Biological Heterogeneity of Chondrosarcoma: From (Epi) Genetics through Stemness and Deregulated Signaling to Immunophenotype
Abstract
Simple Summary
Abstract
1. Introduction
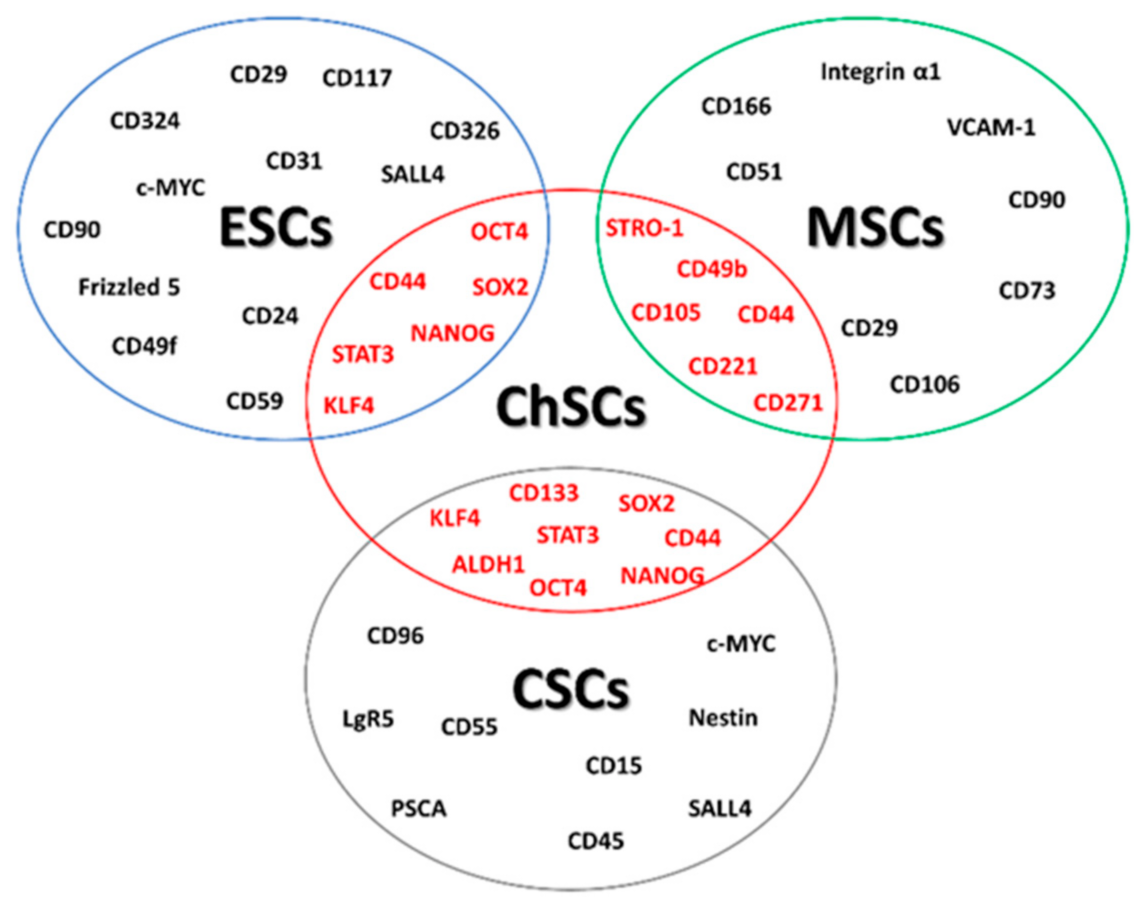
2. Chondrosarcoma Stem Cell
3. Genomic Abnormalities in Chondrosarcoma
4. Mutations in Chondrosarcoma
5. Epigenetics of Chondrosarcoma
5.1. Hypermethylation of DNA
5.2. Hypomethylation of DNA
6. Deregulated Signaling Pathways in Chondrosarcoma
6.1. Tumor Suppressor pRb and p53 Pathways
6.2. PI3K–AKT Signal Transduction
6.3. The SRC Signaling Pathway
6.4. The mTOR-S6K Pathway
6.5. Hedgehog Signaling Pathway
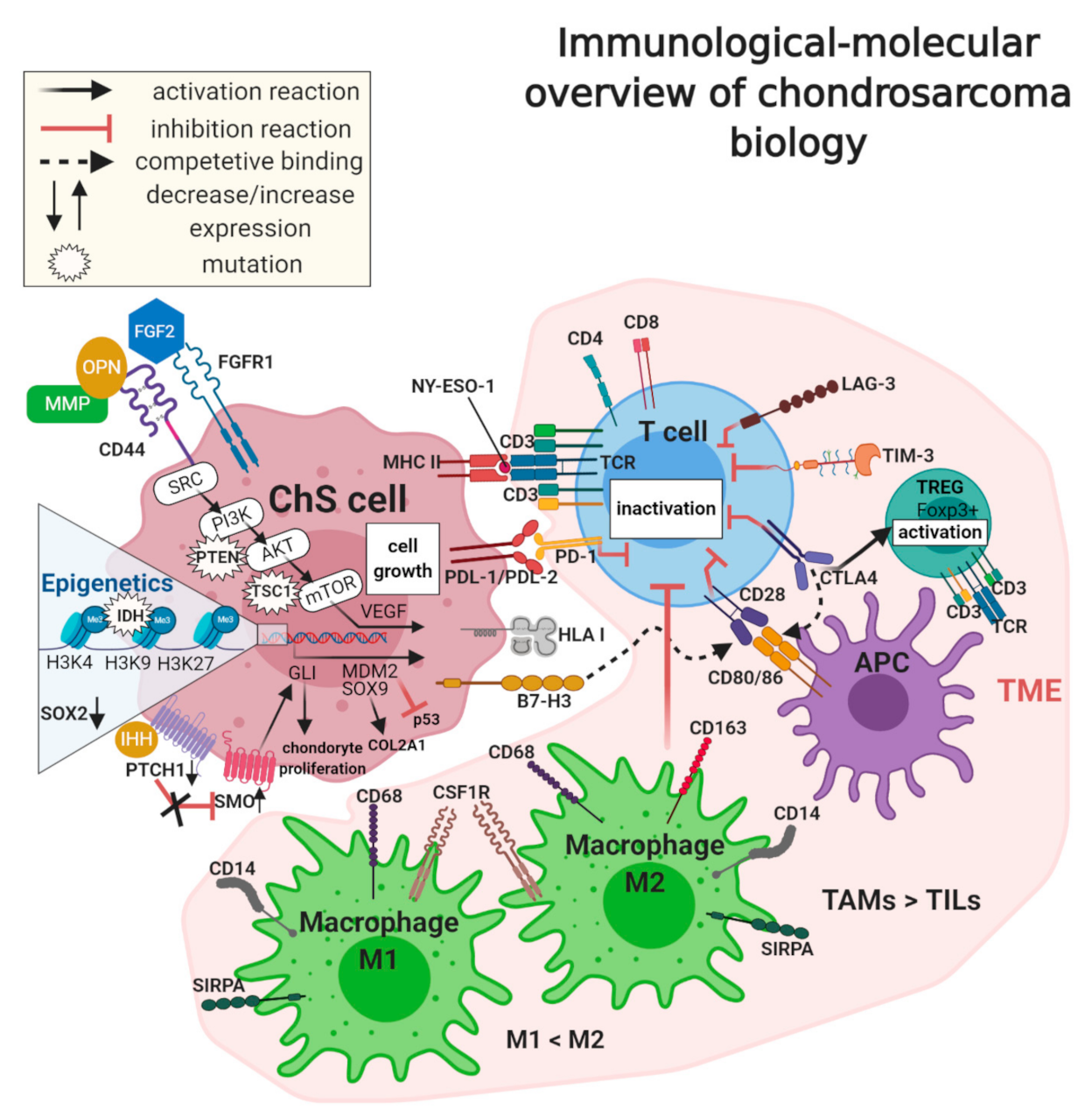
7. Chondrosarcoma Microenvironment
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Leddy, L.R.; Holmes, R.E. Chondrosarcoma of Bone. In Orthopaedic Oncology; Springer: Berlin/Heidelberg, Germany, 2014; pp. 117–130. [Google Scholar]
- Murphey, M.D.; Walker, E.A.; Wilson, A.J.; Kransdorf, M.J.; Temple, H.T.; Gannon, F.H. From the Archives of the AFIP. RadioGraphics 2003, 23, 1245–1278. [Google Scholar] [CrossRef]
- Fletcher, C.D.M.; Unni, K.K.; Mertens, F. World Health Organization Classification of Tumors: Pathology and Genetics of Tumors of Soft Tissue and Bone; IARC Press: Lyon, France, 2002. [Google Scholar]
- Limaiem, F.; Davis, D.D.; Sticco, K.L. Cancer, Chondrosarcoma. 2020. Available online: https://www.ncbi.nlm.nih.gov/books/NBK538132/ (accessed on 3 September 2020).
- Heck, R.K.; Peabody, T.D.; Simon, M.A. Staging of Primary Malignancies of Bone. CA Cancer J. Clin. 2006, 56, 366–375. [Google Scholar] [CrossRef]
- Kim, M.-J.; Cho, K.-J.; Ayala, A.G.; Ro, J.Y. Chondrosarcoma: With updates on molecular genetics. Sarcoma 2011, 2011, 405437. [Google Scholar] [CrossRef]
- Lokuhetty, D.; White, V.A.; Cree, I.A. Soft Tissue and Bone Tumours WHO Classification of Tumours, 5th ed.; IARC Press: Lyon, France, 2020. [Google Scholar]
- Evans, H.L.; Ayala, A.G.; Romsdahl, M.M. Prognostic factors in chondrosarcoma of bone.A clinicopathologic analysis with emphasis on histologic grading. Cancer 1977, 40, 818–831. [Google Scholar] [CrossRef]
- Grimer, R.J.; Gosheger, G.; Taminiau, A.; Biau, D.; Matejovsky, Z.; Kollender, Y.; San-Julian, M.; Gherlinzoni, F.; Ferrari, C. Dedifferentiated chondrosarcoma: Prognostic factors and outcome from a European group. Eur. J. Cancer 2007, 43, 2060–2065. [Google Scholar] [CrossRef] [PubMed]
- Chow, W.A. Chondrosarcoma: Biology, genetics, and epigenetics. F1000Research 2018, 7, 1826. [Google Scholar] [CrossRef]
- Shakked, R.J.; Geller, D.S.; Gorlick, R.; Dorfman, H.D. Mesenchymal Chondrosarcoma: Clinicopathologic Study of 20 Cases. Arch. Pathol. Lab. Med. 2012, 136, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Italiano, A.; Mir, O.; Cioffi, A.; Palmerini, E.; Piperno-Neumann, S.; Perrin, C.; Chaigneau, L.; Penel, N.; Duffaud, F.; Kurtz, J.E.; et al. Advanced chondrosarcomas: Role of chemotherapy and survival. Ann. Oncol. 2013, 24, 2916–2922. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, A.D.; Ayoub, K.; Mangham, D.C.; Grimer, R.J.; Carter, S.R.; Tillman, R.M. Experience in the treatment of dedifferentiated chondrosarcoma. J. Bone Jt. Surg. 2000, 82, 55–61. [Google Scholar] [CrossRef]
- Rutkowski, P.; Mazurkiewicz, T.; Kotrych, D.; Krzakowski, M.; Klepacka, T.; Ługowska, I.; Grzesiakowska, U.; Falkowski, S.; Świtaj, T.; Szacht, M.M.; et al. Zalecenia postępowania diagnostyczno-terapeutycznego u chorych na pierwotne nowotwory złośliwe kości Recommendations for diagnostics and therapy of patients with primary malignant bone tumors. Chir. Narz. Ruchu Ortop. Pol. 2016, 81, 183–194. [Google Scholar]
- Tawbi, H.A.; Burgess, M.; Bolejack, V.; Van Tine, B.A.; Schuetze, S.M.; Hu, J.; D’Angelo, S.; Attia, S.; Riedel, R.F.; Priebat, D.A.; et al. Pembrolizumab in advanced soft-tissue sarcoma and bone sarcoma (SARC028): A multicentre, two-cohort, single-arm, open-label, phase 2 trial. Lancet Oncol. 2017, 18, 1493–1501. [Google Scholar] [CrossRef]
- Pan, Y.; Ma, S.; Cao, K.; Zhou, S.; Zhao, A.; Li, M.; Qian, F.; Zhu, C. Therapeutic approaches targeting cancer stem cells. J. Cancer Res. Ther. 2018, 14, 1525–1534. [Google Scholar]
- Kuşoğlu, A.; Biray Avcı, Ç. Cancer stem cells: A brief review of the current status. Gene 2019, 681, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Barbato, L.; Bocchetti, M.; Di Biase, A.; Regad, T. Cancer Stem Cells and Targeting Strategies. Cells 2019, 8, 926. [Google Scholar] [CrossRef]
- Erkisa, M.; Karakas, D.; Ulukayad, E. Cancer stem cells: Root of the evil. Crit. Rev. Oncog. 2019, 24, 69–87. [Google Scholar] [CrossRef] [PubMed]
- Ayob, A.Z.; Ramasamy, T.S. Cancer stem cells as key drivers of tumour progression. J. Biomed. Sci. 2018, 25, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Delgado, P.; Lacerenza, S.; Obrador-Hevia, A.; Lopez-Alvarez, M.; Mondaza-Hernandez, J.L.; Blanco-Alcaina, E.; Sanchez-Bustos, P.; Hindi, N.; Moura, D.S.; Martin-Broto, J. Cancer Stem Cells in Soft-Tissue Sarcomas. Cells 2020, 9, 1449. [Google Scholar] [CrossRef] [PubMed]
- Genadry, K.C.; Pietrobono, S.; Rota, R.; Linardic, C.M. Soft Tissue Sarcoma Cancer Stem Cells: An Overview. Front. Oncol. 2018, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Trucco, M.; Loeb, D. Sarcoma Stem Cells: Do We Know What We Are Looking for? Sarcoma 2012, 2012, 1–8. [Google Scholar] [CrossRef]
- Najafi, M.; Farhood, B.; Mortezaee, K. Cancer stem cells (CSCs) in cancer progression and therapy. J. Cell. Physiol. 2019, 234, 8381–8395. [Google Scholar] [CrossRef]
- Was, H.; Krol, S.K.; Rotili, D.; Mai, A.; Wojtas, B.; Kaminska, B.; Maleszewska, M. Histone deacetylase inhibitors exert anti-tumor effects on human adherent and stem-like glioma cells 11 Medical and Health Sciences 1112 Oncology and Carcinogenesis. Clin. Epigenet. 2019, 11, 1–13. [Google Scholar]
- Król, S.K.; Kaczmarczyk, A.; Wojnicki, K.; Wojtas, B.; Gielniewski, B.; Grajkowska, W.; Kotulska, K.; Szczylik, C.; Czepko, R.; Banach, M.; et al. Aberrantly Expressed RECQL4 Helicase Supports Proliferation and Drug Resistance of Human Glioma Cells and Glioma Stem Cells. Cancers 2020, 12, 2919. [Google Scholar] [CrossRef]
- Shibata, M.; Hoque, M.O. Targeting Cancer Stem Cells: A Strategy for Effective Eradication of Cancer. Cancers 2019, 11, 732. [Google Scholar] [CrossRef]
- Yang, L.; Shi, P.; Zhao, G.; Xu, J.; Peng, W.; Zhang, J.; Zhang, G.; Wang, X.; Dong, Z.; Chen, F.; et al. Targeting Cancer Stem Cell Pathways for Cancer Therapy; Springer: New York, NY, USA, 2020; Volume 5, ISBN 4139202001105. [Google Scholar]
- David, E.; Blanchard, F.; Heymann, M.F.; De Pinieux, G.; Gouin, F.; Rédini, F.; Heymann, D. The Bone Niche of Chondrosarcoma: A Sanctuary for Drug Resistance, Tumour Growth and also a Source of New Therapeutic Targets. Sarcoma 2011, 2011, 932451. [Google Scholar] [CrossRef] [PubMed]
- Diaz-Romero, J.; Romeo, S.; Bovée, J.V.M.G.; Hogendoorn, P.C.W.; Heini, P.F.; Mainil-Varlet, P. Hierarchical clustering of flow cytometry data for the study of conventional central chondrosarcoma. J. Cell. Physiol. 2010, 225, 601–611. [Google Scholar] [CrossRef] [PubMed]
- Dominici, M.; Le Blanc, K.; Mueller, I.; Slaper-Cortenbach, I.; Marini, F.C.; Krause, D.S.; Deans, R.J.; Keating, A.; Prockop, D.J.; Horwitz, E.M. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy 2006, 8, 315–317. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Ozaki, T. Overcoming Therapeutic Resistance of Bone Sarcomas: Overview of the Molecular Mechanisms and Therapeutic Targets for Bone Sarcoma Stem Cells. Stem Cells Int. 2016, 2016, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Boehme, K.; Schleicher, S.; Traub, F.; Rolauffs, B. Chondrosarcoma: A Rare Misfortune in Aging Human Cartilage? The Role of Stem and Progenitor Cells in Proliferation, Malignant Degeneration and Therapeutic Resistance. Int. J. Mol. Sci. 2018, 19, 311. [Google Scholar] [CrossRef]
- Alexander, D.; Schäfer, F.; Munz, A.; Friedrich, B.; Klein, C.; Hoffmann, J.; Bühring, H.J.; Reinert, S. LNGFR induction during Osteogenesis of human jaw periosteum-derived cells. Cell. Physiol. Biochem. 2009, 24, 283–290. [Google Scholar] [CrossRef]
- Wirths, S.; Malenke, E.; Kluba, T.; Rieger, S.; Müller, M.R.; Schleicher, S.; von Weyhern, C.H.; Nagl, F.; Fend, F.; Vogel, W.; et al. Shared Cell Surface Marker Expression in Mesenchymal Stem Cells and Adult Sarcomas. Stem Cells Transl. Med. 2013, 2, 53–60. [Google Scholar] [CrossRef]
- Tian, J.; Li, X.; Si, M.; Liu, T.; Li, J. CD271+ Osteosarcoma Cells Display Stem-Like Properties. PLoS ONE 2014, 9, e98549. [Google Scholar] [CrossRef]
- Andrzejewska, A.; Lukomska, B.; Janowski, M. Concise Review: Mesenchymal Stem Cells: From Roots to Boost. Stem Cells 2019, 37, 855–864. [Google Scholar] [CrossRef]
- Gibbs, C.P.; Kukekov, V.G.; Reith, J.D.; Tchigrinova, O.; Suslov, O.N.; Scott, E.W.; Ghivizzani, S.C.; Ignatova, T.N.; Steindler, D.A. Stem-like cells in bone sarcomas: Implications for tumorigenesis. Neoplasia 2005, 7, 967–976. [Google Scholar] [CrossRef]
- Senbanjo, L.T.; Chellaiah, M.A. CD44: A Multifunctional Cell Surface Adhesion Receptor Is a Regulator of Progression and Metastasis of Cancer Cells. Front. Cell Dev. Biol. 2017, 5, 18. [Google Scholar] [CrossRef] [PubMed]
- Heyse, T.J.; Malcherczyk, D.; Moll, R.; Timmesfeld, N.; Wapelhorst, J.; Fuchs-Winkelmann, S.; Paletta, J.R.J.; Schofer, M.D. CD44: Survival and metastasis in chondrosarcoma. Osteoarthr. Cartil. 2010, 18, 849–856. [Google Scholar] [CrossRef] [PubMed]
- Tirino, V.; Desiderio, V.; D’Aquino, R.; De Francesco, F.; Pirozzi, G.; Galderisi, U.; Cavaliere, C.; De Rosa, A.; Papaccio, G. Detection and characterization of CD133+ cancer stem cells in human solid tumours. PLoS ONE 2008, 3, e3469. [Google Scholar] [CrossRef]
- Tirino, V.; Desiderio, V.; Paino, F.; De Rosa, A.; Papaccio, F.; Fazioli, F.; Pirozzi, G.; Papaccio, G. Human primary bone sarcomas contain CD133 + cancer stem cells displaying high tumorigenicity in vivo. FASEB J. 2011, 25, 2022–2030. [Google Scholar] [CrossRef]
- Clark, D.W.; Palle, K. Aldehyde dehydrogenases in cancer stem cells: Potential as therapeutic targets. Ann. Transl. Med. 2016, 4, 1–8. [Google Scholar] [CrossRef]
- Menendez, S.T.; Rey, V.; Martinez-Cruzado, L.; Gonzalez, M.V.; Morales-Molina, A.; Santos, L.; Blanco, V.; Alvarez, C.; Estupiñan, O.; Allonca, E.; et al. SOX2 Expression and Transcriptional Activity Identifies a Subpopulation of Cancer Stem Cells in Sarcoma with Prognostic Implications. Cancers 2020, 12, 964. [Google Scholar] [CrossRef]
- Silva, J.; Nichols, J.; Theunissen, T.W.; Guo, G.; van Oosten, A.L.; Barrandon, O.; Wray, J.; Yamanaka, S.; Chambers, I.; Smith, A. Nanog Is the Gateway to the Pluripotent Ground State. Cell 2009, 138, 722–737. [Google Scholar] [CrossRef]
- Baek, K.H.; Choi, J.; Pei, C.Z. Cellular functions of OCT-3/4 regulated by ubiquitination in proliferating cells. Cancers 2020, 12, 663. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Shen, J.; Wang, K.; Hornicek, F.; Duan, Z. The Roles of Sox Family Genes in Sarcoma. Curr. Drug Targets 2016, 17, 1761–1772. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Shen, J.K.; Xu, J.; Trahan, C.A.; Hornicek, F.J.; Duan, Z. Aberrant DNA methylations in chondrosarcoma. Epigenomics 2016, 8, 1519–1525. [Google Scholar] [CrossRef]
- Armstrong, L.; Stojkovic, M.; Dimmick, I.; Ahmad, S.; Stojkovic, P.; Hole, N.; Lako, M. Phenotypic Characterization of Murine Primitive Hematopoietic Progenitor Cells Isolated on Basis of Aldehyde Dehydrogenase Activity. Stem Cells 2004, 22, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Vassalli, G. Aldehyde dehydrogenases: Not just markers, but functional regulators of stem cells. Stem Cells Int. 2019, 2019, 3904645. [Google Scholar] [CrossRef] [PubMed]
- Sládek, N.E. Human aldehyde dehydrogenases: Potential pathological, pharmacological, and toxicological impact. J. Biochem. Mol. Toxicol. 2003, 17, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Rey, V.; Menendez, S.T.; Estupiñan, O.; Rodriguez, A.; Santos, L.; Tornin, J.; Martinez-Cruzado, L.; Castillo, D.; Ordoñez, G.R.; Costilla, S.; et al. New Chondrosarcoma Cell Lines with Preserved Stem Cell Properties to Study the Genomic Drift During In Vitro/In Vivo Growth. J. Clin. Med. 2019, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Hoyt, A.K.; Moran, A.; Granger, C.; Sedani, A.; Saigh, S.; Brown, J.; Galoian, K.A. PRP-1 significantly decreases the ALDHhigh cancer stem cell population and regulates the aberrant Wnt/β-catenin pathway in human chondrosarcoma JJ012 cells. Oncol. Rep. 2019, 42, 103–114. [Google Scholar] [CrossRef]
- Vares, G.; Ahire, V.; Sunada, S.; Ho Kim, E.; Sai, S.; Chevalier, F.; Romeo, P.H.; Yamamoto, T.; Nakajima, T.; Saintigny, Y. A multimodal treatment of carbon ions irradiation, miRNA-34 and mTOR inhibitor specifically control high-grade chondrosarcoma cancer stem cells. Radiother. Oncol. 2020, 150, 253–261. [Google Scholar] [CrossRef]
- Misso, G.; Di Martino, M.T.; De Rosa, G.; Farooqi, A.A.; Lombardi, A.; Campani, V.; Zarone, M.R.; Gullà, A.; Tagliaferri, P.; Tassone, P.; et al. Mir-34: A new weapon against cancer? Mol. Ther. Nucleic Acids 2014, 3, e195. [Google Scholar] [CrossRef]
- Guessous, F.; Zhang, Y.; Kofman, A.; Catania, A.; Li, Y.; Schiff, D.; Purow, B.; Abounader, R. microRNA-34a is tumor suppressive in brain tumors and glioma stem cells. Cell Cycle 2010, 9, 1031–1036. [Google Scholar] [CrossRef]
- Liu, C.; Kelnar, K.; Liu, B.; Chen, X.; Calhoun-Davis, T.; Li, H.; Patrawala, L.; Yan, H.; Jeter, C.; Honorio, S.; et al. Identification of miR-34a as a potent inhibitor of prostate cancer progenitor cells and metastasis by directly repressing CD44 HHS Public Access. Nat. Med. 2011, 17, 211–215. [Google Scholar] [CrossRef] [PubMed]
- Nalls, D.; Tang, S.-N.; Rodova, M.; Srivastava, R.K.; Shankar, S. Targeting Epigenetic Regulation of miR-34a for Treatment of Pancreatic Cancer by Inhibition of Pancreatic Cancer Stem Cells. PLoS ONE 2011, 6, e24099. [Google Scholar] [CrossRef]
- Kim, H.; Cho, Y.; Kim, H.-S.; Kang, D.; Cheon, D.; Kim, Y.-J.; Chang, M.J.; Lee, K.M.; Chang, C.B.; Kang, S.-B.; et al. A system-level approach identifies HIF-2α as a critical regulator of chondrosarcoma progression. Nat. Commun. 2020, 11, 5023. [Google Scholar] [CrossRef]
- Chen, C.; Ma, Q.; Ma, X.; Liu, Z.; Liu, X. Association of Elevated HIF-2α Levels with Low Beclin 1 Expression and Poor Prognosis in Patients with Chondrosarcoma. Ann. Surg. Oncol. 2011, 18, 2364–2372. [Google Scholar] [CrossRef]
- Chen, C.; Zhou, H.; Wei, F.; Jiang, L.; Liu, X.; Liu, Z.; Ma, Q. Increased levels of hypoxia-inducible factor-1α are associated with Bcl-xL expression, tumor apoptosis, and clinical outcome in chondrosarcoma. J. Orthop. Res. 2011, 29, 143–151. [Google Scholar] [CrossRef]
- Oshiro, Y.; Chaturvedi, V.; Hayden, D.; Nazeer, T.; Johnson, M.; Johnston, D.A.; Ordóñez, N.G.; Ayala, A.G.; Czerniak, B. Altered p53 is associated with aggressive behavior of chondrosarcoma. Cancer 1998, 83, 2324–2334. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Toguchida, J.; Wadayama, B.; Kanoe, H.; Nakayama, T.; Ishizaki, K.; Ikenaga, M.; Kotoura, Y.; Sasaki, M.S. Loss of heterozygosity and tumor suppressor gene mutations in chondrosarcomas. Anticancer Res. 1996, 16, 2009–2015. [Google Scholar] [PubMed]
- Terek, R.M.; Healey, J.H.; Garin-Chesa, P.; Mak, S.; Huvos, A.; Albino, A.P. p53 mutations in chondrosarcoma. Diagn. Mol. Pathol. 1998, 7, 51–56. [Google Scholar] [CrossRef] [PubMed]
- van Beerendonk, H.M.; Rozeman, L.B.; Taminiau, A.H.; Sciot, R.; Bovée, J.V.; Cleton-Jansen, A.-M.; Hogendoorn, P.C. Molecular analysis of the INK4A/INK4A-ARF gene locus in conventional(central) chondrosarcomas and enchondromas: Indication of an important gene for tumour progression. J. Pathol. 2004, 202, 359–366. [Google Scholar] [CrossRef]
- Fiedorowicz, M.; Bartnik, E.; Sobczuk, P.; Teterycz, P.; Czarnecka, A.M. Molecular biology of sarcoma. Oncol. Clin. Pract. 2018, 14, 307–330. [Google Scholar] [CrossRef]
- Hallor, K.H.; Staaf, J.; Bovee, J.V.M.G.; Hogendoorn, P.C.W.; Cleton-Jansen, A.-M.; Knuutila, S.; Savola, S.; Niini, T.; Brosjo, O.; Bauer, H.C.F.; et al. Genomic Profiling of Chondrosarcoma: Chromosomal Patterns in Central and Peripheral Tumors. Clin. Cancer Res. 2009, 15, 2685–2694. [Google Scholar] [CrossRef]
- Sandberg, A.A.; Bridge, J.A. Updates on the cytogenetics and molecular genetics of bone and soft tissue tumors. Cancer Genet. Cytogenet. 2003, 143, 1–31. [Google Scholar] [CrossRef]
- Miettinen, M.; Felisiak-Golabek, A.; Luiña Contreras, A.; Glod, J.; Kaplan, R.N.; Killian, J.K.; Lasota, J. New fusion sarcomas: Histopathology and clinical significance of selected entities. Hum. Pathol. 2019, 86, 57–65. [Google Scholar] [CrossRef]
- Sandberg, A.A. Genetics of chondrosarcoma and related tumors. Curr. Opin. Oncol. 2004, 342–354. [Google Scholar] [CrossRef]
- Agaram, N.P.; Zhang, L.; Sung, Y.-S.; Singer, S.; Antonescu, C.R. Extraskeletal myxoid chondrosarcoma with non–EWSR1-NR4A3 variant fusions correlate with rhabdoid phenotype and high-grade morphology. Hum. Pathol. 2014, 45, 1084–1091. [Google Scholar] [CrossRef]
- Broehm, C.J.; Wu, J.; Gullapalli, R.R.; Bocklage, T. Extraskeletal myxoid chondrosarcoma with a t(9;16)(q22;p11.2) resulting in a NR4A3-FUS fusion. Cancer Genet. 2014, 207, 276–280. [Google Scholar] [CrossRef]
- Paioli, A.; Stacchiotti, S.; Campanacci, D.; Palmerini, E.; Frezza, A.M.; Longhi, A.; Radaelli, S.; Donati, D.M.; Beltrami, G.; Bianchi, G.; et al. Extraskeletal Myxoid Chondrosarcoma with Molecularly Confirmed Diagnosis: A Multicenter Retrospective Study Within the Italian Sarcoma Group. Ann. Surg. Oncol. 2020, 28, 1142–1150. [Google Scholar] [CrossRef] [PubMed]
- Stacchiotti, S.; Baldi, G.G.; Morosi, C.; Gronchi, A.; Maestro, R. Extraskeletal Myxoid Chondrosarcoma: State of the Art and Current Research on Biology and Clinical Management. Cancers 2020, 12, 2703. [Google Scholar] [CrossRef] [PubMed]
- Sbaraglia, M.; Dei Tos, A.P. The pathology of soft tissue sarcomas. Radiol. Med. 2019, 124, 266–281. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Motoi, T.; Khanin, R.; Olshen, A.; Mertens, F.; Bridge, J.; Cin, P.D.; Antonescu, C.R.; Singer, S.; Hameed, M.; et al. Identification of a novel, recurrent HEY1-NCOA2 fusion in mesenchymal chondrosarcoma based on a genome-wide screen of exon-level expression data. Genes Chromosomes Cancer 2012, 51, 127–139. [Google Scholar] [CrossRef]
- Li, L.; Hu, X.; Eid, J.E.; Rosenberg, A.E.; Wilky, B.A.; Ban, Y.; Sun, X.; Galoian, K.; DeSalvo, J.; Yue, J.; et al. Mutant IDH1 Depletion Downregulates Integrins and Impairs Chondrosarcoma Growth. Cancers 2020, 12, 141. [Google Scholar] [CrossRef] [PubMed]
- Lugowska, I.; Teterycz, P.; Mikula, M.; Kulecka, M.; Kluska, A.; Balabas, A.; Piatkowska, M.; Wagrodzki, M.; Pienkowski, A.; Rutkowski, P.; et al. IDH1/2 Mutations Predict Shorter Survival in Chondrosarcoma. J. Cancer 2018, 9, 998–1005. [Google Scholar] [CrossRef] [PubMed]
- Dang, L.; White, D.W.; Gross, S.; Bennett, B.D.; Bittinger, M.A.; Driggers, E.M.; Fantin, V.R.; Jang, H.G.; Jin, S.; Keenan, M.C.; et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature 2009, 462, 739–744. [Google Scholar] [CrossRef] [PubMed]
- Venneker, S.; Kruisselbrink, A.B.; Baranski, Z.; Palubeckaite, I.; Briaire-de Bruijn, I.H.; Oosting, J.; French, P.J.; Danen, E.H.J.; Bovée, J.V.M.G. Beyond the Influence of IDH Mutations: Exploring Epigenetic Vulnerabilities in Chondrosarcoma. Cancers 2020, 12, 3589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Wei, Q.; Tsushima, H.; Puviindran, V.; Tang, Y.J.; Pathmanapan, S.; Poon, R.; Ramu, E.; Al-Jazrawe, M.; Wunder, J.; et al. Intracellular cholesterol biosynthesis in enchondroma and chondrosarcoma. JCI Insight 2019, 4, e127232. [Google Scholar] [CrossRef]
- Amary, M.F.; Bacsi, K.; Maggiani, F.; Damato, S.; Halai, D.; Berisha, F.; Pollock, R.; O’Donnell, P.; Grigoriadis, A.; Diss, T.; et al. IDH1 and IDH2 mutations are frequent events in central chondrosarcoma and central and periosteal chondromas but not in other mesenchymal tumours. J. Pathol. 2011, 224, 334–343. [Google Scholar] [CrossRef]
- Tarpey, P.S.; Behjati, S.; Cooke, S.L.; Van Loo, P.; Wedge, D.C.; Pillay, N.; Marshall, J.; O’Meara, S.; Davies, H.; Nik-Zainal, S.; et al. Frequent mutation of the major cartilage collagen gene COL2A1 in chondrosarcoma. Nat. Genet. 2013, 45, 923–926. [Google Scholar] [CrossRef]
- Nicolle, R.; Ayadi, M.; Gomez-Brouchet, A.; Armenoult, L.; Banneau, G.; Elarouci, N.; Tallegas, M.; Decouvelaere, A.V.; Aubert, S.; Rédini, F.; et al. Integrated molecular characterization of chondrosarcoma reveals critical determinants of disease progression. Nat. Commun. 2019, 10, 1–11. [Google Scholar] [CrossRef]
- Cote, G.M.; He, J.; Choy, E. Next-Generation Sequencing for Patients with Sarcoma: A Single Center Experience. Oncologist 2018, 23, 234–242. [Google Scholar] [CrossRef]
- Olivier, M.; Hollstein, M.; Hainaut, P. TP53 Mutations in Human Cancers: Origins, Consequences, and Clinical Use. Cold Spring Harb. Perspect. Biol. 2010, 2, a001008. [Google Scholar] [CrossRef] [PubMed]
- Baugh, E.H.; Ke, H.; Levine, A.J.; Bonneau, R.A.; Chan, C.S. Why are there hotspot mutations in the TP53 gene in human cancers? Cell Death Differ. 2018, 25, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Nazeri, E.; Gouran Savadkoohi, M.; Majidzadeh-A, K.; Esmaeili, R. Chondrosarcoma: An overview of clinical behavior, molecular mechanisms mediated drug resistance and potential therapeutic targets. Crit. Rev. Oncol. Hematol. 2018, 131, 102–109. [Google Scholar] [CrossRef] [PubMed]
- Lam, S.W.; Langevelde, K.; Suurmeijer, A.J.H.; Cleven, A.H.G.; Bovée, J.V.M.G. Conventional chondrosarcoma with focal clear cell change: A clinicopathological and molecular analysis. Histopathology 2019, 75, 843–852. [Google Scholar] [CrossRef]
- Bovée, J.V.M.G.; Hogendoorn, P.C.W.; Wunder, J.S.; Alman, B.A. Cartilage tumours and bone development: Molecular pathology and possible therapeutic targets. Nat. Rev. Cancer 2010, 10, 481–488. [Google Scholar] [CrossRef]
- Kaminska, B.; Czapski, B.; Guzik, R.; Król, S.K.; Gielniewski, B. Consequences of IDH1/2 mutations in gliomas and an assessment of inhibitors targeting mutated IDH proteins. Molecules 2019, 24, 968. [Google Scholar] [CrossRef]
- Cleven, A.H.G.; Suijker, J.; Agrogiannis, G.; Briaire-de Bruijn, I.H.; Frizzell, N.; Hoekstra, A.S.; Wijers-Koster, P.M.; Cleton-Jansen, A.-M.; Bovée, J.V.M.G. IDH1 or -2 mutations do not predict outcome and do not cause loss of 5-hydroxymethylcytosine or altered histone modifications in central chondrosarcomas. Clin. Sarcoma Res. 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Suijker, J.; Oosting, J.; Koornneef, A.; Struys, E.A.; Salomons, G.S.; Schaap, F.G.; Waaijer, C.J.F.; Wijers-Koster, P.M.; Briaire-de Bruijn, I.H.; Haazen, L.; et al. Inhibition of mutant IDH1 decreases D-2-HG levels without affecting tumorigenic properties of chondrosarcoma cell lines. Oncotarget 2015, 6, 12505–12519. [Google Scholar] [CrossRef]
- Hamm, C.A.; Stevens, J.W.; Xie, H.; Vanin, E.F.; Morcuende, J.A.; Abdulkawy, H.; Seftor, E.A.; Sredni, S.T.; Bischof, J.M.; Wang, D.; et al. Microenvironment alters epigenetic and gene expression profiles in Swarm rat chondrosarcoma tumors. BMC Cancer 2010, 10, 1–16. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Asp, J.; Angiorgi, L.S.; Nerot, S.E.I.; Indahl, A.L.; Olendini, L.M.; Enassi, M.S.B.; Icci, P.P. Changes of the p16 gene but not the p53 gene in human chondrosarcoma tissues. Int. J. Cancer 2000, 786, 782–786. [Google Scholar] [CrossRef]
- Röpke, M.; Boltze, C.; Neumann, H.W.; Roessner, A.; Schneider-Stock, R. Genetic and epigenetic alterations in tumor progression in a dedifferentiated chondrosarcoma. Pathol. Res. Pract. 2003, 199, 437–444. [Google Scholar] [CrossRef] [PubMed]
- Jin, Z.; Han, Y.X.; Han, X.R. Loss of RUNX3 expression may contribute to poor prognosis in patients with chondrosarcoma. J. Mol. Histol. 2013, 44, 645–652. [Google Scholar] [CrossRef] [PubMed]
- Bui, C.; Ouzzine, M.; Talhaoui, I.; Sharp, S.; Prydz, K.; Coughtrie, M.W.H.; Fournel-Gigleux, S. Epigenetics: Methylation-associated repression of heparan sulfate 3- O -sulfotransferase gene expression contributes to the invasive phenotype of H-EMC-SS chondrosarcoma cells. FASEB J. 2010, 24, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Peterse, E.F.P.; Van Den Akker, B.E.W.M.; Niessen, B.; Oosting, J.; Suijker, J.; De Jong, Y.; Danen, E.H.J.; Cleton-Jansen, A.M.; Bovée, J.V.M.G. NAD Synthesis Pathway Interference Is a Viable Therapeutic Strategy for Chondrosarcoma. Mol. Cancer Res. 2017, 15, 1714–1721. [Google Scholar] [CrossRef]
- Bovolenta, P.; Esteve, P.; Ruiz, J.M.; Cisneros, E.; Lopez-Rios, J. Beyond Wnt inhibition: New functions of secreted Frizzled-related proteins in development and disease. J. Cell Sci. 2008, 121, 737–746. [Google Scholar] [CrossRef]
- Sheng, W.; Zhang, Z.C.; Shi, D.Y.; Wang, B.C.; Wu, Q.; Shao, Z.W.; Yang, S.H.; He, T.C.; Liu, J.X. Epigenetic silencing of SFRP5 promotes the metastasis and invasion of chondrosarcoma by expression inhibition and Wnt signaling pathway activation. Chem. Biol. Interact. 2018, 296, 1–8. [Google Scholar] [CrossRef]
- Hamm, C.A.; Xie, H.; Costa, F.F.; Vanin, E.F.; Seftor, E.A.; Sredni, S.T.; Bischof, J.; Wang, D.; Bonaldo, M.F.; Hendrix, M.J.C.; et al. Global demethylation of rat chondrosarcoma cells after treatment with 5-Aza-2′-deoxycytidine results in increased tumorigenicity. PLoS ONE 2009, 4, e8340. [Google Scholar] [CrossRef]
- Domann, F.E.; Fitzgerald, M.P.; Gourronc, F.; Teoh, M.L.T.; Provenzano, M.J.; Case, A.J.; Martin, J.A. Human chondrosarcoma cells acquire an epithelial-like gene expression pattern via an epigenetic switch: Evidence for mesenchymal-epithelial transition during sarcomagenesis. Sarcoma 2011, 2011, 598218. [Google Scholar]
- Jancalek, R. The role of the TP73 gene and its transcripts in neuro-oncology. Br. J. Neurosurg. 2014, 28, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Garbutt, C.; Hornicek, F.J.; Liu, F.; Duan, Z. Aberration of p73 promoter methylation in chondrosarcoma. Anticancer Res. 2017, 37, 2939–2945. [Google Scholar] [PubMed]
- Hosseini, A.; Minucci, S. Alterations of Histone Modifications in Cancer. In Epigenetics in Human Disease; Elsevier: Amsterdam, The Netherlands, 2018; pp. 141–217. [Google Scholar]
- Shanmugam, M.K.; Arfuso, F.; Arumugam, S.; Chinnathambi, A.; Jinsong, B.; Warrier, S.; Wang, L.Z.; Kumar, A.P.; Ahn, K.S.; Sethi, G.; et al. Role of novel histone modifications in cancer. Oncotarget 2018, 9, 11414–11426. [Google Scholar] [CrossRef]
- Zhao, Z.; Shilatifard, A. Epigenetic modifications of histones in cancer. Genome Biol. 2019, 20, 1–16. [Google Scholar] [CrossRef]
- Ververis, K.; Hiong, A.; Karagiannis, T.C.; Licciardi, P. V Histone deacetylase inhibitors (HDACIs): Multitargeted anticancer agents. Biologics 2013, 7, 47–60. [Google Scholar]
- Mottamal, M.; Zheng, S.; Huang, T.; Wang, G. Histone Deacetylase Inhibitors in Clinical Studies as Templates for New Anticancer Agents. Molecules 2015, 20, 3898–3941. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Wang, J.; Xu, J.; Xie, C.; Gao, F.; Li, Z. The expression of SIRT1 regulates the metastaticplasticity of chondrosarcoma cells by inducing epithelial-mesenchymal transition. Sci. Rep. 2017, 7, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Gu, J.; Ma, J.; Xu, Z.; Tao, H. Histone deacetylase inhibitors repress chondrosarcoma cell proliferation. J. Buon. 2015, 20, 269–274. [Google Scholar]
- Yamamoto, S.; Tanaka, K.; Sakimura, R.; Okada, T.; Nakamura, T.; Li, Y.; Takasaki, M.; Nakabeppu, Y.; Iwamoto, Y. Suberoylanilide hydroxamic acid (SAHA) induces apoptosis or autophagy-associated cell death in chondrosarcoma cell lines. Anticancer Res. 2008, 28, 1585–1591. [Google Scholar] [PubMed]
- Sakimura, R.; Tanaka, K.; Yamamoto, S.; Matsunobu, T.; Li, X.; Hanada, M.; Okada, T.; Nakamura, T.; Li, Y.; Iwamoto, Y. The Effects of Histone Deacetylase Inhibitors on the Induction of Differentiation in Chondrosarcoma Cells. Clin. Cancer Res. 2007, 13, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Röpke, M.; Boltze, C.; Meyer, B.; Neumann, H.W.; Roessner, A.; Schneider-Stock, R. Rb-loss is associated with high malignancy in chondrosarcoma. Oncol. Rep. 2006, 15, 89–95. [Google Scholar] [CrossRef][Green Version]
- Schrage, Y.M.; Lam, S.; Jochemsen, A.G.; Cleton-Jansen, A.-M.; Taminiau, A.H.M.; Hogendoorn, P.C.W.; Bovée, J.V.M.G. Central chondrosarcoma progression is associated with pRb pathway alterations: CDK4 down-regulation and p16 overexpression inhibit cell growth in vitro. J. Cell. Mol. Med. 2009, 13, 2843–2852. [Google Scholar] [CrossRef]
- Ho, L.; Stojanovski, A.; Whetstone, H.; Wei, Q.X.; Mau, E.; Wunder, J.S.; Alman, B. Gli2 and p53 Cooperate to Regulate IGFBP-3- Mediated Chondrocyte Apoptosis in the Progression from Benign to Malignant Cartilage Tumors. Cancer Cell 2009, 16, 126–136. [Google Scholar] [CrossRef] [PubMed]
- Ray-Coquard, I.; Blay, J.Y.; Italiano, A.; Le Cesne, A.; Penel, N.; Zhi, J.; Heil, F.; Rueger, R.; Graves, B.; Ding, M.; et al. Effect of the MDM2 antagonist RG7112 on the P53 pathway in patients with MDM2-amplified, well-differentiated or dedifferentiated liposarcoma: An exploratory proof-of-mechanism study. Lancet Oncol. 2012, 13, 1133–1140. [Google Scholar] [CrossRef]
- Sherr, C.J.; McCormick, F. The RB and p53 pathways in cancer. Cancer Cell 2002, 2, 103–112. [Google Scholar] [CrossRef]
- Ianari, A.; Natale, T.; Calo, E.; Ferretti, E.; Alesse, E.; Screpanti, I.; Haigis, K.; Gulino, A.; Lees, J.A. Proapoptotic Function of the Retinoblastoma Tumor Suppressor Protein. Cancer Cell 2009, 15, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Roberts, P.J.; Kumarasamy, V.; Witkiewicz, A.K.; Knudsen, E.S. Chemotherapy and CDK4/6 Inhibitors: Unexpected Bedfellows. Mol. Cancer Ther. 2020, 19, 1575–1588. [Google Scholar] [CrossRef]
- Engelman, J.A. Targeting PI3K signalling in cancer: Opportunities, challenges and limitations. Nat. Rev. Cancer 2009, 9, 550–562. [Google Scholar] [CrossRef]
- Schrage, Y.M.; Briaire-de Bruijn, I.H.; De Miranda, N.F.C.C.; Van Oosterwijk, J.; Taminiau, A.H.M.; Van Wezel, T.; Hogendoorn, P.C.W.; Bovée, J.V.M.G. Kinome profiling of chondrosarcoma reveals Src-pathway activity and dasatinib as option for treatment. Cancer Res. 2009, 69, 6216–6222. [Google Scholar] [CrossRef]
- Jang, J.H.; Chung, C.P. Tenascin-C promotes cell survival by activation of Akt in human chondrosarcoma cell. Cancer Lett. 2005, 229, 101–105. [Google Scholar] [CrossRef]
- Lin, C.; Meitner, P.A.; Terek, R.M. PTEN mutation is rare in chondrosarcoma. Diagnostic Mol. Pathol. 2002, 11, 22–26. [Google Scholar] [CrossRef]
- LoRusso, P.M. Inhibition of the PI3K/AKT/mTOR pathway in solid tumors. J. Clin. Oncol. 2016, 34, 3803–3815. [Google Scholar] [CrossRef]
- Van Oosterwijk, J.G.; Van Ruler, M.A.J.H.; Briaire-De Bruijn, I.H.; Herpers, B.; Gelderblom, H.; Van De Water, B.; Bovée, J.V.M.G. Src kinases in chondrosarcoma chemoresistance and migration: Dasatinib sensitises to doxorubicin in TP53 mutant cells. Br. J. Cancer 2013, 109, 1214–1222. [Google Scholar] [CrossRef]
- Schuetze, S.; Wathen, J.K.; Lucas, D.R.; Choy, E.; Samuels, B.L. SARC009: Phase 2 study of dasatinib in patients with previously treated, high-grade, advanced sarcoma. Cancer 2016, 122, 868–874. [Google Scholar] [CrossRef]
- Gibbons, J.J.; Abraham, R.T.; Yu, K. Mammalian Target of Rapamycin: Discovery of Rapamycin Reveals a Signaling Pathway Important for Normal and Cancer Cell Growth. Semin. Oncol. 2009, 36, S3–S17. [Google Scholar] [CrossRef]
- Sun, S.Y.E. mTOR kinase inhibitors as potential cancer therapeutic drugs. Cancer Lett. 2013, 340, 1–8. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Van Oosterwijk, J.G.; Sicinska, E.; Moss, S.; Remillard, S.P.; Van Wezel, T.; Bühnemann, C.; Hassan, A.B.; Demetri, G.D.; Bovée, J.V.M.G.; et al. Functional profiling of receptor tyrosine kinases and downstream signaling in human chondrosarcomas identifies pathways for rational targeted therapy. Clin. Cancer Res. 2013, 19, 3796–3807. [Google Scholar] [CrossRef]
- Galoian, K.; Temple, T.H.; Galoyan, A. Cytostatic effect of the hypothalamic cytokine prp-1 is mediated by mTOR and cMYc inhibition in high grade chondrosarcoma. Neurochem. Res. 2011, 36, 812–818. [Google Scholar] [CrossRef]
- Wu, M.H.; Lo, J.F.; Kuo, C.H.; Lin, J.A.; Lin, Y.M.; Chen, L.M.; Tsai, F.J.; Tsai, C.H.; Huang, C.Y.; Tang, C.H. Endothelin-1 promotes MMP-13 production and migration in human chondrosarcoma cells through FAK/PI3K/Akt/mTOR pathways. J. Cell. Physiol. 2012, 227, 3016–3026. [Google Scholar] [CrossRef] [PubMed]
- Perez, J.; Decouvelaere, A.V.; Pointecouteau, T.; Pissaloux, D.; Michot, J.P.; Besse, A.; Blay, J.Y.; Dutour, A. Inhibition of chondrosarcoma growth by mTOR inhibitor in an in vivo syngeneic rat model. PLoS ONE 2012, 7, e32458. [Google Scholar] [CrossRef] [PubMed]
- Merimsky, O.; Bernstein-Molho, R.; Sagi-Eisenberg, R. Targeting the mammalian target of rapamycin in myxoid chondrosarcoma. Anticancer. Drugs 2008, 19, 1019–1021. [Google Scholar] [CrossRef] [PubMed]
- Bernstein-Molho, R.; Kollender, Y.; Issakov, J.; Bickels, J.; Dadia, S.; Flusser, G.; Meller, I.; Sagi-Eisenberg, R.; Merimsky, O. Clinical activity of mTOR inhibition in combination with cyclophosphamide in the treatment of recurrent unresectable chondrosarcomas. Cancer Chemother. Pharmacol. 2012, 70, 855–860. [Google Scholar] [CrossRef] [PubMed]
- Cannonier, S.; Sterling, J. The Role of Hedgehog Signaling in Tumor Induced Bone Disease. Cancers 2015, 7, 1658–1683. [Google Scholar] [CrossRef]
- Sakamoto, A. The molecular pathogenesis of dedifferentiated chondrosarcoma. Indian J. Orthop. 2014, 48, 262. [Google Scholar] [CrossRef]
- Campbell, V.T.; Nadesan, P.; Ali, S.A.; Wang, C.Y.Y.; Whetstone, H.; Poon, R.; Wei, Q.; Keilty, J.; Proctor, J.; Wang, L.W.; et al. Hedgehog Pathway Inhibition in Chondrosarcoma Using the Smoothened Inhibitor IPI-926 Directly Inhibits Sarcoma Cell Growth. Mol. Cancer Ther. 2014, 13, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.J.; Hohenberger, P.; Okuno, S.; Eriksson, M.; Patel, S.; Ferrari, S.; Casali, P.G.; Chawla, S.P.; Woehr, M.; Ross, R.; et al. Results from a Phase 2 Randomized, Placebo-Controlled, Double Blind Study of the Hedgehog Pathway Antagonist IPI-926 in Patients with Advanced Chondrosarcoma. In Proceedings of the Connective Tissue Oncology Society 18th Annual Meeting, New York, NY, USA, 30 October 2013. [Google Scholar]
- Simard, F.A.; Richert, I.; Vandermoeten, A.; Decouvelaere, A.-V.; Michot, J.-P.; Caux, C.; Blay, J.-Y.; Dutour, A. Description of the immune microenvironment of chondrosarcoma and contribution to progression. Oncoimmunology 2017, 6, e1265716. [Google Scholar] [CrossRef] [PubMed]
- Boxberg, M.; Steiger, K.; Lenze, U.; Rechl, H.; von Eisenhart-Rothe, R.; Wörtler, K.; Weichert, W.; Langer, R.; Specht, K. PD-L1 and PD-1 and characterization of tumor-infiltrating lymphocytes in high grade sarcomas of soft tissue—Prognostic implications and rationale for immunotherapy. Oncoimmunology 2018, 7, e1389366. [Google Scholar] [CrossRef] [PubMed]
- Richert, I.; Gomez-Brouchet, A.; Bouvier, C.; Du Bouexic De Pinieux, G.; Karanian, M.; Blay, J.-Y.; Dutour, A. The immune landscape of chondrosarcoma—Potential for therapeutic targeting of CSFR1+ macrophages. J. Bone Oncol. 2020, 20, 100271. [Google Scholar] [CrossRef]
- Aras, S.; Zaidi, M.R. TAMeless traitors: Macrophages in cancer progression and metastasis. Br. J. Cancer 2017, 117, 1583–1591. [Google Scholar] [CrossRef]
- Pardoll, D.M. The blockade of immune checkpoints in cancer immunotherapy. Nat. Rev. Cancer 2012, 12, 252–264. [Google Scholar] [CrossRef]
- Simard, F. Implication of Immune System in Chondrosarcoma Progression and Therapeutic Response: Could Immunotherapy Play a Role in Chondrosarcoma Treatment? Ph.D. Thesis, Université de Lyon, Lyon, France, 2016. [Google Scholar]
- Hadrup, S.; Donia, M.; Thor Straten, P. Effector CD4 and CD8 T cells and their role in the tumor microenvironment. Cancer Microenviron. 2013, 6, 123–133. [Google Scholar] [CrossRef]
- Kostine, M.; Cleven, A.H.; de Miranda, N.F.C.C.; Italiano, A.; Cleton-Jansen, A.-M.; Bovée, J.V.M.G. Analysis of PD-L1, T-cell infiltrate and HLA expression in chondrosarcoma indicates potential for response to immunotherapy specifically in the dedifferentiated subtype. Mod. Pathol. 2016, 29, 1028–1037. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Shoushtari, A.N.; Agaram, N.P.; Kuk, D.; Qin, L.-X.; Carvajal, R.D.; Dickson, M.A.; Gounder, M.; Keohan, M.L.; Schwartz, G.K.; et al. Prevalence of tumor-infiltrating lymphocytes and PD-L1 expression in the soft tissue sarcoma microenvironment. Hum. Pathol. 2015, 46, 357–365. [Google Scholar] [CrossRef]
- Thanindratarn, P.; Dean, D.C.; Nelson, S.D.; Hornicek, F.J.; Duan, Z. Advances in immune checkpoint inhibitors for bone sarcoma therapy. J. Bone Oncol. 2019, 15, 100221. [Google Scholar] [CrossRef]
- Zhu, Z.; Jin, Z.; Zhang, M.; Tang, Y.; Yang, G.; Yuan, X.; Yao, J.; Sun, D. Prognostic value of programmed death-ligand 1 in sarcoma: A meta-analysis. Oncotarget 2017, 8, 59570–59580. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhu, G.; Yang, Z.; Zeng, K.; Liu, F.; Sun, J. Expression of PD-L1/PD-L2 is associated with high proliferation index of Ki-67 but not with TP53 overexpression in chondrosarcoma. Int. J. Biol. Markers 2018, 33, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.J.; Ricciotti, R.W.; Mantilla, J.; Loggers, E.T.; Pollack, S.M.; Cranmer, L.D. Response to PD1 inhibition in conventional chondrosarcoma. J. Immunother. Cancer 2018, 6, 94. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Moon, Y.J.; Kwon, K.S.; Bae, J.S.; Wagle, S.; Kim, K.M.; Park, H.S.; Lee, H.; Moon, W.S.; Chung, M.J.; et al. Tumor infiltrating PD1-positive lymphocytes and the expression of PD-L1 predict poor prognosis of soft tissue sarcomas. PLoS ONE 2013, 8, e82870. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Cao, J.; Zhao, C.; Li, X.; Zhou, C.; Hirsch, F. TIM-3, a promising target for cancer immunotherapy. Onco. Targets. Ther. 2018, 11, 7005–7009. [Google Scholar] [CrossRef]
- Gao, X.; Zhu, Y.; Li, G.; Huang, H.; Zhang, G.; Wang, F.; Sun, J.; Yang, Q.; Zhang, X.; Lu, B. TIM-3 Expression Characterizes Regulatory T Cells in Tumor Tissues and Is Associated with Lung Cancer Progression. PLoS ONE 2012, 7, e30676. [Google Scholar] [CrossRef]
- Komohara, Y.; Morita, T.; Annan, D.A.; Horlad, H.; Ohnishi, K.; Yamada, S.; Nakayama, T.; Kitada, S.; Suzu, S.; Kinoshita, I.; et al. The Coordinated Actions of TIM-3 on Cancer and Myeloid Cells in the Regulation of Tumorigenicity and Clinical Prognosis in Clear Cell Renal Cell Carcinomas. Cancer Immunol. Res. 2015, 3, 999–1007. [Google Scholar] [CrossRef]
- Andrews, L.P.; Marciscano, A.E.; Drake, C.G.; Vignali, D.A.A. LAG3 (CD223) as a cancer immunotherapy target. Immunol. Rev. 2017, 276, 80–96. [Google Scholar] [CrossRef]
- D’Angelo, S.P.; Tap, W.D.; Schwartz, G.K.; Carvajal, R.D. Sarcoma Immunotherapy: Past Approaches and Future Directions. Sarcoma 2014, 2014, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Leach, D.R.; Krummel, M.F.; Allison, J.P. Enhancement of Antitumor Immunity by CTLA-4 Blockade. Science 1996, 271, 1734–1736. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, Q.; Chen, W.; Shan, B.; Ding, Y.; Zhang, G.; Cao, N.; Liu, L.; Zhang, Y. B7-H3 is Overexpressed in Patients Suffering Osteosarcoma and Associated with Tumor Aggressiveness and Metastasis. PLoS ONE 2013, 8, e70689. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, M.; Huang, C.; Lin, C.; Tang, C.; Lin, C.; Lee, I.; Huang, H.; Chen, J. Basic fibroblast growth factor promotes doxorubicin resistance in chondrosarcoma cells by affecting XRCC5 expression. Mol. Carcinog. 2020, 59, 293–303. [Google Scholar] [CrossRef]
- Thomas, R.; Al-Khadairi, G.; Roelands, J.; Hendrickx, W.; Dermime, S.; Bedognetti, D.; Decock, J. NY-ESO-1 Based Immunotherapy of Cancer: Current Perspectives. Front. Immunol. 2018, 9, 947. [Google Scholar] [CrossRef]
- Endo, M.; de Graaff, M.A.; Ingram, D.R.; Lim, S.; Lev, D.C.; Briaire-de Bruijn, I.H.; Somaiah, N.; Bovée, J.V.; Lazar, A.J.; Nielsen, T.O. NY-ESO-1 (CTAG1B) expression in mesenchymal tumors. Mod. Pathol. 2015, 28, 587–595. [Google Scholar] [CrossRef] [PubMed]
- Pollack, S.M.; Li, Y.; Blaisdell, M.J.; Farrar, E.A.; Chou, J.; Hoch, B.L.; Loggers, E.T.; Rodler, E.; Eary, J.F.; Conrad, E.U.; et al. NYESO-1/LAGE-1s and PRAME Are Targets for Antigen Specific T Cells in Chondrosarcoma following Treatment with 5-Aza-2-Deoxycitabine. PLoS ONE 2012, 7, e32165. [Google Scholar] [CrossRef] [PubMed]
| Sarcoma Type | Genes | Chromosomal Aberrations |
|---|---|---|
| Soft tissue chondroma | HMGA2-LPPa | t (3;12) (q28; 214) |
| Mesenchymal chondrosarcoma | HEY1-NCOA2b | del (8) (q13; q21) |
| IRFBP2-CDX1c | t (1;5) (q42; q32) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zając, A.; Król, S.K.; Rutkowski, P.; Czarnecka, A.M. Biological Heterogeneity of Chondrosarcoma: From (Epi) Genetics through Stemness and Deregulated Signaling to Immunophenotype. Cancers 2021, 13, 1317. https://doi.org/10.3390/cancers13061317
Zając A, Król SK, Rutkowski P, Czarnecka AM. Biological Heterogeneity of Chondrosarcoma: From (Epi) Genetics through Stemness and Deregulated Signaling to Immunophenotype. Cancers. 2021; 13(6):1317. https://doi.org/10.3390/cancers13061317
Chicago/Turabian StyleZając, Agnieszka, Sylwia K. Król, Piotr Rutkowski, and Anna M. Czarnecka. 2021. "Biological Heterogeneity of Chondrosarcoma: From (Epi) Genetics through Stemness and Deregulated Signaling to Immunophenotype" Cancers 13, no. 6: 1317. https://doi.org/10.3390/cancers13061317
APA StyleZając, A., Król, S. K., Rutkowski, P., & Czarnecka, A. M. (2021). Biological Heterogeneity of Chondrosarcoma: From (Epi) Genetics through Stemness and Deregulated Signaling to Immunophenotype. Cancers, 13(6), 1317. https://doi.org/10.3390/cancers13061317








