DNA Damage Response during Replication Correlates with CIN70 Score and Determines Survival in HNSCC Patients
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
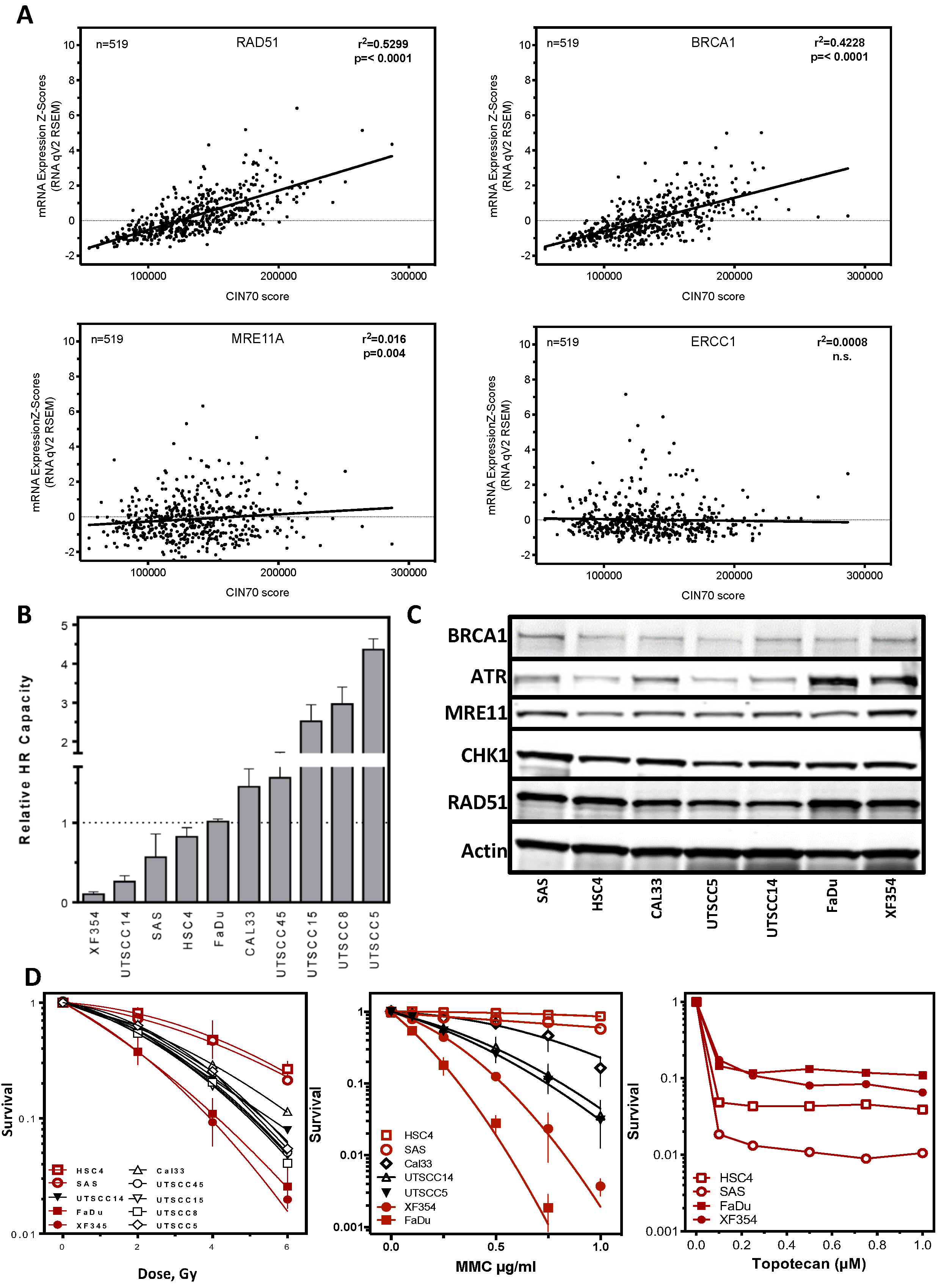
2.1. Functional Aneuploidy (CIN70) Inconclusively Determines Poorer Survival in HNSCC Patients
2.2. DNA Repair Protein Expression Does Not Reflect Cellular Resistance
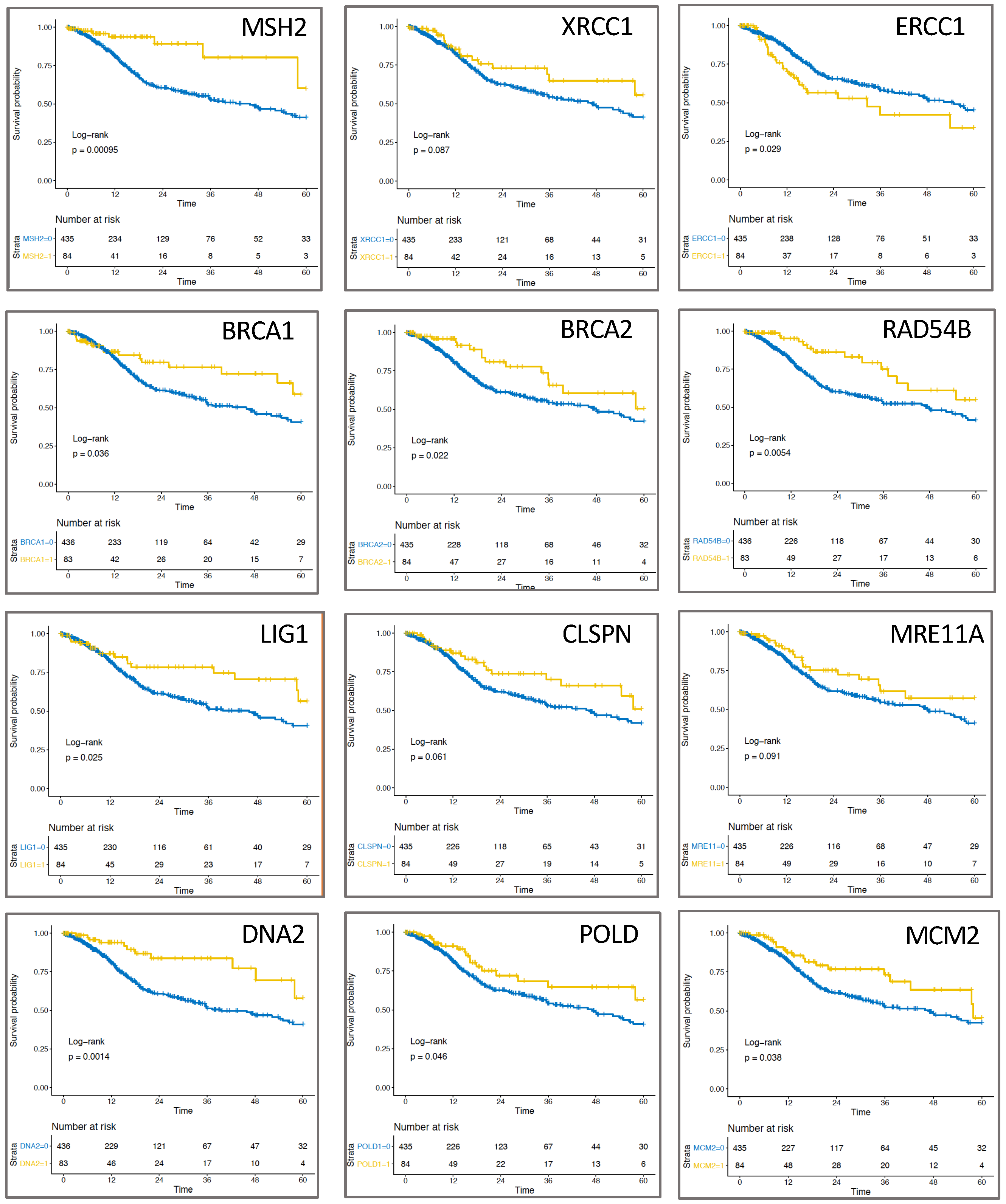
2.3. Expression of Replication-Associated Proteins Impacts Survival of HNSCC Patients
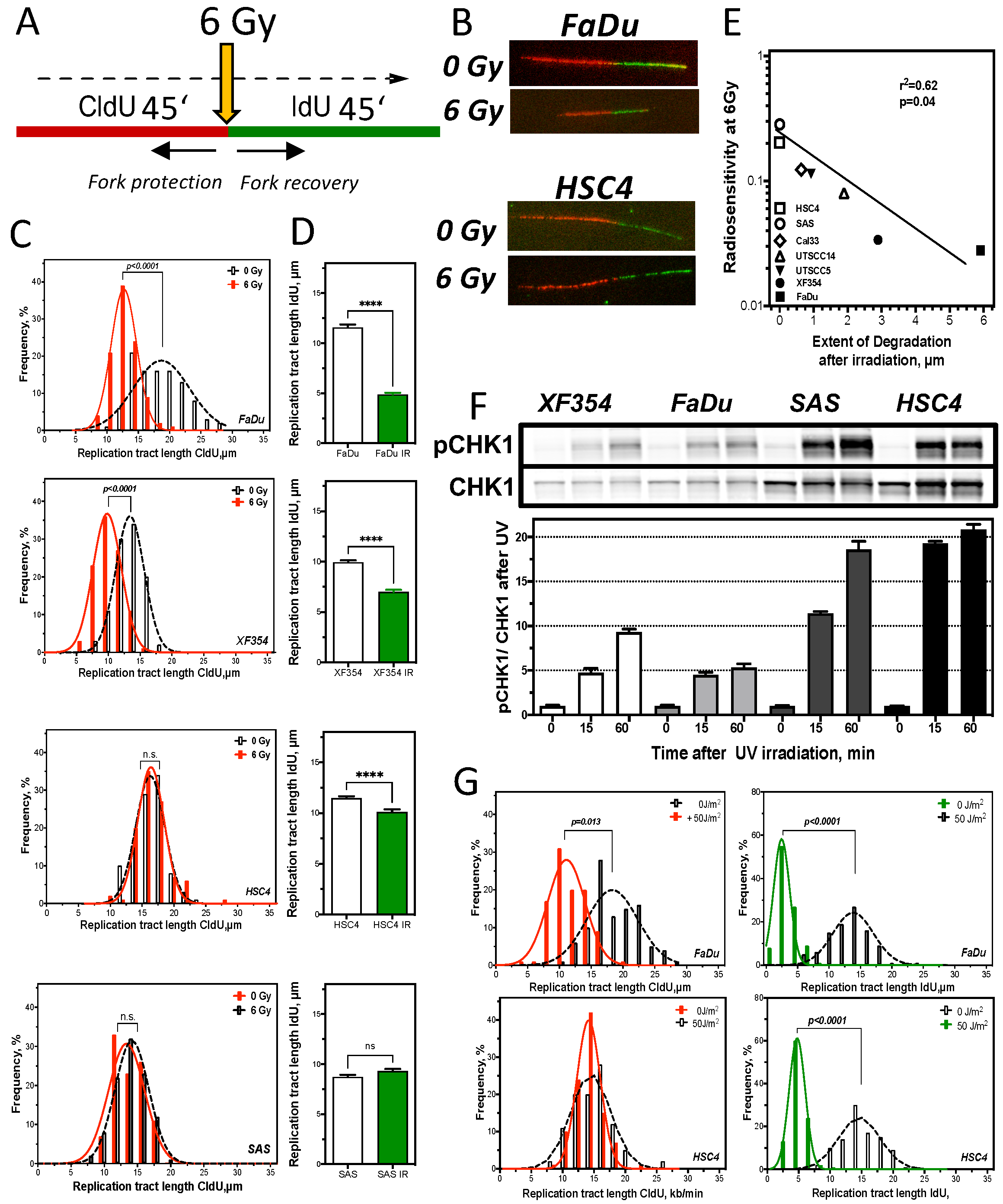
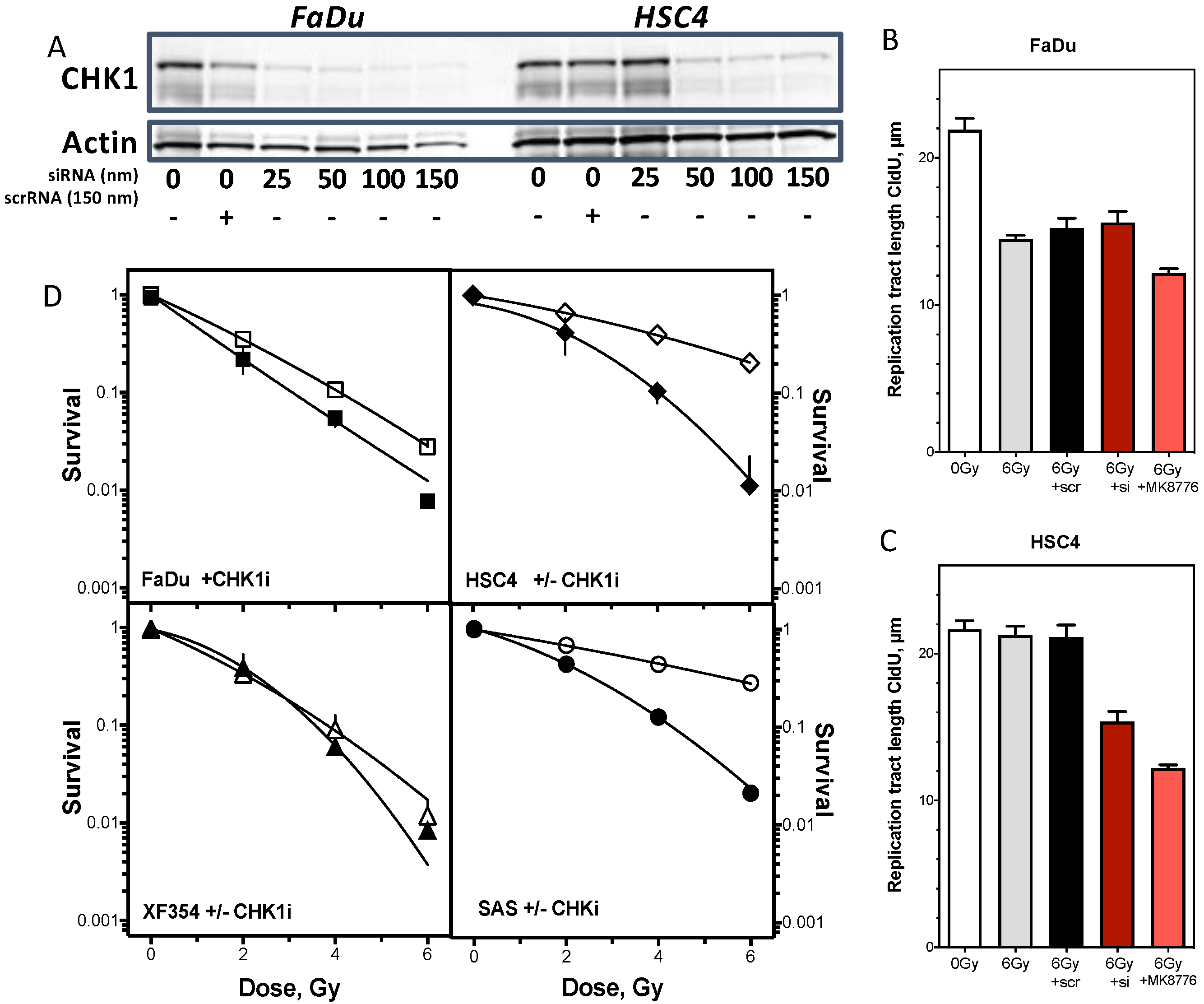
2.4. Resistance Is Mediated by Replication Fork Stability and Active DNA Damage Response of S Phase
3. Discussion
3.1. CIN70 Score in HNSCC
3.2. Replication Fork Stability Determines Resistance in HNSCC
4. Materials and Methods
4.1. Clinical in Silico Analysis
4.2. Cell Culture and Treatments
4.3. Western Blot
4.4. DNA Fiber Assay
4.5. Clonogenic Survival
4.6. Patient Survival and Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cancer Genome Atlas Network. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Dubot, C.; Bernard, V.; Sablin, M.P.; Vacher, S.; Chemlali, W.; Schnitzler, A.; Pierron, G.; Ait Rais, K.; Bessoltane, N.; Jeannot, E.; et al. Comprehensive genomic profiling of head and neck squamous cell carcinoma reveals FGFR1 amplifications and tumour genomic alterations burden as prognostic biomarkers of survival. Eur. J. Cancer 2018, 91, 47–55. [Google Scholar] [CrossRef]
- Verhagen, C.V.M.; Vossen, D.M.; Borgmann, K.; Hageman, F.; Grenman, R.; Verwijs-Janssen, M.; Mout, L.; Kluin, R.J.C.; Nieuwland, M.; Severson, T.M.; et al. Fanconi anemia and homologous recombination gene variants are associated with functional DNA repair defects in vitro and poor outcome in patients with advanced head and neck squamous cell carcinoma. Oncotarget 2018, 9, 18198–18213. [Google Scholar] [CrossRef]
- Wurster, S.; Hennes, F.; Parplys, A.C.; Seelbach, J.I.; Mansour, W.Y.; Zielinski, A.; Petersen, C.; Clauditz, T.S.; Munscher, A.; Friedl, A.A.; et al. PARP1 inhibition radiosensitizes HNSCC cells deficient in homologous recombination by disabling the DNA replication fork elongation response. Oncotarget 2016, 7, 9732–9741. [Google Scholar] [CrossRef] [PubMed]
- Connell, P.P.; Jayathilaka, K.; Haraf, D.J.; Weichselbaum, R.R.; Vokes, E.E.; Lingen, M.W. Pilot study examining tumor expression of RAD51 and clinical outcomes in human head cancers. Int. J. Oncol. 2006, 28, 1113–1119. [Google Scholar] [CrossRef]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in cancer. Cell 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Andor, N.; Graham, T.A.; Jansen, M.; Xia, L.C.; Aktipis, C.A.; Petritsch, C.; Ji, H.P.; Maley, C.C. Pan-cancer analysis of the extent and consequences of intratumor heterogeneity. Nat. Med. 2016, 22, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Knijnenburg, T.A.; Wang, L.; Zimmermann, M.T.; Chambwe, N.; Gao, G.F.; Cherniack, A.D.; Fan, H.; Shen, H.; Way, G.P.; Greene, C.S.; et al. Genomic and molecular landscape of DNA damage repair deficiency across the cancer genome atlas. Cell Rep. 2018, 23, 239–254.e6. [Google Scholar] [CrossRef]
- Lengauer, C.; Kinzler, K.W.; Vogelstein, B. Genetic instabilities in human cancers. Nature 1998, 396, 643–649. [Google Scholar] [CrossRef]
- Geigl, J.B.; Obenauf, A.C.; Schwarzbraun, T.; Speicher, M.R. Defining ‘chromosomal instability’. Trends Genet. 2008, 24, 64–69. [Google Scholar] [CrossRef]
- Carter, S.L.; Eklund, A.C.; Kohane, I.S.; Harris, L.N.; Szallasi, Z. A signature of chromosomal instability inferred from gene expression profiles predicts clinical outcome in multiple human cancers. Nat. Genet. 2006, 38, 1043–1048. [Google Scholar] [CrossRef] [PubMed]
- Begg, A.C.; Stewart, F.A.; Vens, C. Strategies to improve radiotherapy with targeted drugs. Nat. Rev. Cancer 2011, 11, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, A.J.; Lakshman, M.; Chan, N.; Bristow, R.G. Poly(ADP-ribose) polymerase inhibition as a model for synthetic lethality in developing radiation oncology targets. Semin. Radiat. Oncol. 2010, 20, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Kotter, A.; Cornils, K.; Borgmann, K.; Dahm-Daphi, J.; Petersen, C.; Dikomey, E.; Mansour, W.Y. Inhibition of PARP1-dependent end-joining contributes to Olaparib-mediated radiosensitization in tumor cells. Mol. Oncol. 2014, 8, 1616–1625. [Google Scholar] [CrossRef] [PubMed]
- Min, W.; Bruhn, C.; Grigaravicius, P.; Zhou, Z.W.; Li, F.; Kruger, A.; Siddeek, B.; Greulich, K.O.; Popp, O.; Meisezahl, C.; et al. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat. Commun. 2013, 4, 2993. [Google Scholar] [CrossRef] [PubMed]
- Southgate, H.E.D.; Chen, L.; Tweddle, D.A.; Curtin, N.J. ATR inhibition potentiates PARP inhibitor cytotoxicity in high risk neuroblastoma cell lines by multiple mechanisms. Cancers 2020, 12, 1095. [Google Scholar] [CrossRef] [PubMed]
- Busch, C.J.; Kroger, M.S.; Jensen, J.; Kriegs, M.; Gatzemeier, F.; Petersen, C.; Munscher, A.; Rothkamm, K.; Rieckmann, T. G2-checkpoint targeting and radiosensitization of HPV/p16-positive HNSCC cells through the inhibition of Chk1 and Wee1. Radiother. Oncol. 2017, 122, 260–266. [Google Scholar] [CrossRef]
- Tennstedt, P.; Fresow, R.; Simon, R.; Marx, A.; Terracciano, L.; Petersen, C.; Sauter, G.; Dikomey, E.; Borgmann, K. RAD51 overexpression is a negative prognostic marker for colorectal adenocarcinoma. Int. J. Cancer 2013, 132, 2118–2126. [Google Scholar] [CrossRef]
- Parplys, A.C.; Petermann, E.; Petersen, C.; Dikomey, E.; Borgmann, K. DNA damage by X-rays and their impact on replication processes. Radiother. Oncol. 2012, 102, 466–471. [Google Scholar] [CrossRef]
- Parplys, A.C.; Seelbach, J.I.; Becker, S.; Behr, M.; Wrona, A.; Jend, C.; Mansour, W.Y.; Joosse, S.A.; Stuerzbecher, H.W.; Pospiech, H.; et al. High levels of RAD51 perturb DNA replication elongation and cause unscheduled origin firing due to impaired CHK1 activation. Cell Cycle 2015, 14, 3190–3202. [Google Scholar] [CrossRef] [PubMed]
- Rieckhoff, J.; Meyer, F.; Classen, S.; Zielinski, A.; Riepen, B.; Wikman, H.; Petersen, C.; Rothkamm, K.; Borgmann, K.; Parplys, A.C. Exploiting chromosomal instability of PTEN-deficient triple-negative breast cancer cell lines for the sensitization against PARP1 inhibition in a replication-dependent manner. Cancers 2020, 12, 2809. [Google Scholar] [CrossRef]
- Limoli, C.L.; Giedzinski, E.; Bonner, W.M.; Cleaver, J.E. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma -H2AX formation, and Mre11 relocalization. Proc. Natl. Acad. Sci. USA 2002, 99, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Smeets, S.J.; Brakenhoff, R.H.; Ylstra, B.; van Wieringen, W.N.; van de Wiel, M.A.; Leemans, C.R.; Braakhuis, B.J. Genetic classification of oral and oropharyngeal carcinomas identifies subgroups with a different prognosis. Cell Oncol. 2009, 31, 291–300. [Google Scholar] [CrossRef]
- Pierssens, D.; Borgemeester, M.C.; van der Heijden, S.J.H.; Peutz-Kootstra, C.J.; Ruland, A.M.; Haesevoets, A.M.; Kessler, P.; Kremer, B.; Speel, E.M. Chromosome instability in tumor resection margins of primary OSCC is a predictor of local recurrence. Oral Oncol. 2017, 66, 14–21. [Google Scholar] [CrossRef]
- Vossen, D.M.; Verhagen, C.V.M.; van der Heijden, M.; Essers, P.B.M.; Bartelink, H.; Verheij, M.; Wessels, L.F.A.; van den Brekel, M.W.M.; Vens, C. Genetic factors associated with a poor outcome in head and neck cancer patients receiving definitive chemoradiotherapy. Cancers 2019, 11, 445. [Google Scholar] [CrossRef]
- Tinhofer, I.; Budach, V.; Saki, M.; Konschak, R.; Niehr, F.; Johrens, K.; Weichert, W.; Linge, A.; Lohaus, F.; Krause, M.; et al. Targeted next-generation sequencing of locally advanced squamous cell carcinomas of the head and neck reveals druggable targets for improving adjuvant chemoradiation. Eur. J. Cancer 2016, 57, 78–86. [Google Scholar] [CrossRef] [PubMed]
- Leemans, C.R.; Snijders, P.J.F.; Brakenhoff, R.H. The molecular landscape of head and neck cancer. Nat. Rev. Cancer 2018, 18, 269–282. [Google Scholar] [CrossRef]
- Ock, C.Y.; Son, B.; Keam, B.; Lee, S.Y.; Moon, J.; Kwak, H.; Kim, S.; Kim, T.M.; Jeon, Y.K.; Kwon, S.K.; et al. Identification of genomic mutations associated with clinical outcomes of induction chemotherapy in patients with head and neck squamous cell carcinoma. J. Cancer Res. Clin. Oncol. 2016, 142, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Birkbak, N.J.; Eklund, A.C.; Li, Q.; McClelland, S.E.; Endesfelder, D.; Tan, P.; Tan, I.B.; Richardson, A.L.; Szallasi, Z.; Swanton, C. Paradoxical relationship between chromosomal instability and survival outcome in cancer. Cancer Res. 2011, 71, 3447–3452. [Google Scholar] [CrossRef]
- Moeller, B.J.; Yordy, J.S.; Williams, M.D.; Giri, U.; Raju, U.; Molkentine, D.P.; Byers, L.A.; Heymach, J.V.; Story, M.D.; Lee, J.J.; et al. DNA repair biomarker profiling of head and neck cancer: Ku80 expression predicts locoregional failure and death following radiotherapy. Clin. Cancer Res. 2011, 17, 2035–2043. [Google Scholar] [CrossRef]
- Pavon, M.A.; Parreno, M.; Leon, X.; Sancho, F.J.; Cespedes, M.V.; Casanova, I.; Lopez-Pousa, A.; Mangues, M.A.; Quer, M.; Barnadas, A.; et al. Ku70 predicts response and primary tumor recurrence after therapy in locally advanced head and neck cancer. Int. J. Cancer 2008, 123, 1068–1079. [Google Scholar] [CrossRef]
- Welsh, J.W.; Ellsworth, R.K.; Kumar, R.; Fjerstad, K.; Martinez, J.; Nagel, R.B.; Eschbacher, J.; Stea, B. Rad51 protein expression and survival in patients with glioblastoma multiforme. Int. J. Radiat. Oncol. Biol. Phys. 2009, 74, 1251–1255. [Google Scholar] [CrossRef] [PubMed]
- Soderlund, K.; Skoog, L.; Fornander, T.; Askmalm, M.S. The BRCA1/BRCA2/Rad51 complex is a prognostic and predictive factor in early breast cancer. Radiother. Oncol. 2007, 84, 242–251. [Google Scholar] [CrossRef]
- Prochnow, S.; Wilczak, W.; Bosch, V.; Clauditz, T.S.; Muenscher, A. ERCC1, XPF and XPA-locoregional differences and prognostic value of DNA repair protein expression in patients with head and neck squamous cell carcinoma. Clin. Oral Investig. 2019, 23, 3319–3329. [Google Scholar] [CrossRef] [PubMed]
- Ang, M.K.; Patel, M.R.; Yin, X.Y.; Sundaram, S.; Fritchie, K.; Zhao, N.; Liu, Y.; Freemerman, A.J.; Wilkerson, M.D.; Walter, V.; et al. High XRCC1 protein expression is associated with poorer survival in patients with head and neck squamous cell carcinoma. Clin. Cancer Res. 2011, 17, 6542–6552. [Google Scholar] [CrossRef] [PubMed]
- Theocharis, S.; Klijanienko, J.; Giaginis, C.; Rodriguez, J.; Jouffroy, T.; Girod, A.; Point, D.; Tsourouflis, G.; Sastre-Garau, X. Expression of DNA repair proteins, MSH2, MLH1 and MGMT in mobile tongue squamous cell carcinoma: Associations with clinicopathological parameters and patients’ survival. J. Oral Pathol. Med. 2011, 40, 218–226. [Google Scholar] [CrossRef]
- Vaezi, A.; Wang, X.; Buch, S.; Gooding, W.; Wang, L.; Seethala, R.R.; Weaver, D.T.; D’Andrea, A.D.; Argiris, A.; Romkes, M.; et al. XPF expression correlates with clinical outcome in squamous cell carcinoma of the head and neck. Clin. Cancer Res. 2011, 17, 5513–5522. [Google Scholar] [CrossRef]
- Schulz, A.; Meyer, F.; Dubrovska, A.; Borgmann, K. Cancer stem cells and radioresistance: DNA repair and beyond. Cancers 2019, 11, 862. [Google Scholar] [CrossRef]
- Schlacher, K.; Christ, N.; Siaud, N.; Egashira, A.; Wu, H.; Jasin, M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell 2011, 145, 529–542. [Google Scholar] [CrossRef]
- Schlacher, K.; Wu, H.; Jasin, M. A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell 2012, 22, 106–116. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Ray Chaudhuri, A.; Lopes, M.; Costanzo, V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010, 17, 1305–1311. [Google Scholar] [CrossRef] [PubMed]
- Moiani, D.; Ronato, D.A.; Brosey, C.A.; Arvai, A.S.; Syed, A.; Masson, J.Y.; Petricci, E.; Tainer, J.A. Targeting allostery with avatars to design inhibitors assessed by cell activity: Dissecting MRE11 endo- and exonuclease activities. Methods Enzymol. 2018, 601, 205–241. [Google Scholar] [CrossRef]
- Mason, J.M.; Chan, Y.L.; Weichselbaum, R.W.; Bishop, D.K. Non-enzymatic roles of human RAD51 at stalled replication forks. Nat. Commun. 2019, 10, 4410. [Google Scholar] [CrossRef] [PubMed]
- Pardo, B.; Moriel-Carretero, M.; Vicat, T.; Aguilera, A.; Pasero, P. Homologous recombination and Mus81 promote replication completion in response to replication fork blockage. EMBO Rep. 2020, 21, e49367. [Google Scholar] [CrossRef] [PubMed]
- Tacconi, E.M.; Lai, X.; Folio, C.; Porru, M.; Zonderland, G.; Badie, S.; Michl, J.; Sechi, I.; Rogier, M.; Matia Garcia, V.; et al. BRCA1 and BRCA2 tumor suppressors protect against endogenous acetaldehyde toxicity. EMBO Mol. Med. 2017, 9, 1398–1414. [Google Scholar] [CrossRef]
- Vallerga, M.B.; Mansilla, S.F.; Federico, M.B.; Bertolin, A.P.; Gottifredi, V. Rad51 recombinase prevents Mre11 nuclease-dependent degradation and excessive PrimPol-mediated elongation of nascent DNA after UV irradiation. Proc. Natl. Acad. Sci. USA 2015, 112, E6624–E6633. [Google Scholar] [CrossRef] [PubMed]
- Calzetta, N.L.; Gonzalez Besteiro, M.A.; Gottifredi, V. Mus81-Eme1-dependent aberrant processing of DNA replication intermediates in mitosis impairs genome integrity. Sci. Adv. 2020, 6. [Google Scholar] [CrossRef]
- Nikkila, J.; Parplys, A.C.; Pylkas, K.; Bose, M.; Huo, Y.; Borgmann, K.; Rapakko, K.; Nieminen, P.; Xia, B.; Pospiech, H.; et al. Heterozygous mutations in PALB2 cause DNA replication and damage response defects. Nat. Commun. 2013, 4, 2578. [Google Scholar] [CrossRef]
- Wurlitzer, M.; Mockelmann, N.; Kriegs, M.; Vens, M.; Omidi, M.; Hoffer, K.; Bargen, C.V.; Moller-Koop, C.; Witt, M.; Droste, C.; et al. Mass spectrometric comparison of HPV-positive and HPV-negative oropharyngeal cancer. Cancers 2020, 12, 1531. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bold, I.T.; Specht, A.-K.; Droste, C.F.; Zielinski, A.; Meyer, F.; Clauditz, T.S.; Münscher, A.; Werner, S.; Rothkamm, K.; Petersen, C.; et al. DNA Damage Response during Replication Correlates with CIN70 Score and Determines Survival in HNSCC Patients. Cancers 2021, 13, 1194. https://doi.org/10.3390/cancers13061194
Bold IT, Specht A-K, Droste CF, Zielinski A, Meyer F, Clauditz TS, Münscher A, Werner S, Rothkamm K, Petersen C, et al. DNA Damage Response during Replication Correlates with CIN70 Score and Determines Survival in HNSCC Patients. Cancers. 2021; 13(6):1194. https://doi.org/10.3390/cancers13061194
Chicago/Turabian StyleBold, Ioan T., Ann-Kathrin Specht, Conrad F. Droste, Alexandra Zielinski, Felix Meyer, Till S. Clauditz, Adrian Münscher, Stefan Werner, Kai Rothkamm, Cordula Petersen, and et al. 2021. "DNA Damage Response during Replication Correlates with CIN70 Score and Determines Survival in HNSCC Patients" Cancers 13, no. 6: 1194. https://doi.org/10.3390/cancers13061194
APA StyleBold, I. T., Specht, A.-K., Droste, C. F., Zielinski, A., Meyer, F., Clauditz, T. S., Münscher, A., Werner, S., Rothkamm, K., Petersen, C., & Borgmann, K. (2021). DNA Damage Response during Replication Correlates with CIN70 Score and Determines Survival in HNSCC Patients. Cancers, 13(6), 1194. https://doi.org/10.3390/cancers13061194







