Neo-Fs Index: A Novel Immunohistochemical Biomarker Panel Predicts Survival and Response to Anti-Angiogenetic Agents in Clear Cell Renal Cell Carcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
2.1. Baseline Characteristics of the Study Population
2.2. Immunohistochemical Marker Expression
2.3. Prognostic Impact of Immunohistochemical Markers
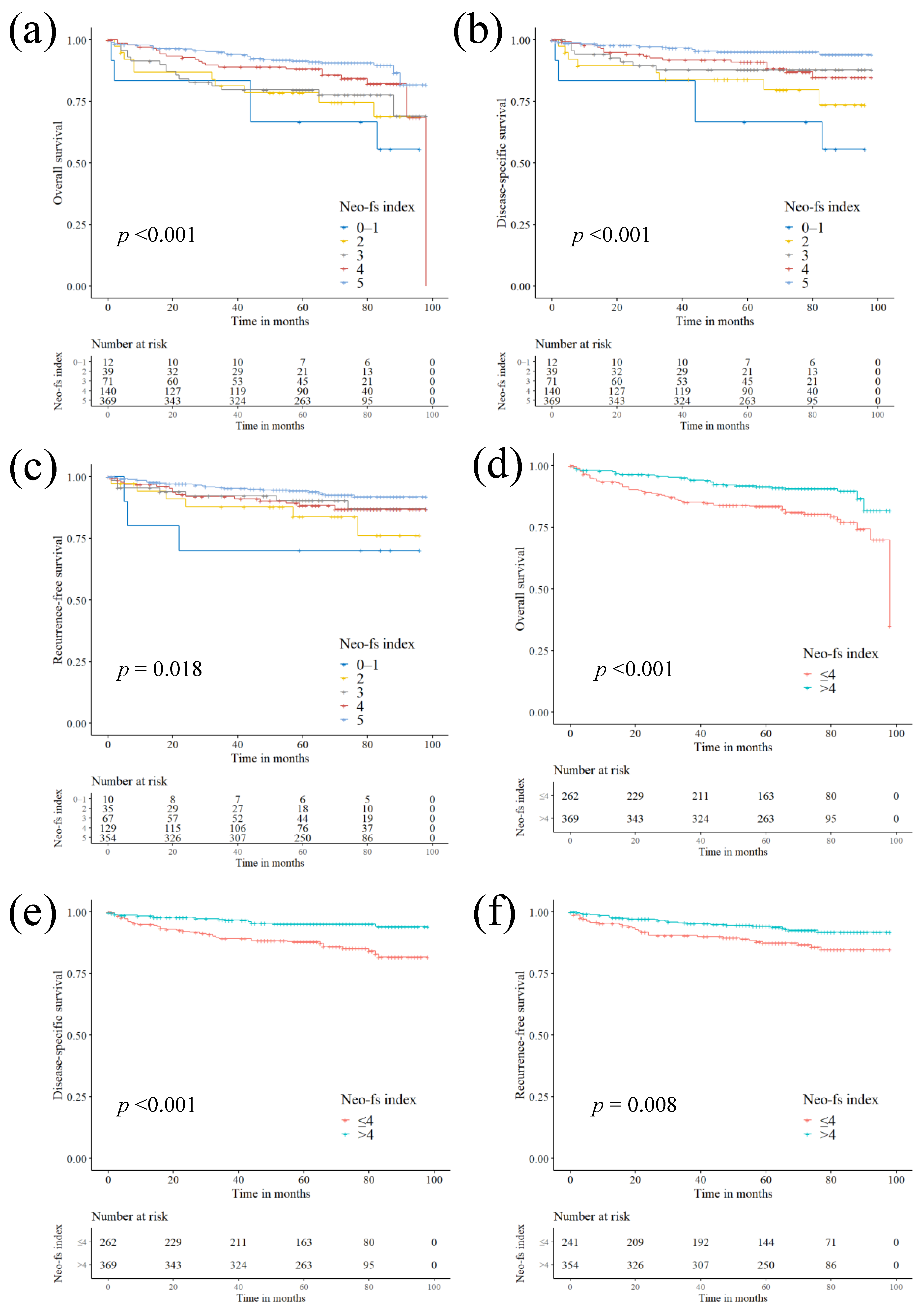
2.4. Prognostic Impact of Neo-Fs Index and its Association with Clinicopathological Characteristics
2.5. Impact of Immunohistochemical Markers and Neo-fs Index on the Treatment Response
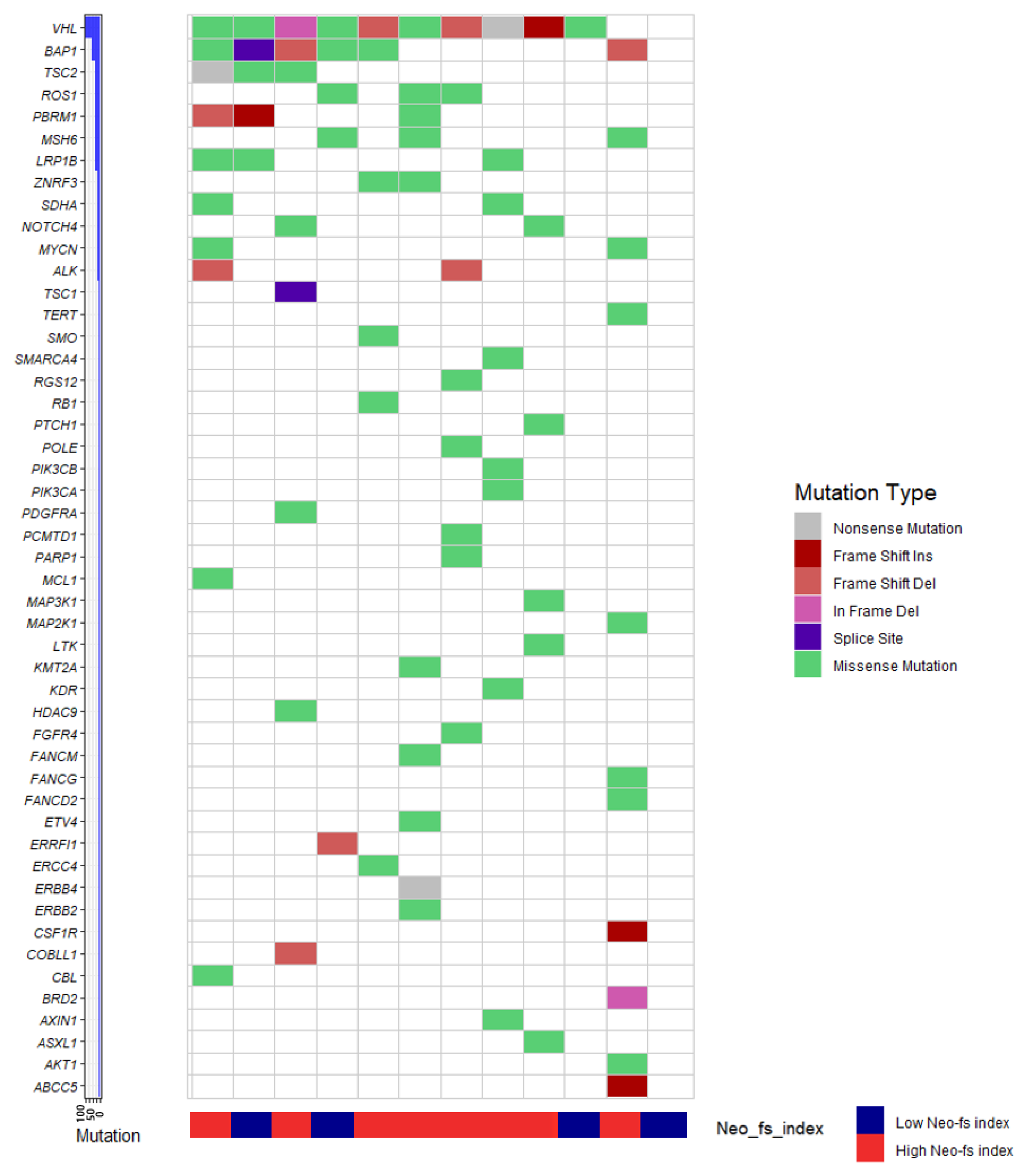
2.6. The Cancer Genome Atlas (TCGA) Gene Expression
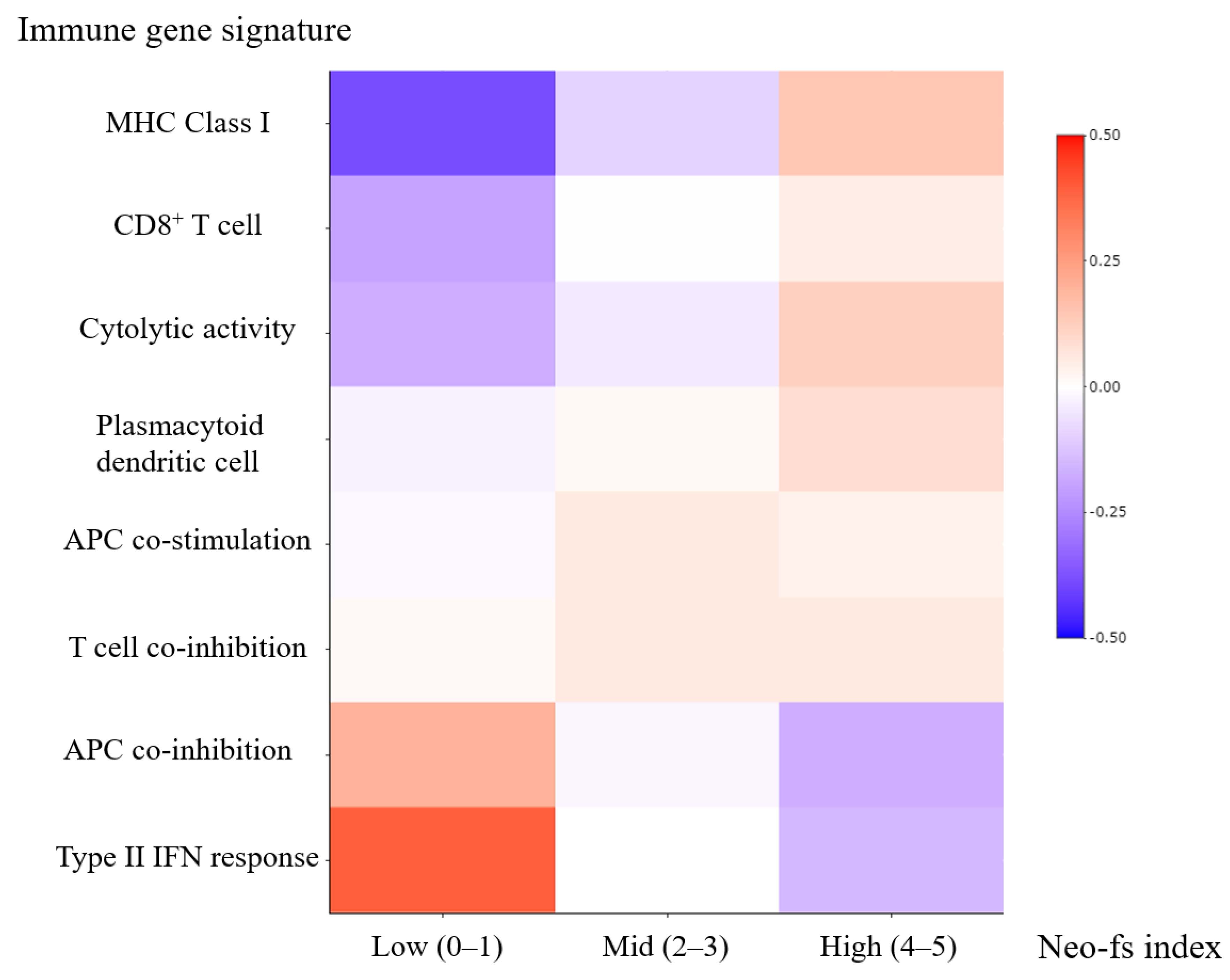
2.7. Molecular Phenotype of Clear Cell Renal Cell Carcinoma and Neo-fs Index
3. Discussion
4. Materials and Methods
4.1. Case Selection
4.2. Pathological Evaluation
4.3. Immunohistochemistry
4.4. Outcome Measures
4.5. TCGA Gene Expression Data
4.6. Targeted Next-Generation Sequencing
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Turajlic, S.; Litchfield, K.; Xu, H.; Rosenthal, R.; McGranahan, N.; Reading, J.L.; Wong, Y.N.S.; Rowan, A.; Kanu, N.; Al Bakir, M.; et al. Insertion-and-deletion-derived tumour-specific neoantigens and the immunogenic phenotype: A pan-cancer analysis. Lancet Oncol. 2017, 18, 1009–1021. [Google Scholar] [CrossRef]
- Moch, H.; Humphrey, P.A.; Ulbright, T.M. WHO Classification of Tumours of the Urinary System and Male Genital Organs; International Agency for Research on Cancer: Lyon, France, 2016. [Google Scholar]
- Bedke, J.; Gauler, T.; Grünwald, V.; Hegele, A.; Herrmann, E.; Hinz, S.; Janssen, J.; Schmitz, S.; Schostak, M.; Tesch, H.; et al. Systemic therapy in metastatic renal cell carcinoma. World J. Urol. 2017, 35, 179–188. [Google Scholar] [CrossRef]
- Choueiri, T.K.; Motzer, R.J. Systemic Therapy for Metastatic Renal-Cell Carcinoma. N. Engl. J. Med. 2017, 376, 354–366. [Google Scholar] [CrossRef] [PubMed]
- Motzer, R.J.; Escudier, B.; McDermott, D.F.; George, S.; Hammers, H.J.; Srinivas, S.; Tykodi, S.S.; Sosman, J.A.; Procopio, G.; Plimack, E.R.; et al. Nivolumab versus Everolimus in Advanced Renal-Cell Carcinoma. N. Engl. J. Med. 2015, 373, 1803–1813. [Google Scholar] [CrossRef]
- National Comprehensive Cancer Network. NCCN Clinical Practice Guidelines in Oncology: Kidney Cancer (Version 2.2020). Available online: https://www.nccn.org/professionals/physician_gls/pdf/kidney.pdf (accessed on 22 June 2020).
- Escudier, B.; Porta, C.; Schmidinger, M.; Rioux-Leclercq, N.; Bex, A.; Khoo, V.; Grünwald, V.; Gillessen, S.; Horwich, A. Renal cell carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up†. Ann. Oncol. 2019, 30, 706–720. [Google Scholar] [CrossRef] [PubMed]
- Rooney, M.S.; Shukla, S.A.; Wu, C.J.; Getz, G.; Hacohen, N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 2015, 160, 48–61. [Google Scholar] [CrossRef]
- Wang, W.; Man, X.; Zhan, X.; Gao, J.; Gong, Y.; Li, Z. Differential expressions of APC protein in pancreatic cancer and its precursor lesions. Chin. J. Gastroenterol. 2012, 17, 266–270. [Google Scholar] [CrossRef]
- Chu, D.; Zhang, Z.; Zhou, Y.; Wang, W.; Li, Y.; Zhang, H.; Dong, G.; Zhao, Q.; Ji, G. Notch1 and Notch2 have opposite prognostic effects on patients with colorectal cancer. Ann. Oncol. 2011, 22, 2440–2447. [Google Scholar] [CrossRef]
- Zhuang, Z.; Lin, J.; Huang, Y.; Lin, T.; Zheng, Z.; Ma, X. Notch 1 is a valuable therapeutic target against cell survival and proliferation in clear cell renal cell carcinoma. Oncol. Lett. 2017, 14, 3437–3444. [Google Scholar] [CrossRef][Green Version]
- Shen, J.; Ju, Z.; Zhao, W.; Wang, L.; Peng, Y.; Ge, Z.; Nagel, Z.D.; Zou, J.; Wang, C.; Kapoor, P.; et al. ARID1A deficiency promotes mutability and potentiates therapeutic antitumor immunity unleashed by immune checkpoint blockade. Nat. Med. 2018, 24, 556–562. [Google Scholar] [CrossRef]
- Lowenthal, B.M.; Nason, K.S.; Pennathur, A.; Luketich, J.D.; Pai, R.K.; Davison, J.M.; Ma, C. Loss of ARID1A expression is associated with DNA mismatch repair protein deficiency and favorable prognosis in advanced stage surgically resected esophageal adenocarcinoma. Hum. Pathol. 2019, 94, 1–10. [Google Scholar] [CrossRef]
- Donadon, M.; Di Tommaso, L.; Soldani, C.; Franceschini, B.; Terrone, A.; Mimmo, A.; Vitali, E.; Roncalli, M.; Lania, A.; Torzilli, G. Filamin A expression predicts early recurrence of hepatocellular carcinoma after hepatectomy. Liver Int. 2018, 38, 303–311. [Google Scholar] [CrossRef]
- Gachechiladze, M.; Skarda, J.; Janikova, M.; Mgebrishvili, G.; Kharaishvili, G.; Kolek, V.; Grygarkova, I.; Klein, J.; Poprachova, A.; Arabuli, M.; et al. Overexpression of filamin-A protein is associated with aggressive phenotype and poor survival outcomes in NSCLC patients treated with platinum-based combination chemotherapy. Neoplasma 2016, 63, 274–281. [Google Scholar] [CrossRef][Green Version]
- McGranahan, N.; Furness, A.J.; Rosenthal, R.; Ramskov, S.; Lyngaa, R.; Saini, S.K.; Jamal-Hanjani, M.; Wilson, G.A.; Birkbak, N.J.; Hiley, C.T.; et al. Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 2016, 351, 1463–1469. [Google Scholar] [CrossRef] [PubMed]
- Miao, D.; Margolis, C.A.; Gao, W.; Voss, M.H.; Li, W.; Martini, D.J.; Norton, C.; Bossé, D.; Wankowicz, S.M.; Cullen, D.; et al. Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 2018, 359, 801–806. [Google Scholar] [CrossRef]
- Voss, M.H.; Buros Novik, J.; Hellmann, M.D.; Ball, M.; Hakimi, A.A.; Miao, D.; Margolis, C.; Horak, C.; Wind-Rotolo, M.; De Velasco, G.; et al. Correlation of degree of tumor immune infiltration and insertion-and-deletion (indel) burden with outcome on programmed death 1 (PD1) therapy in advanced renal cell cancer (RCC). J. Clin. Oncol. 2018, 36 (Suppl. 15), 4518. [Google Scholar] [CrossRef]
- Yang, J.; Yan, J.; Liu, B. Targeting VEGF/VEGFR to Modulate Antitumor Immunity. Front. Immunol. 2018, 9, 978. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, L.M.A.; Fernandez, I.P.; Cassinello, J. Tyrosine kinase inhibitors reprogramming immunity in renal cell carcinoma: Rethinking cancer immunotherapy. Clin. Transl. Oncol. 2017, 19, 1175–1182. [Google Scholar] [CrossRef] [PubMed]
- Greifenberg, V.; Ribechini, E.; Rössner, S.; Lutz, M.B. Myeloid-derived suppressor cell activation by combined LPS and IFN-gamma treatment impairs DC development. Eur. J. Immunol. 2009, 39, 2865–2876. [Google Scholar] [CrossRef] [PubMed]
- Tartour, E.; Pere, H.; Maillere, B.; Terme, M.; Merillon, N.; Taieb, J.; Sandoval, F.; Quintin-Colonna, F.; Lacerda, K.; Karadimou, A.; et al. Angiogenesis and immunity: A bidirectional link potentially relevant for the monitoring of antiangiogenic therapy and the development of novel therapeutic combination with immunotherapy. Cancer Metastasis Rev. 2011, 30, 83–95. [Google Scholar] [CrossRef]
- Pal, S.K.; Hossain, D.M.; Zhang, Q.; Frankel, P.H.; Jones, J.O.; Carmichael, C.; Ruel, C.; Lau, C.; Kortylewski, M. Pazopanib as third line therapy for metastatic renal cell carcinoma: Clinical efficacy and temporal analysis of cytokine profile. J. Urol. 2015, 193, 1114–1121. [Google Scholar] [CrossRef]
- Finke, J.; Ko, J.; Rini, B.; Rayman, P.; Ireland, J.; Cohen, P. MDSC as a mechanism of tumor escape from sunitinib mediated anti-angiogenic therapy. Int. Immunopharmacol. 2011, 11, 856–861. [Google Scholar] [CrossRef]
- Snyder, A.; Makarov, V.; Merghoub, T.; Yuan, J.; Zaretsky, J.M.; Desrichard, A.; Walsh, L.A.; Postow, M.A.; Wong, P.; Ho, T.S.; et al. Genetic Basis for Clinical Response to CTLA-4 Blockade in Melanoma. N. Engl. J. Med. 2014, 371, 2189–2199. [Google Scholar] [CrossRef]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef]
- Hansen, U.K.; Ramskov, S.; Bjerregaard, A.M.; Borch, A.; Andersen, R.; Draghi, A.; Donia, M.; Bentzen, A.K.; Marquard, A.M.; Szallasi, Z.; et al. Tumor-Infiltrating T Cells From Clear Cell Renal Cell Carcinoma Patients Recognize Neoepitopes Derived From Point and Frameshift Mutations. Front. Immunol. 2020, 11, 373. [Google Scholar] [CrossRef] [PubMed]
- D’Aniello, C.; Berretta, M.; Cavaliere, C.; Rossetti, S.; Facchini, B.A.; Iovane, G.; Mollo, G.; Capasso, M.; Pepa, C.D.; Pesce, L.; et al. Biomarkers of Prognosis and Efficacy of Anti-angiogenic Therapy in Metastatic Clear Cell Renal Cancer. Front. Oncol. 2019, 9, 1400. [Google Scholar] [CrossRef] [PubMed]
- Amin, M.B.; Edge, S.; Greene, F.; Byrd, D.R.; Brookland, R.K.; Washington, M.K.; Gershenwald, J.E.; Compton, C.C.; Hess, K.R. (Eds.) AJCC Cancer Staging Manual, 8th ed.; Springer International Publishing: New York, NY, USA; American Joint Commission on Cancer: Chicago, IL, USA, 2017. [Google Scholar]
- Jeon, M.J.; Chun, S.M.; Lee, J.Y.; Choi, K.W.; Kim, D.; Kim, T.Y.; Jang, S.J.; Kim, W.B.; Shong, Y.K.; Song, D.E.; et al. Mutational profile of papillary thyroid microcarcinoma with extensive lymph node metastasis. Endocrine 2019, 64, 130–138. [Google Scholar] [CrossRef]
- Chun, S.M.; Sung, C.O.; Jeon, H.; Kim, T.I.; Lee, J.Y.; Park, H.; Kim, Y.; Kim, D.; Jang, S.J. Next-Generation Sequencing Using S1 Nuclease for Poor-Quality Formalin-Fixed, Paraffin-Embedded Tumor Specimens. J. Mol. Diagn. 2018, 20, 802–811. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.; Markovets, A.; Ahdesmaki, M.; Chapman, B.; Hofmann, O.; McEwen, R.; Johnson, J.; Dougherty, B.; Barrett, J.C.; Dry, J.R. VarDict: A novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016, 44, e108. [Google Scholar] [CrossRef]
- Cibulskis, K.; Lawrence, M.S.; Carter, S.L.; Sivachenko, A.; Jaffe, D.; Sougnez, C.; Gabriel, S.; Meyerson, M.; Lander, E.S.; Getz, G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 2013, 31, 213–219. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef] [PubMed]
- Lek, M.; Karczewski, K.J.; Minikel, E.V.; Samocha, K.E.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.H.; Ware, J.S.; Hill, A.J.; Cummings, B.B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef]
- McLaren, W.; Pritchard, B.; Rios, D.; Chen, Y.; Flicek, P.; Cunningham, F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics 2010, 26, 2069–2070. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.T.; Thorvaldsdóttir, H.; Winckler, W.; Guttman, M.; Lander, E.S.; Getz, G.; Mesirov, J.P. Integrative genomics viewer. Nat. Biotechnol. 2011, 29, 24–26. [Google Scholar] [CrossRef]
| Clinicopathological Characteristics | N (%) | Immunohistochemistry | N (%) |
|---|---|---|---|
| Sex | APC (0–1 vs. 2–3) | ||
| Male | 480 (75.2%) | Low expression | 548 (86.7%) |
| Female | 158 (24.8%) | High expression | 84 (13.3%) |
| Age (years) | NOTCH1 (0–1 vs. 2–3) | ||
| <55 years | 316 (49.5%) | Low expression | 436 (69.0%) |
| ≥55 years | 322 (50.5%) | High expression | 196 (31.0%) |
| Procedure | ARID1A (0–2 vs. 3) | ||
| Partial nephrectomy | 340 (53.3%) | Low expression | 627 (99.1%) |
| Radical nephrectomy | 298 (46.7%) | High expression | 6 (0.9%) |
| WHO/ISUP nuclear grade * | FAT1 (0–1 vs. 2–3) | ||
| 1‒2 | 331 (51.9%) | Low expression | 474 (74.9%) |
| 3‒4 | 307 (48.1%) | High expression | 159 (25.1%) |
| Tumor size (cm) | VHL (0 vs. 1–3) | ||
| <4 cm | 388 (60.8%) | Low expression | 177 (28.0%) |
| ≥4 cm | 250 (39.2%) | High expression | 455 (72.0%) |
| pT stage | EYS (0–1 vs. 2–3) | ||
| pT1‒2 | 496 (77.7%) | Low expression | 527 (83.0%) |
| pT3‒4 | 142 (22.3%) | High expression | 108 (17.0%) |
| pN stage | KMT2D (0–1 vs. 2–3) | ||
| pN0/pNx | 623 (97.6%) | Low expression | 296 (46.8%) |
| pN1 | 15 (2.4%) | High expression | 337 (53.2%) |
| Lymphovascular invasion | Filamin A (0–2 vs. 3) | ||
| Absent | 537 (84.2%) | Low expression | 579 (91.5%) |
| Present | 101 (15.8%) | High expression | 54 (8.5%) |
| Resection margin | PTEN (0 vs. 1–3) | ||
| Clear | 624 (97.8%) | Low expression | 112 (17.6%) |
| Involved | 14 (2.2%) | High expression | 526 (82.4%) |
| Necrosis | p53 (0 vs. 1–3) | ||
| Absent | 538 (84.3%) | Low expression | 36 (5.6%) |
| Present | 100 (15.7%) | High expression | 602 (94.4%) |
| Sarcomatoid change | |||
| Absent | 603 (94.5%) | ||
| Present | 35 (5.5%) | ||
| Anti-angiogenetic agent | |||
| Not received | 573 (89.8%) | ||
| Received | 65 (10.2%) | ||
| mTOR inhibitor | |||
| Not received | 600 (94.0%) | ||
| Received | 38 (6.0%) | ||
| Variables | Overall Survival | Disease-Specific Survival | Recurrence-Free Survival | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| Clinicopathologic variables | ||||||
| Female (vs. Male) | 0.729 (0.430–1.238) | 0.242 | 0.726 (0.376–1.402) | 0.341 | 0.800 (0.422–1.515) | 0.493 |
| Age ≥ 55 years | 3.328 (2.057–5.387) | <0.001 | 2.702 (1.516–4.817) | 0.001 | 2.316 (1.322–4.059) | 0.003 |
| Radical nephrectomy (vs. partial nephrectomy) | 3.797 (2.345–6.146) | <0.001 | 16.769 (6.069–46.335) | <0.001 | 3.915 (2.167–7.073) | <0.001 |
| ISUP grade 3–4 | 4.052 (2.464–6.663) | <0.001 | 12.064 (4.818–30.210) | <0.001 | 5.385 (2.785–10.414) | <0.001 |
| Tumor size ≥ 4 cm | 5.622 (3.474–9.097) | <0.001 | 19.062 (7.610–47.747) | <0.001 | 4.818 (2.724–8.520) | <0.001 |
| pT3–4 | 6.281 (4.117–9.584) | <0.001 | 16.709 (8.807–31.699) | <0.001 | 8.920 (5.205–15.289) | <0.001 |
| pN1 (vs. pN0/pNx) | 15.837 (8.688–28.868) | <0.001 | 26.214 (13.893–49.463) | <0.001 | 69.925 (25.878–188.944) | <0.001 |
| Lymphovascular invasion | 7.281 (4.777–11.097) | <0.001 | 12.505 (7.250–21.569) | <0.001 | 6.041 (3.522–10.360) | <0.001 |
| Margin involvement | 5.792 (2.793–12.010) | <0.001 | 7.757 (3.511–17.136) | <0.001 | 9.450 (4.038–22.113) | <0.001 |
| Necrosis | 7.462 (4.926–11.304) | <0.001 | 23.111 (12.436–42.951) | <0.001 | 8.777 (5.172–14.893) | <0.001 |
| Sarcomatoid change | 7.289 (4.416–12.031) | <0.001 | 12.974 (7.516–22.396) | <0.001 | 9.280 (4.792–17.970) | <0.001 |
| AAA recipient | 11.146 (7.334–16.938) | <0.001 | 36.948 (20.155–67.735) | <0.001 | 56.860 (32.589–99.207) | <0.001 |
| mTOR inhibitor recipient | 13.798 (8.881–21.438) | <0.001 | 32.525 (19.109–55.362) | <0.001 | 46.282 (24.568–87.191) | <0.001 |
| Immunohistochemistry | ||||||
| High APC expression | 1.663 (0.979–2.827) | 0.060 | 2.129 (1.143–3.966) | 0.017 | 1.537 (0.774–3.049) | 0.219 |
| High NOTCH1 expression | 1.806 (1.182–2.758) | 0.006 | 2.029 (1.195–3.447) | 0.009 | 1.835 (1.077–3.128) | 0.026 |
| High ARID1A expression | 4.634 (1.675–12.820) | 0.003 | 6.290 (1.954–20.251) | 0.002 | 2.307 (0.319–16.687) | 0.408 |
| High FAT1 expression | 0.659 (0.383–1.134) | 0.132 | 0.415 (0.188–0.916) | 0.029 | 0.627 (0.315–1.245) | 0.182 |
| High VHL expression | 0.573 (0.375–0.877) | 0.010 | 0.482 (0.286–0.814) | 0.006 | 0.527 (0.307–0.904) | 0.020 |
| High EYS expression | 2.416 (1.540–3.789) | <0.001 | 3.294 (1.911–5.676) | <0.001 | 1.710 (0.919–3.180) | 0.090 |
| High KMT2D expression | 0.859 (0.562–1.313) | 0.483 | 0.967 (0.566–1.653) | 0.904 | 0.705 (0.413–1.205) | 0.201 |
| High Filamin A expression | 2.439 (1.417–4.198) | 0.001 | 3.826 (2.080–7.040) | <0.001 | 3.217 (1.659–6.236) | 0.001 |
| High PTEN expression | 0.438 (0.280–0.686) | <0.001 | 0.284 (0.167–0.482) | <0.001 | 0.637 (0.337–1.207) | 0.167 |
| High p53 expression | 0.719 (0.329–1.570) | 0.408 | 0.387 (0.176–0.854) | 0.019 | 0.361 (0.164–0.798) | 0.012 |
| Neo-fs index | ||||||
| 0–1 | 4.497 (1.759–11.498) | 0.002 | 8.655 (3.206–23.369) | <0.001 | 4.715 (1.418–15.679) | 0.011 |
| 2 | 2.811 (1.388–5.694) | 0.004 | 4.553 (1.978–10.478) | <0.001 | 2.797 (1.143–6.843) | 0.024 |
| 3 | 2.424 (1.337–4.395) | 0.004 | 2.496 (1.085–5.741) | 0.031 | 1.647 (0.710–3.822) | 0.246 |
| 4 | 1.673 (0.981–2.853) | 0.059 | 2.392 (1.220–4.691) | 0.011 | 1.804 (0.946–3.439) | 0.073 |
| 5 (reference) | 1 | - | 1 | - | 1 | - |
| p-for trend | 0.690 (0.584–0.815) | <0.001 | 0.608 (0.499–0.741) | <0.001 | 0.711 (0.573–0.883) | 0.002 |
| Neo-fs index | ||||||
| Low (≤4) | 1 | - | 1 | - | 1 | - |
| High (>4) | 0.461 (0.301–0.708) | <0.001 | 0.331 (0.188–0.581) | <0.001 | 0.495 (0.291–0.844) | 0.010 |
| Variables | Overall Survival (OS) | Disease-Specific Survival (DSS) | Recurrence-Free Survival (RFS) | |||
|---|---|---|---|---|---|---|
| HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | |
| Clinicopathologic variables | ||||||
| Age ≥ 55 years | 3.005 (1.833–4.925) | <0.001 | 2.501 (1.365–4.585) | 0.003 | 1.671 (0.923–3.027) | 0.090 |
| Radical nephrectomy (vs. partial nephrectomy) | 0.915 (0.484–1.729) | 0.784 | 1.919 (0.593–6.207) | 0.276 | 0.811 (0.394–1.671) | 0.571 |
| ISUP grade 3–4 | 1.271 (0.704–2.296) | 0.426 | 1.799 (0.634–5.105) | 0.269 | 2.396 (1.098–5.227) | 0.028 |
| Tumor size ≥ 4 cm | 2.374 (1.220–4.620) | 0.011 | 3.516 (1.160–10.653) | 0.026 | 2.348 (1.056–5.220) | 0.036 |
| pT3–4 | 0.874 (0.442–1.729) | 0.699 | 0.824 (0.321–2.116) | 0.687 | 1.545 (0.707–3.379) | 0.276 |
| pN1 (vs. pN0/pNx) | 1.270 (0.585–2.757) | 0.546 | 1.112 (0.503–2.458) | 0.792 | 3.916 (1.075–14.266) | 0.038 |
| Lymphovascular invasion | 1.537 (0.657–3.593) | 0.322 | 1.409 (0.561–3.537) | 0.465 | 2.162 (1.101–4.245) | 0.025 |
| Margin involvement | 2.552 (1.441–4.519) | 0.001 | 2.527 (1.222–5.225) | 0.012 | 3.193 (1.033–9.870) | 0.044 |
| Necrosis | 1.633 (0.885–3.012) | 0.116 | 2.186 (0.948–5.038) | 0.066 | 1.386 (0.642–2.994) | 0.406 |
| Sarcomatoid change | 1.311 (0.710–2.420) | 0.387 | 1.396 (0.739–2.636) | 0.304 | 0.912 (0.392–2.122) | 0.830 |
| AAA recipient | 2.796 (1.342–5.825) | 0.006 | 6.642 (2.642–16.699) | <0.001 | 29.152 (13.253–64.125) | <0.001 |
| mTOR inhibitor recipient | 1.429 (0.696–2.934) | 0.330 | 1.219 (0.586–2.537) | 0.596 | 1.176 (0.540–2.562) | 0.683 |
| Immunohistochemistry | ||||||
| High APC expression | NA | NA | 2.717 (1.333–5.539) | 0.006 | NA | NA |
| High NOTCH1 expression | 1.694 (1.057–2.714) | 0.028 | 1.782 (0.963–3.298) | 0.066 | 2.021 (1.116–3.659) | 0.020 |
| High ARID1A expression | 4.558 (1.568–13.252) | 0.005 | 6.303 (1.726–23.016) | 0.005 | NA | NA |
| High FAT1 expression | 1.231 (0.690–2.197) | 0.483 | 1.287 (0.542–3.053) | 0.567 | NA | NA |
| High VHL expression | 1.131 (0.712–1.797) | 0.601 | 1.273 (0.712–2.276) | 0.415 | 1.003 (0.558–1.801) | 0.992 |
| High EYS expression | 1.806 (1.108–2.945) | 0.018 | 2.212 (1.188–4.119) | 0.012 | NA | NA |
| High Filamin A expression | 1.524 (0.795–2.920) | 0.204 | 2.108 (1.007–4.415) | 0.048 | 1.243 (0.497–3.112) | 0.642 |
| High PTEN expression | 0.977 (0.602–1.585) | 0.924 | 0.999 (0.562–1.775) | 0.998 | NA | NA |
| High p53 expression | NA | NA | 0.745 (0.325–1.707) | 0.486 | 0.930 (0.364–2.377) | 0.880 |
| Neo-fs index | ||||||
| 0–1 | 2.099 (0.775–5.688) | 0.145 | 3.135 (1.029–9.556) | 0.044 | 1.840 (0.454–7.457) | 0.393 |
| 2 | 2.285 (1.011–5.162) | 0.047 | 4.494 (1.578–12.800) | 0.005 | 2.935 (1.038–8.296) | 0.042 |
| 3 | 2.774 (1.486–5.177) | 0.001 | 3.007 (1.238–7.300) | 0.015 | 1.475 (0.564–3.86) | 0.428 |
| 4 | 1.128 (0.628–2.025) | 0.688 | 1.665 (0.749–3.699) | 0.211 | 2.265 (1.105–4.642) | 0.026 |
| 5 (reference) | 1 | - | 1 | - | 1 | - |
| p-for trend | 0.757 (0.632–0.907) | 0.003 | 0.690 (0.552–0.863) | 0.001 | 0.787 (0.615–1.007) | 0.057 |
| Neo-fs index | ||||||
| Low (≤4) | 1 | - | 1 | - | 1 | - |
| High (>4) | 0.595 (0.372–0.951) | 0.030 | 0.430 (0.225–0.825) | 0.011 | 0.480 (0.259–0.890) | 0.020 |
| Variables | Anti-Angiogenic Agent | mTOR Inhibitor | ||||
|---|---|---|---|---|---|---|
| PR/SD,PD | ORR | p | SD/PD | DCR | p | |
| Low APC | 17/36 | 32.1% | 0.092 | 7/18 | 28.0% | 0.999 |
| High APC | 0/8 | 0% | 1/2 | 33.3% | ||
| Low NOTCH1 | 12/26 | 31.6% | 0.406 | 5/14 | 26.3% | 0.573 |
| High NOTCH1 | 5/18 | 21.7% | 3/6 | 33.3% | ||
| Low ARID1A | 17/44 | 27.9% | 0.999 | 8/19 | 29.6% | 0.999 |
| High ARID1A | 0/1 | 0% | 0/1 | 0.0% | ||
| Low EYS | 15/29 | 34.1% | 0.114 | 8/13 | 38.1% | 0.075 |
| High EYS | 2/15 | 11.8% | 0/7 | 0% | ||
| Low Filamin A | 16/32 | 33.3% | 0.088 | 6/16 | 27.3% | 0.999 |
| High Filamin A | 1/12 | 7.7% | 2/4 | 33.3% | ||
| Indel signatures (positive number) | ||||||
| 0–1 | 0/3 | 0% | 0.027 | 0/2 | 0% | 0.742 |
| 2 | 1/4 | 20.0% | 0/1 | 0% | ||
| 3 | 1/8 | 11.1% | 2/1 | 66.7% | ||
| 4 | 3/14 | 17.6% | 2/6 | 25.0% | ||
| 5 | 12/15 | 44.4% | 4/10 | 28.6% | ||
| Indel signatures (positive number) | ||||||
| Neo-fs index ≤ 4 | 5/29 | 14.7% | 0.010 | 4/10 | 28.6% | 0.999 |
| Neo-fs index > 4 | 12/15 | 44.4% | 4/10 | 28.6% | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Park, J.-Y.; Shin, S.-J.; Lim, B.J.; Go, H. Neo-Fs Index: A Novel Immunohistochemical Biomarker Panel Predicts Survival and Response to Anti-Angiogenetic Agents in Clear Cell Renal Cell Carcinoma. Cancers 2021, 13, 1199. https://doi.org/10.3390/cancers13061199
Kim J, Park J-Y, Shin S-J, Lim BJ, Go H. Neo-Fs Index: A Novel Immunohistochemical Biomarker Panel Predicts Survival and Response to Anti-Angiogenetic Agents in Clear Cell Renal Cell Carcinoma. Cancers. 2021; 13(6):1199. https://doi.org/10.3390/cancers13061199
Chicago/Turabian StyleKim, Jisup, Jee-Young Park, Su-Jin Shin, Beom Jin Lim, and Heounjeong Go. 2021. "Neo-Fs Index: A Novel Immunohistochemical Biomarker Panel Predicts Survival and Response to Anti-Angiogenetic Agents in Clear Cell Renal Cell Carcinoma" Cancers 13, no. 6: 1199. https://doi.org/10.3390/cancers13061199
APA StyleKim, J., Park, J.-Y., Shin, S.-J., Lim, B. J., & Go, H. (2021). Neo-Fs Index: A Novel Immunohistochemical Biomarker Panel Predicts Survival and Response to Anti-Angiogenetic Agents in Clear Cell Renal Cell Carcinoma. Cancers, 13(6), 1199. https://doi.org/10.3390/cancers13061199






