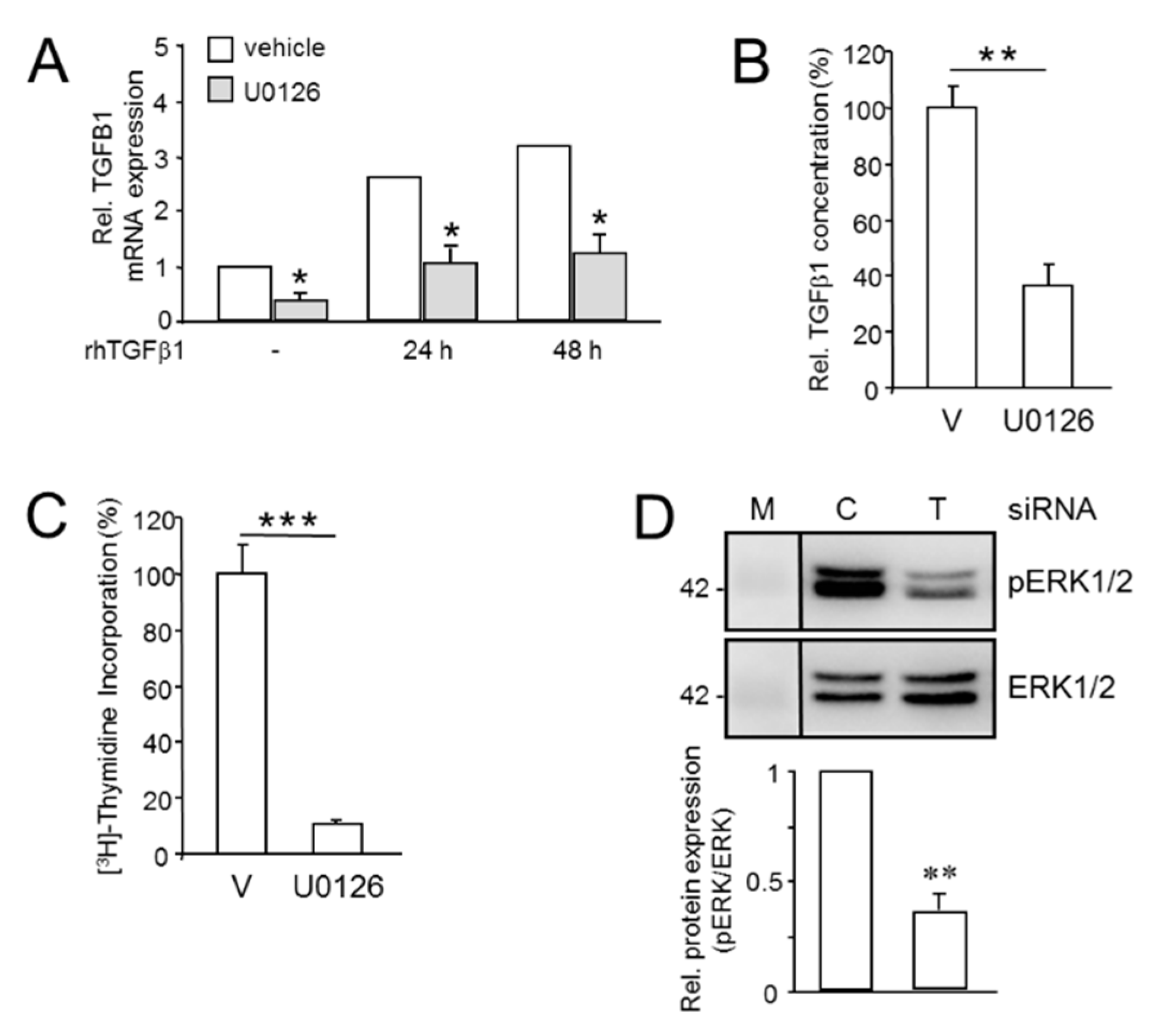
2.1. Divergent Effects of Endogenous and Exogenous TGFβ1 on TGFβ Target Genes
2.2. Knockdown of TGFB1 Enhances the Migratory Response to rhTGFβ1
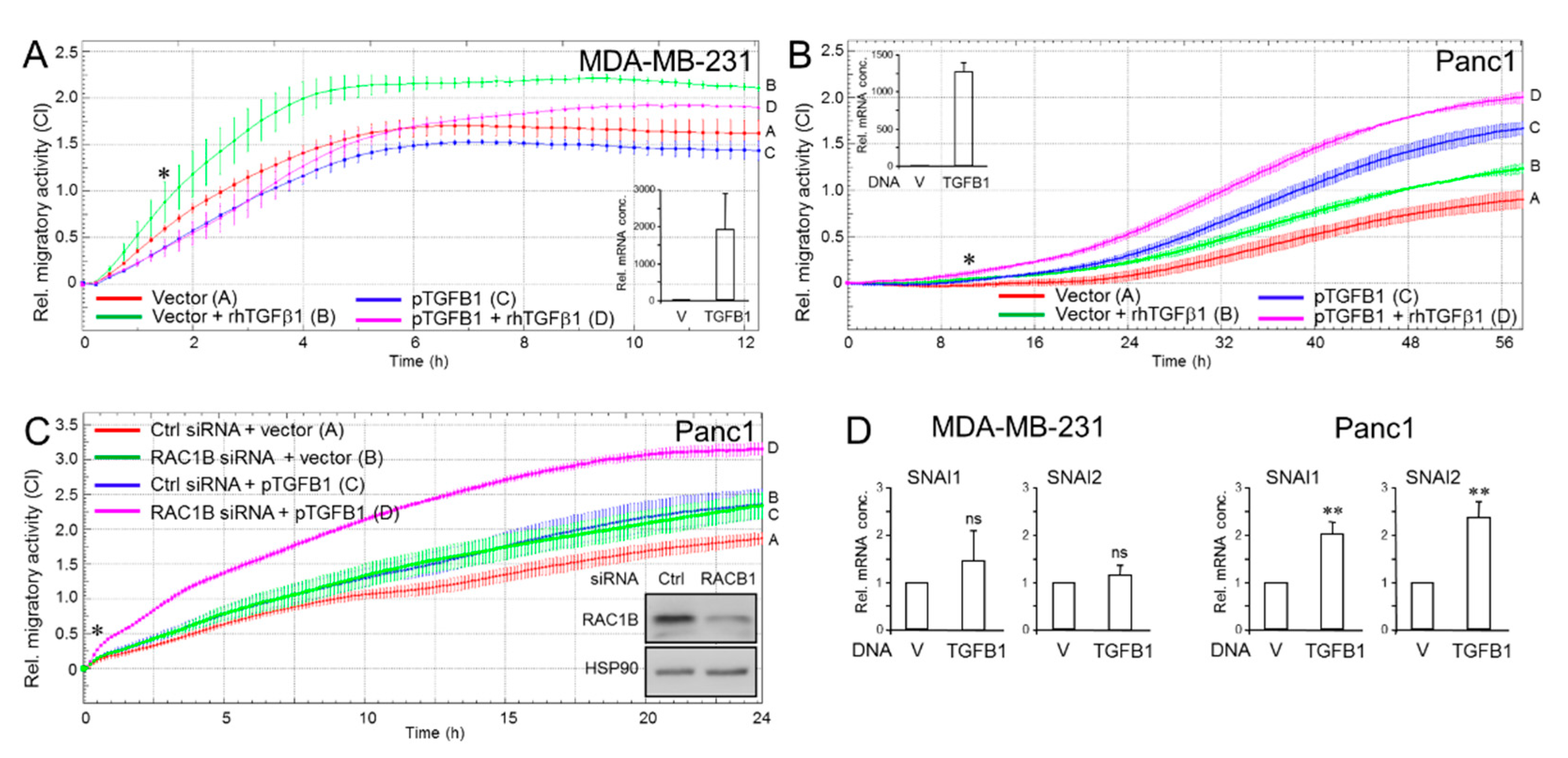
2.3. Ectopic TGFB1 Antagonizes Basal and rhTGFβ1-Induced Migration in MDA-MB-231 Cells but Synergizes with rhTGFβ1-Induced Migration in Panc1 Cells
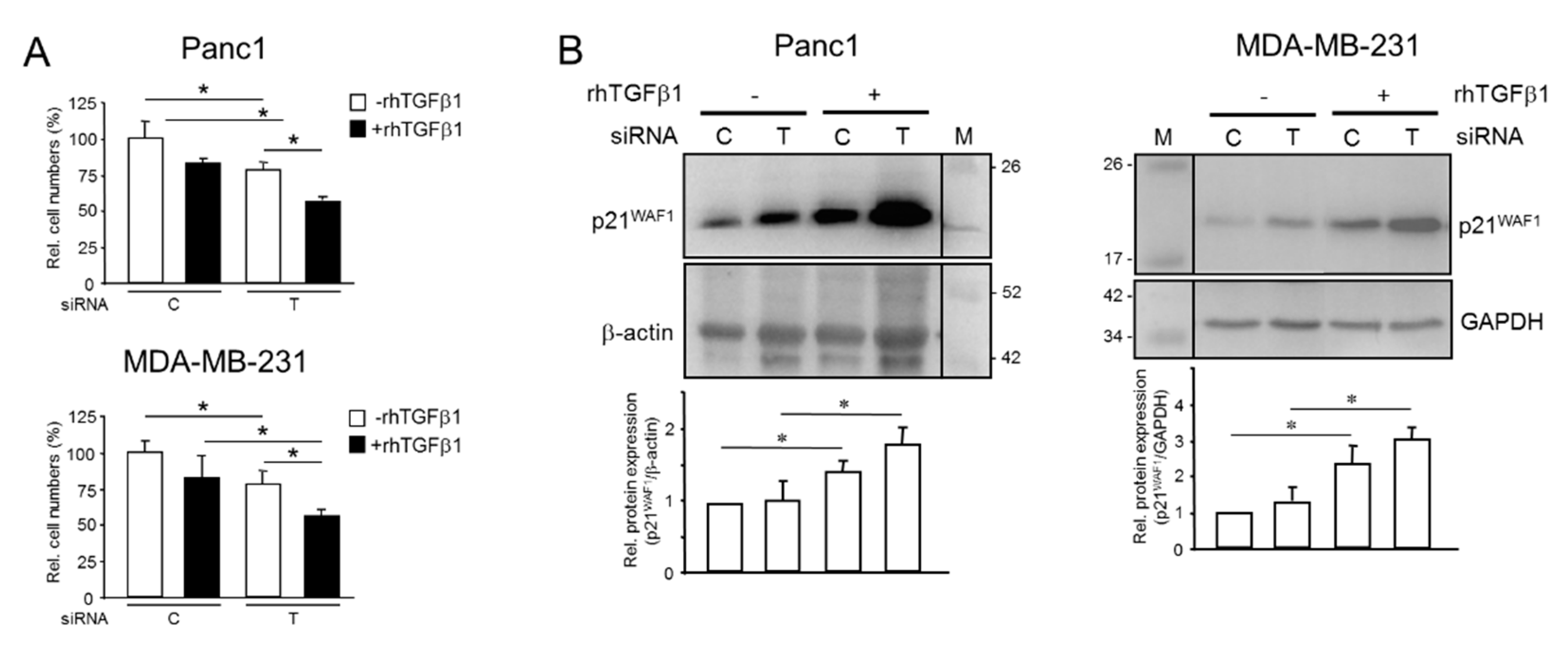
2.4. Endogenous TGFB1 Opposes rhTGFβ1-Induced Growth Arrest and Induction of p21WAF1 Expression
2.5. Mutual Induction of aTGFβ1 or ERK Activation Sustains Proliferation
2.6. Effect of Inhibition of the ALK5 Kinase on TGFB1 Expression, Cell Migration, and ERK2 Activation
3. Discussion
4. Materials and Methods
4.1. Cell Lines and Treatments
4.2. Transient Transfections
4.3. Quantitative Real-Time PCR Analysis
4.4. Western Blotting
4.5. TGFβ1 ELISA
4.6. (3H)-Thymidine Incorporation Assay
4.7. Real-Time Cell Migration Assays
4.8. Proliferation Assays
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hezel, A.F.; Kimmelman, A.C.; Stanger, B.Z.; Bardeesy, N.; Depinho, R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006, 20, 1218–1249. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.M.; Ansari, J.A.K.; El-Aziz, N.M.A.; Abozeed, W.N.; Warith, A.M.A.; Alsaleh, K.; Nabholtz, J.M. Triple Negative Breast Cancer: A Tale of Two Decades. Anticancer Agents Med. Chem. 2017, 17, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Brancaccio, M.; Natale, F.; Falco, G.; Angrisano, T. Cell-Free DNA Methylation: The New Frontiers of Pancreatic Cancer Biomarkers’ Discovery. Genes 2019, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.; Rosso, K.; Nathanson, S.D. Pathogenesis, prevention, diagnosis and treatment of breast cancer. World J. Clin. Oncol. 2014, 5, 283–298. [Google Scholar] [CrossRef]
- Torres, C.; Grippo, P.J. Pancreatic cancer subtypes: A roadmap for precision medicine. Ann. Med. 2018, 50, 277–287. [Google Scholar] [CrossRef]
- Shao, F.; Sun, H.; Deng, C.X. Potential therapeutic targets of triple-negative breast cancer based on its intrinsic subtype. Oncotarget 2017, 8, 73329–73344. [Google Scholar] [CrossRef]
- Dardare, J.; Witz, A.; Merlin, J.L.; Gilson, P.; Harlé, A. SMAD4 and the TGFβ Pathway in Patients with Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2020, 21, 3534. [Google Scholar] [CrossRef]
- Derynck, R.; Turley, S.J.; Akhurst, R.J. TGFβ biology in cancer progression and immunotherapy. Nat. Rev. Clin. Oncol. 2021, 18, 9–34. [Google Scholar] [CrossRef]
- Xu, X.; Zhang, L.; He, X.; Zhang, P.; Sun, C.; Xu, X.; Lu, Y.; Li, F. TGF-β plays a vital role in triple-negative breast cancer (TNBC) drug-resistance through regulating stemness, EMT and apoptosis. Biochem. Biophys. Res. Commun. 2018, 502, 160–165. [Google Scholar] [CrossRef]
- Neuzillet, C.; Hammel, P.; Tijeras-Raballand, A.; Couvelard, A.; Raymond, E. Targeting the Ras-ERK pathway in pancreatic adenocarcinoma. Cancer Metastasis Rev. 2013, 32, 147–162. [Google Scholar] [CrossRef]
- Gupta, G.K.; Collier, A.L.; Lee, D.; Hoefer, R.A.; Zheleva, V.; Siewertsz van Reesema, L.L.; Tang-Tan, A.M.; Guye, M.L.; Chang, D.Z.; Winston, J.S.; et al. Perspectives on Triple-Negative Breast Cancer: Current Treatment Strategies, Unmet Needs, and Potential Targets for Future Therapies. Cancers 2020, 12, 2392. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, K.; Janda, E.; Pierreux, C.E.; Rytömaa, M.; Schulze, A.; McMahon, M.; Hill, C.S.; Beug, H.; Downward, J. Raf induces TGFbeta production while blocking its apoptotic but not invasive responses: A mechanism leading to increased malignancy in epithelial cells. Genes Dev. 2000, 14, 2610–2622. [Google Scholar] [CrossRef] [PubMed]
- Pardoux, C.; Derynck, R. JNK regulates expression and autocrine signaling of TGF-beta1. Mol. Cell 2004, 15, 170–171. [Google Scholar] [CrossRef]
- Van Obberghen-Schilling, E.; Roche, N.S.; Flanders, K.C.; Sporn, M.B.; Roberts, A.B. Transforming growth factor beta 1 positively regulates its own expression in normal and transformed cells. J. Biol. Chem. 1988, 263, 7741–7746. [Google Scholar] [CrossRef]
- Zhang, M.; Fraser, D.; Phillips, A. ERK, p38, and Smad signaling pathways differentially regulate transforming growth factor-beta1 autoinduction in proximal tubular epithelial cells. Am. J. Pathol. 2006, 169, 1282–1293. [Google Scholar] [CrossRef]
- Ungefroren, H.; Otterbein, H.; Wellner, U.F.; Keck, T.; Lehnert, H.; Marquardt, J.U. RAC1B Regulation of TGFB1 Reveals an Unexpected Role of Autocrine TGFβ1 in the Suppression of Cell Motility. Cancers 2020, 12, 3570. [Google Scholar] [CrossRef] [PubMed]
- Witte, D.; Otterbein, H.; Forster, M.; Giehl, K.; Zeiser, R.; Lehnert, H.; Ungefroren, H. Negative regulation of TGF-beta1-induced MKK6-p38 and MEK-ERK signalling and epithelial-mesenchymal transition by Rac1b. Sci. Rep. 2017, 7, 17313. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Sergina, N.; Ko, T.C.; Gong, J.; Brattain, M.G. Autocrine and exogenous transforming growth factor beta control cell cycle inhibition through pathways with different sensitivity. J. Biol. Chem. 2004, 279, 40237–40244. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.P.; Sun, L.-Z.; Willson, J.K.V.; Fields, K.; Humphrey, L.E.; Brattain, M.G. Repression of autocrine TGF-β1 and β2 in quiescent CBS colon carcinoma cells leads to progression of tumorigenic properties. Cell Growth Diff. 1993, 4, 115–123. [Google Scholar] [PubMed]
- Lei, X.; Yang, J.; Nichols, R.W.; Sun, L.Z. Abrogation of TGFbeta signaling induces apoptosis through the modulation of MAP kinase pathways in breast cancer cells. Exp. Cell Res. 2007, 313, 1687–1695. [Google Scholar] [CrossRef]
- Safina, A.; Vandette, E.; Bakin, A.V. ALK5 promotes tumor angiogenesis by upregulating matrix metalloproteinase-9 in tumor cells. Oncogene 2007, 26, 2407–2422. [Google Scholar] [CrossRef]
- Yu, N.; Kozlowski, J.M.; Park, I.I.; Chen, L.; Zhang, Q.; Xu, D.; Doll, J.A.; Crawford, S.E.; Brendler, C.B.; Lee, C. Overexpression of transforming growth factor β1 in malignant prostate cells is partly caused by a runaway of TGF-β1 auto-induction mediated through a defective recruitment of protein phosphatase 2A by TGF-β type I receptor. Urology 2010, 76, e8–e13. [Google Scholar] [CrossRef]
- Liu, Z.; Bandyopadhyay, A.; Nichols, R.W.; Wang, L.; Hinck, A.P.; Wang, S.; Sun, L.Z. Blockade of Autocrine TGF-beta Signaling Inhibits Stem Cell Phenotype, Survival, and Metastasis of Murine Breast Cancer Cells. J. Stem Cell Res. Ther. 2012, 2, 1–8. [Google Scholar] [CrossRef]
- Yang, H.; Kyo, S.; Takatura, M.; Sun, L. Autocrine transforming growth factor beta suppresses telomerase activity and transcription of human telomerase reverse transcriptase in human cancer cells. Cell Growth Diff. 2001, 12, 119–127. [Google Scholar] [PubMed]
- Baker, K.; Raut, P.; Jass, J.R. Microsatellite unstable colorectal cancer cell lines with truncating TGFbetaRII mutations remain sensitive to endogenous TGFbeta. J. Pathol. 2007, 213, 257–265. [Google Scholar] [CrossRef]
- Daroqui, M.C.; Vazquez, P.; Bal de Kier Joffé, E.; Bakin, A.V.; Puricelli, L.I. TGF-β autocrine pathway and MAPK signaling promote cell invasiveness and in vivo mammary adenocarcinoma tumor progression. Oncol. Rep. 2012, 28, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Handler, J.; Cullis, J.; Avanzi, A.; Vucic, E.A.; Bar-Sagi, D. Pre-neoplastic pancreas cells enter a partially mesenchymal state following transient TGF-beta exposure. Oncogene 2018, 37, 4334–4342. [Google Scholar] [CrossRef]
- Ungefroren, H. Autocrine TGF-β in Cancer: Review of the Literature and Caveats in Experimental Analysis. Int. J. Mol. Sci. 2021, 22, 977. [Google Scholar] [CrossRef]
- Voss, M.; Wolff, B.; Savitskaia, N.; Ungefroren, H.; Deppert, W.; Schmiegel, W.; Kalthoff, H.; Naumann, M. TGFbeta-induced growth inhibition involves cell cycle inhibitor p21 and pRb independent from p15 expression. Int. J. Oncol. 1999, 14, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Koo, B.H.; Kim, Y.; Je Cho, Y.; Kim, D.S. Distinct roles of transforming growth factor-beta signaling and transforming growth factor-beta receptor inhibitor SB431542 in the regulation of p21 expression. Eur. J. Pharmacol. 2015, 764, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Hobbs, F.; Copeland, R.A.; Magolda, R.L.; Scherle, P.A.; Trzaskos, J.M. Identification of a novel inhibitor of mitogen-activated protein kinase kinase. J. Biol. Chem. 1998, 273, 18623–18632. [Google Scholar]
- Horiguchi, K.; Shirakihara, T.; Nakano, A.; Imamura, T.; Miyazono, K.; Saitoh, M. Role of Ras signaling in the induction of snail by transforming growth factor-beta. J. Biol. Chem. 2009, 284, 245–253. [Google Scholar] [CrossRef]
- Giehl, K.; Seidel, B.; Gierschik, P.; Adler, G.; Menke, A. TGFbeta1 represses proliferation of pancreatic carcinoma cells which correlates with Smad4-independent inhibition of ERK activation. Oncogene 2000, 19, 4531–4541. [Google Scholar] [CrossRef]
- Riesco-Eizaguirre, G.; Rodríguez, I.; De la Vieja, A.; Costamagna, E.; Carrasco, N.; Nistal, M.; Santisteban, P. The BRAFV600E oncogene induces transforming growth factor beta secretion leading to sodium iodide symporter repression and increased malignancy in thyroid cancer. Cancer Res. 2009, 69, 8317–8325. [Google Scholar] [CrossRef] [PubMed]
- Park, B.J.; Park, J.I.; Byun, D.S.; Park, J.H.; Chi, S.G. Mitogenic conversion of transforming growth factor-beta1 effect by oncogenic Ha-Ras-induced activation of the mitogen-activated protein kinase signaling pathway in human prostate cancer. Cancer Res. 2000, 60, 3031–3038. [Google Scholar] [PubMed]
- Ungefroren, H.; Witte, D.; Fiedler, C.; Gädeken, T.; Kaufmann, R.; Lehnert, H.; Gieseler, F.; Rauch, B.H. The Role of PAR2 in TGF-β1-Induced ERK Activation and Cell Motility. Int. J. Mol. Sci. 2017, 18, 2776. [Google Scholar] [CrossRef]
- Lee, M.K.; Pardoux, C.; Hall, M.C.; Lee, P.S.; Warburton, D.; Qing, J.; Smith, S.M.; Derynck, R. TGF-beta activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 2007, 26, 3957–3967. [Google Scholar] [CrossRef] [PubMed]
- Muthusamy, B.P.; Budi, E.H.; Katsuno, Y.; Lee, M.K.; Smith, S.M.; Mirza, A.M.; Akhurst, R.J.; Derynck, R. ShcA Protects against Epithelial-Mesenchymal Transition through Compartmentalized Inhibition of TGF-beta-Induced Smad Activation. PLoS Biol. 2015, 13, e1002325. [Google Scholar] [CrossRef]
- Inman, G.J.; Nicolás, F.J.; Callahan, J.F.; Harling, J.D.; Gaster, L.M.; Reith, A.D.; Laping, N.J.; Hill, C.S. SB-431542 is a potent and specific inhibitor of transforming growth factor-beta superfamily type I activin receptor-like kinase (ALK) receptors ALK4, ALK5, and ALK7. Mol. Pharmacol. 2002, 62, 65–74. [Google Scholar] [CrossRef]
- Principe, D.R.; Diaz, A.M.; Torres, C.; Mangan, R.J.; DeCant, B.; McKinney, R.; Tsao, M.S.; Lowy, A.; Munshi, H.G.; Jung, B.; et al. TGFβ engages MEK/ERK to differentially regulate benign and malignant pancreas cell function. Oncogene 2017, 36, 4336–4348. [Google Scholar] [CrossRef]
- Ungefroren, H.; Sebens, S.; Growth, S.; Gieseler, F.; Fändrich, F. The Src family kinase inhibitors PP2 and PP1 block TGF-beta1-mediated cellular responses by direct and differential inhibition of type I and type II TGF-beta receptors. Curr. Cancer Drug Targets 2011, 11, 524–535. [Google Scholar] [CrossRef] [PubMed]
- Cuenda, A.; Rouse, J.; Doza, Y.N.; Meier, R.; Cohen, P.; Gallagher, T.F.; Young, P.R.; Lee, J.C. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 1995, 364, 229–233. [Google Scholar]
- Ungefroren, H.; Otterbein, H.; Fiedler, C.; Mihara, K.; Hollenberg, M.D.; Gieseler, F.; Lehnert, H.; Witte, D. RAC1B Suppresses TGF-β1-Dependent Cell Migration in Pancreatic Carcinoma Cells through Inhibition of the TGF-β Type I Receptor ALK5. Cancers 2019, 11, 691. [Google Scholar] [CrossRef]
- Kretschmer, A.; Moepert, K.; Dames, S.; Sternberger, M.; Kaufmann, J.; Klippel, A. Differential regulation of TGF-beta signaling through Smad2, Smad3 and Smad4. Oncogene 2003, 22, 6748–6763. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ko, C.Y.; Meyers, E.E.; Pedroja, B.S.; Pelaez, N.; Bernstein, A.M. Concentration-dependent effects of transforming growth factor β1 on corneal wound healing. Mol. Vis. 2011, 17, 2835–2846. [Google Scholar]
- de Ceuninck van Capelle, C.; Spit, M.; Ten Dijke, P. Current perspectives on inhibitory SMAD7 in health and disease. Crit. Rev. Biochem. Mol. Biol. 2020, 55, 691–715. [Google Scholar] [CrossRef]
- Lei, X.; Bandyopadhyay, A.; Le, T.; Sun, L. Autocrine TGFbeta supports growth and survival of human breast cancer MDA-MB-231 cells. Oncogene 2002, 21, 7514–7523. [Google Scholar] [CrossRef]
- Dunfield, L.D.; Nachtigal, M.W. Inhibition of the antiproliferative effect of TGFbeta by EGF in primary human ovarian cancer cells. Oncogene 2003, 22, 4745–4751. [Google Scholar] [CrossRef]
- Wang, S.E.; Yu, Y.; Criswell, T.L.; Debusk, L.M.; Lin, P.C.; Zent, R.; Johnson, D.H.; Ren, X.; Arteaga, C.L. Oncogenic mutations regulate tumor microenvironment through induction of growth factors and angiogenic mediators. Oncogene 2010, 29, 3335–3348. [Google Scholar] [CrossRef]
- Geiser, A.G.; Busam, K.J.; Kim, S.J.; Lafyatis, R.; O’Reilly, M.A.; Webbink, R.; Roberts, A.B.; Sporn, M.B. Regulation of the transforming growth factor-beta 1 and -beta 3 promoters by transcription factor Sp1. Gene 1993, 129, 223–228. [Google Scholar] [CrossRef]
- Tiwari, A.; Iida, M.; Kosnopfel, C.; Abbariki, M.; Menegakis, A.; Fehrenbacher, B.; Maier, J.; Schaller, M.; Brucker, S.Y.; Wheeler, D.L.; et al. Blocking Y-Box Binding Protein-1 through Simultaneous Targeting of PI3K and MAPK in Triple Negative Breast Cancers. Cancers 2020, 12, 2795. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K. Transforming growth factor-beta signaling in epithelial-mesenchymal transition and progression of cancer. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2009, 85, 314–323. [Google Scholar] [CrossRef] [PubMed]
- Zinn, R.; Otterbein, H.; Lehnert, H.; Ungefroren, H. RAC1B: A guardian of the epithelial phenotype and protector against epithelial-mesenchymal transition. Cells 2019, 8, 1569. [Google Scholar] [CrossRef] [PubMed]
- Lowenfels, A.B.; Maisonneuve, P.; Cavallini, G.; Ammann, R.W.; Lankisch, P.G.; Andersen, J.R.; Dimagno, E.P.; Andrén-Sandberg, A.; Domellöf, L. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N. Engl. J. Med. 1993, 328, 1433–1437. [Google Scholar] [CrossRef]
- Hass, R.; von der Ohe, J.; Ungefroren, H. The Intimate Relationship Among EMT, MET and TME: A T(ransdifferentiation) E(nhancing) M(ix) to Be Exploited for Therapeutic Purposes. Cancers 2020, 12, 3674. [Google Scholar] [CrossRef]
- Aiello, N.M.; Kang, Y. Context-dependent EMT programs in cancer metastasis. J. Exp. Med. 2019, 216, 1016–1026. [Google Scholar] [CrossRef]
- Gao, Y.; Dickerson, J.B.; Guo, F.; Zheng, J.; Zheng, Y. Rational design and characterization of a Rac GTPase-specific small molecule inhibitor. Proc. Natl. Acad. Sci. USA 2004, 101, 7618–7623. [Google Scholar] [CrossRef] [PubMed]
- Regala, R.P.; Weems, C.; Jamieson, L.; Copland, J.A.; Thompson, E.A.; Fields, A.P. Atypical protein kinase Ciota plays a critical role in human lung cancer cell growth and tumorigenicity. J. Biol. Chem. 2005, 280, 31109–31115. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ungefroren, H.; Christl, J.; Eiden, C.; Wellner, U.F.; Lehnert, H.; Marquardt, J.-U. Autocrine TGFβ1 Opposes Exogenous TGFβ1-Induced Cell Migration and Growth Arrest through Sustainment of a Feed-Forward Loop Involving MEK-ERK Signaling. Cancers 2021, 13, 1357. https://doi.org/10.3390/cancers13061357
Ungefroren H, Christl J, Eiden C, Wellner UF, Lehnert H, Marquardt J-U. Autocrine TGFβ1 Opposes Exogenous TGFβ1-Induced Cell Migration and Growth Arrest through Sustainment of a Feed-Forward Loop Involving MEK-ERK Signaling. Cancers. 2021; 13(6):1357. https://doi.org/10.3390/cancers13061357
Chicago/Turabian StyleUngefroren, Hendrik, Jessica Christl, Caroline Eiden, Ulrich F. Wellner, Hendrik Lehnert, and Jens-Uwe Marquardt. 2021. "Autocrine TGFβ1 Opposes Exogenous TGFβ1-Induced Cell Migration and Growth Arrest through Sustainment of a Feed-Forward Loop Involving MEK-ERK Signaling" Cancers 13, no. 6: 1357. https://doi.org/10.3390/cancers13061357
APA StyleUngefroren, H., Christl, J., Eiden, C., Wellner, U. F., Lehnert, H., & Marquardt, J.-U. (2021). Autocrine TGFβ1 Opposes Exogenous TGFβ1-Induced Cell Migration and Growth Arrest through Sustainment of a Feed-Forward Loop Involving MEK-ERK Signaling. Cancers, 13(6), 1357. https://doi.org/10.3390/cancers13061357









