Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review
Abstract
Simple Summary
Abstract
1. Introduction
2. Increasing Genetic Knowledge to Improve Clinical Practice
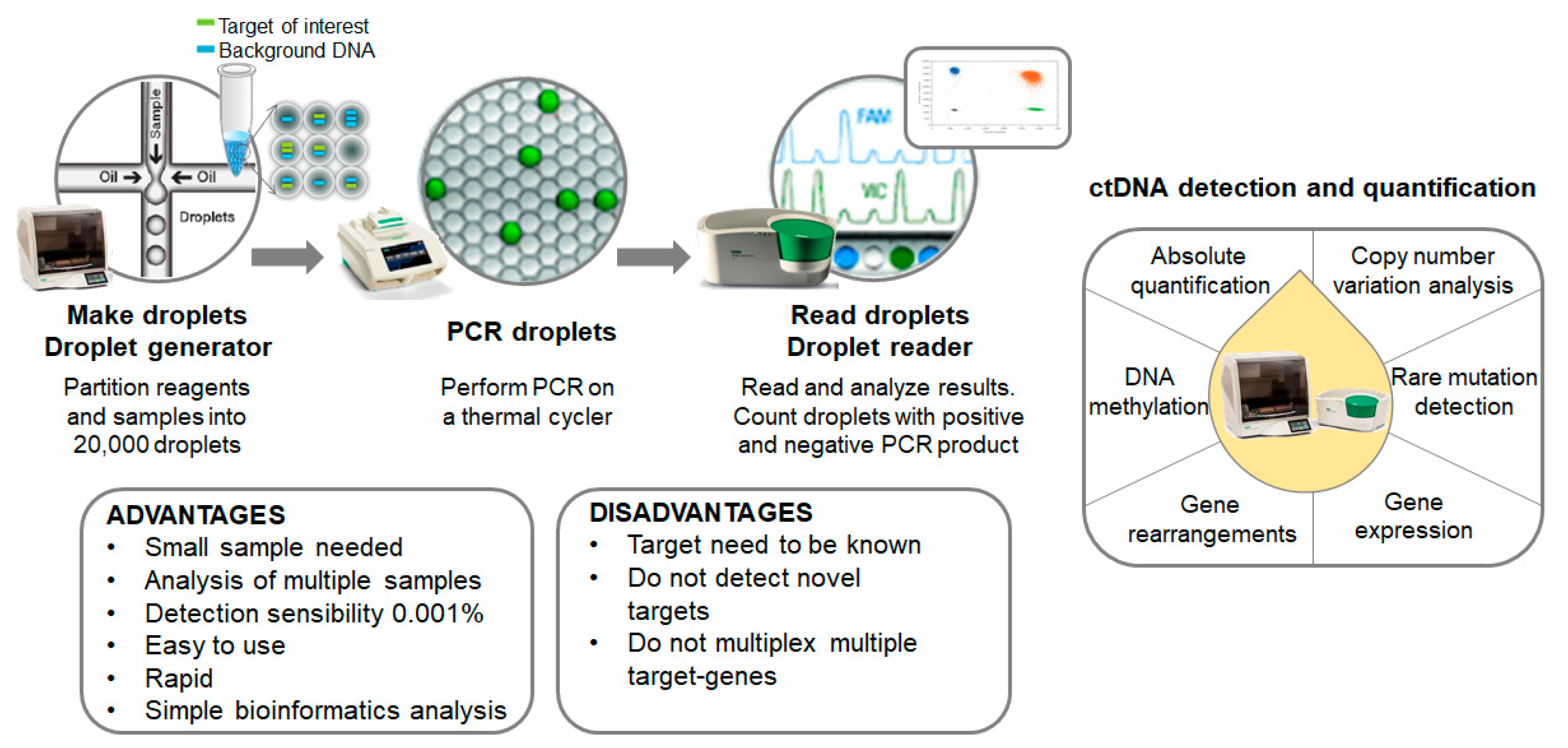
2.1. Digital-Droplet PCR Technology
2.1.1. ddPCR Research Approach
2.1.2. ddPCR Validation and Monitoring Approach
3. Future Perspectives
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Maisonneuve, P. Epidemiology and burden of pancreatic cancer. Presse Med. 2019, 48, e113–e123. [Google Scholar] [CrossRef]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef]
- Wolfgang, C.L.; Herman, J.M.; Laheru, D.A.; Klein, A.P.; Erdek, M.A.; Fishman, E.K.; Hruban, R.H. Recent progress in pancreatic cancer. CA Cancer J. Clin. 2013, 63, 318–348. [Google Scholar] [CrossRef] [PubMed]
- Neoptolemos, J.P.; Palmer, D.H.; Ghaneh, P.; Psarelli, E.E.; Valle, J.W.; Halloran, C.M.; Faluyi, O.; O’Reilly, D.A.; Cunningham, D.; Wadsley, J.; et al. Comparison of adjuvant gemcitabine and capecitabine with gemcitabine monotherapy in patients with resected pancreatic cancer (ESPAC-4): A multicentre, open-label, randomised, phase 3 trial. Lancet 2017, 389, 1011–1024. [Google Scholar] [CrossRef]
- Heitzer, E.; Ulz, P.; Geigl, J.B. Circulating tumor DNA as a liquid biopsy for cancer. Clin. Chem. 2015, 61, 112–123. [Google Scholar] [CrossRef] [PubMed]
- Ignatiadis, M.; Lee, M.; Jeffrey, S.S. Circulating Tumor Cells and Circulating Tumor DNA: Challenges and Opportunities on the Path to Clinical Utility. Clin. Cancer Res. 2015, 21, 4786–4800. [Google Scholar] [CrossRef]
- Diehl, F.; Schmidt, K.; Choti, M.A.; Romans, K.; Goodman, S.; Li, M.; Thornton, K.; Agrawal, N.; Sokoll, L.; Szabo, S.A.; et al. Circulating mutant DNA to assess tumor dynamics. Nat. Med. 2008, 14, 985–990. [Google Scholar] [CrossRef] [PubMed]
- Sausen, M.; Phallen, J.; Adleff, V.; Jones, S.; Leary, R.J.; Barrett, M.T.; Anagnostou, V.; Parpart-Li, S.; Murphy, D.; Kay Li, Q.; et al. Clinical implications of genomic alterations in the tumour and circulation of pancreatic cancer patients. Nat. Commun. 2015, 6, 7686. [Google Scholar] [CrossRef]
- Hadano, N.; Murakami, Y.; Uemura, K.; Hashimoto, Y.; Kondo, N.; Nakagawa, N.; Sueda, T.; Hiyama, E. Prognostic value of circulating tumour DNA in patients undergoing curative resection for pancreatic cancer. Br. J. Cancer 2016, 115, 59–65. [Google Scholar] [CrossRef]
- Abbosh, C.; Birkbak, N.J.; Wilson, G.A.; Jamal-Hanjani, M.; Constantin, T.; Salari, R.; Le Quesne, J.; Moore, D.A.; Veeriah, S.; Rosenthal, R.; et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 2017, 545, 446–451. [Google Scholar] [CrossRef]
- Husain, H.; Velculescu, V.E. Cancer DNA in the Circulation: The Liquid Biopsy. JAMA 2017, 318, 1272–1274. [Google Scholar] [CrossRef]
- Pietrasz, D.; Pecuchet, N.; Garlan, F.; Didelot, A.; Dubreuil, O.; Doat, S.; Imbert-Bismut, F.; Karoui, M.; Vaillant, J.C.; Taly, V.; et al. Plasma Circulating Tumor DNA in Pancreatic Cancer Patients Is a Prognostic Marker. Clin. Cancer Res. 2017, 23, 116–123. [Google Scholar] [CrossRef]
- Cohen, J.D.; Li, L.; Wang, Y.; Thoburn, C.; Afsari, B.; Danilova, L.; Douville, C.; Javed, A.A.; Wong, F.; Mattox, A.; et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018, 359, 926–930. [Google Scholar] [CrossRef] [PubMed]
- Isbell, J.M.; Jones, D.R.; Li, B.T. Circulating tumor DNA: A promising biomarker to guide postoperative treatment and surveillance of non-small cell lung cancer. J. Thorac. Cardiovasc. Surg. 2018, 155, 2628–2631. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.; Cohen, J.D.; Wang, Y.; Li, L.; Christie, M.; Simons, K.; Elsaleh, H.; Kosmider, S.; Wong, R.; Yip, D.; et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: A prospective biomarker study. Gut 2019, 68, 663–671. [Google Scholar] [CrossRef]
- Yachida, S.; Jones, S.; Bozic, I.; Antal, T.; Leary, R.; Fu, B.; Kamiyama, M.; Hruban, R.H.; Eshleman, J.R.; Nowak, M.A.; et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature 2010, 467, 1114–1117. [Google Scholar] [CrossRef]
- Iacobuzio-Donahue, C.A. Genetic evolution of pancreatic cancer: Lessons learnt from the pancreatic cancer genome sequencing project. Gut 2012, 61, 1085–1094. [Google Scholar] [CrossRef] [PubMed]
- Goonetilleke, K.S.; Siriwardena, A.K. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncol. 2007, 33, 266–270. [Google Scholar] [CrossRef] [PubMed]
- Poruk, K.E.; Gay, D.Z.; Brown, K.; Mulvihill, J.D.; Boucher, K.M.; Scaife, C.L.; Firpo, M.A.; Mulvihill, S.J. The clinical utility of CA 19-9 in pancreatic adenocarcinoma: Diagnostic and prognostic updates. Curr. Mol. Med. 2013, 13, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Cohen, J.D.; Javed, A.A.; Thoburn, C.; Wong, F.; Tie, J.; Gibbs, P.; Schmidt, C.M.; Yip-Schneider, M.T.; Allen, P.J.; Schattner, M.; et al. Combined circulating tumor DNA and protein biomarker-based liquid biopsy for the earlier detection of pancreatic cancers. Proc. Natl. Acad. Sci. USA 2017, 114, 10202–10207. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Desseigne, F.; Ychou, M.; Bouche, O.; Guimbaud, R.; Becouarn, Y.; Adenis, A.; Raoul, J.L.; Gourgou-Bourgade, S.; de la Fouchardiere, C.; et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N. Engl. J. Med. 2011, 364, 1817–1825. [Google Scholar] [CrossRef]
- Von Hoff, D.D.; Ervin, T.; Arena, F.P.; Chiorean, E.G.; Infante, J.; Moore, M.; Seay, T.; Tjulandin, S.A.; Ma, W.W.; Saleh, M.N.; et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013, 369, 1691–1703. [Google Scholar] [CrossRef] [PubMed]
- Wang-Gillam, A.; Li, C.P.; Bodoky, G.; Dean, A.; Shan, Y.S.; Jameson, G.; Macarulla, T.; Lee, K.H.; Cunningham, D.; Blanc, J.F.; et al. Nanoliposomal irinotecan with fluorouracil and folinic acid in metastatic pancreatic cancer after previous gemcitabine-based therapy (NAPOLI-1): A global, randomised, open-label, phase 3 trial. Lancet 2016, 387, 545–557. [Google Scholar] [CrossRef]
- Castillo, C.F.F. A Changing Landscape in Pancreatic Cancer. Ann. Surg. 2018, 268, 9–10. [Google Scholar] [CrossRef] [PubMed]
- Conroy, T.; Hammel, P.; Hebbar, M.; Ben Abdelghani, M.; Wei, A.C.; Raoul, J.L.; Chone, L.; Francois, E.; Artru, P.; Biagi, J.J.; et al. FOLFIRINOX or Gemcitabine as Adjuvant Therapy for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2395–2406. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Locker, G.Y.; Kindler, H.L. Maintenance Olaparib for Metastatic Pancreatic Cancer. Reply. N. Engl. J. Med. 2019, 381, 1492–1493. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, B.; Chakrabarty, M.; Cohn, E.M.; Leon, S.A. Determination of circulating DNA levels in patients with benign or malignant gastrointestinal disease. Cancer 1983, 51, 2116–2120. [Google Scholar] [CrossRef]
- Buscail, L.; Bournet, B.; Cordelier, P. Role of oncogenic KRAS in the diagnosis, prognosis and treatment of pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 153–168. [Google Scholar] [CrossRef] [PubMed]
- Bournet, B.; Buscail, C.; Muscari, F.; Cordelier, P.; Buscail, L. Targeting KRAS for diagnosis, prognosis, and treatment of pancreatic cancer: Hopes and realities. Eur. J. Cancer 2016, 54, 75–83. [Google Scholar] [CrossRef]
- Yamada, T.; Nakamori, S.; Ohzato, H.; Oshima, S.; Aoki, T.; Higaki, N.; Sugimoto, K.; Akagi, K.; Fujiwara, Y.; Nishisho, I.; et al. Detection of K-ras gene mutations in plasma DNA of patients with pancreatic adenocarcinoma: Correlation with clinicopathological features. Clin. Cancer Res. 1998, 4, 1527–1532. [Google Scholar]
- Castells, A.; Puig, P.; Mora, J.; Boadas, J.; Boix, L.; Urgell, E.; Sole, M.; Capella, G.; Lluis, F.; Fernandez-Cruz, L.; et al. K-ras mutations in DNA extracted from the plasma of patients with pancreatic carcinoma: Diagnostic utility and prognostic significance. J. Clin. Oncol. 1999, 17, 578–584. [Google Scholar] [CrossRef]
- Tjensvoll, K.; Lapin, M.; Buhl, T.; Oltedal, S.; Steen-Ottosen Berry, K.; Gilje, B.; Soreide, J.A.; Javle, M.; Nordgard, O.; Smaaland, R. Clinical relevance of circulating KRAS mutated DNA in plasma from patients with advanced pancreatic cancer. Mol. Oncol. 2016, 10, 635–643. [Google Scholar] [CrossRef] [PubMed]
- Del Re, M.; Vivaldi, C.; Rofi, E.; Vasile, E.; Miccoli, M.; Caparello, C.; d’Arienzo, P.D.; Fornaro, L.; Falcone, A.; Danesi, R. Early changes in plasma DNA levels of mutant KRAS as a sensitive marker of response to chemotherapy in pancreatic cancer. Sci. Rep. 2017, 7, 7931. [Google Scholar] [CrossRef]
- Yong, E. Cancer biomarkers: Written in blood. Nature 2014, 511, 524–526. [Google Scholar] [CrossRef]
- Postel, M.; Roosen, A.; Laurent-Puig, P.; Taly, V.; Wang-Renault, S.F. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: A cancer diagnostic perspective. Expert Rev. Mol. Diagn. 2018, 18, 7–17. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef] [PubMed]
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535. [Google Scholar] [CrossRef]
- Newman, A.M.; Bratman, S.V.; To, J.; Wynne, J.F.; Eclov, N.C.; Modlin, L.A.; Liu, C.L.; Neal, J.W.; Wakelee, H.A.; Merritt, R.E.; et al. An ultrasensitive method for quantitating circulating tumor DNA with broad patient coverage. Nat. Med. 2014, 20, 548–554. [Google Scholar] [CrossRef]
- Newman, A.M.; Lovejoy, A.F.; Klass, D.M.; Kurtz, D.M.; Chabon, J.J.; Scherer, F.; Stehr, H.; Liu, C.L.; Bratman, S.V.; Say, C.; et al. Integrated digital error suppression for improved detection of circulating tumor DNA. Nat. Biotechnol. 2016, 34, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Pecuchet, N.; Rozenholc, Y.; Zonta, E.; Pietrasz, D.; Didelot, A.; Combe, P.; Gibault, L.; Bachet, J.B.; Taly, V.; Fabre, E.; et al. Analysis of Base-Position Error Rate of Next-Generation Sequencing to Detect Tumor Mutations in Circulating DNA. Clin. Chem. 2016, 62, 1492–1503. [Google Scholar] [CrossRef] [PubMed]
- Vogelstein, B.; Kinzler, K.W. Digital PCR. Proc. Natl. Acad. Sci. USA 1999, 96, 9236–9241. [Google Scholar] [CrossRef]
- Dressman, D.; Yan, H.; Traverso, G.; Kinzler, K.W.; Vogelstein, B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl. Acad. Sci. USA 2003, 100, 8817–8822. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Diehl, F.; Dressman, D.; Vogelstein, B.; Kinzler, K.W. BEAMing up for detection and quantification of rare sequence variants. Nat. Methods 2006, 3, 95–97. [Google Scholar] [CrossRef] [PubMed]
- Perkins, G.; Lu, H.; Garlan, F.; Taly, V. Droplet-Based Digital PCR: Application in Cancer Research. Adv. Clin. Chem. 2017, 79, 43–91. [Google Scholar] [CrossRef] [PubMed]
- Alcaide, M.; Cheung, M.; Bushell, K.; Arthur, S.E.; Wong, H.L.; Karasinska, J.; Renouf, D.; Schaeffer, D.F.; McNamara, S.; Tertre, M.C.D.; et al. A Novel Multiplex Droplet Digital PCR Assay to Identify and Quantify KRAS Mutations in Clinical Specimens. J. Mol. Diagn. 2019, 21, 214–227. [Google Scholar] [CrossRef]
- Ako, S.; Nouso, K.; Kinugasa, H.; Dohi, C.; Matushita, H.; Mizukawa, S.; Muro, S.; Akimoto, Y.; Uchida, D.; Tomoda, T.; et al. Utility of serum DNA as a marker for KRAS mutations in pancreatic cancer tissue. Pancreatology 2017, 17, 285–290. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.W.; Schwerdel, D.; Costa, I.G.; Hackert, T.; Strobel, O.; Lam, S.; Barth, T.F.; Schroppel, B.; Meining, A.; Buchler, M.W.; et al. Detection of Hot-Spot Mutations in Circulating Cell-Free DNA From Patients With Intraductal Papillary Mucinous Neoplasms of the Pancreas. Gastroenterology 2016, 151, 267–270. [Google Scholar] [CrossRef]
- Bernard, V.; Kim, D.U.; San Lucas, F.A.; Castillo, J.; Allenson, K.; Mulu, F.C.; Stephens, B.M.; Huang, J.; Semaan, A.; Guerrero, P.A.; et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology 2019, 156, 108–118. [Google Scholar] [CrossRef]
- Cheng, H.; Liu, C.; Jiang, J.; Luo, G.; Lu, Y.; Jin, K.; Guo, M.; Zhang, Z.; Xu, J.; Liu, L.; et al. Analysis of ctDNA to predict prognosis and monitor treatment responses in metastatic pancreatic cancer patients. Int. J. Cancer 2017, 140, 2344–2350. [Google Scholar] [CrossRef]
- Cheng, H.; Luo, G.; Jin, K.; Fan, Z.; Huang, Q.; Gong, Y.; Xu, J.; Yu, X.; Liu, C. Kras mutation correlating with circulating regulatory T cells predicts the prognosis of advanced pancreatic cancer patients. Cancer Med. 2020, 9, 2153–2159. [Google Scholar] [CrossRef] [PubMed]
- Earl, J.; Garcia-Nieto, S.; Martinez-Avila, J.C.; Montans, J.; Sanjuanbenito, A.; Rodriguez-Garrote, M.; Lisa, E.; Mendia, E.; Lobo, E.; Malats, N.; et al. Circulating tumor cells (Ctc) and kras mutant circulating free Dna (cfdna) detection in peripheral blood as biomarkers in patients diagnosed with exocrine pancreatic cancer. BMC Cancer 2015, 15, 797. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Shi, X.; Shen, J.; Gao, S.; Wang, H.; Shen, S.; Pan, Y.; Li, B.; Xu, X.; Shao, Z.; et al. Preoperative detection of KRAS G12D mutation in ctDNA is a powerful predictor for early recurrence of resectable PDAC patients. Br. J. Cancer 2020, 122, 857–867. [Google Scholar] [CrossRef]
- Kim, M.K.; Woo, S.M.; Park, B.; Yoon, K.A.; Kim, Y.H.; Joo, J.; Lee, W.J.; Han, S.S.; Park, S.J.; Kong, S.Y. Prognostic Implications of Multiplex Detection of KRAS Mutations in Cell-Free DNA from Patients with Pancreatic Ductal Adenocarcinoma. Clin. Chem. 2018, 64, 726–734. [Google Scholar] [CrossRef] [PubMed]
- Kinugasa, H.; Nouso, K.; Miyahara, K.; Morimoto, Y.; Dohi, C.; Tsutsumi, K.; Kato, H.; Matsubara, T.; Okada, H.; Yamamoto, K. Detection of K-ras gene mutation by liquid biopsy in patients with pancreatic cancer. Cancer 2015, 121, 2271–2280. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Alnaggar, M.; Liang, S.; Chen, J.; Xu, K.; Dong, S.; Du, D.; Niu, L. Circulating Tumor DNA as a Sensitive Marker in Patients Undergoing Irreversible Electroporation for Pancreatic Cancer. Cell. Physiol. Biochem. 2018, 47, 1556–1564. [Google Scholar] [CrossRef]
- Mohan, S.; Ayub, M.; Rothwell, D.G.; Gulati, S.; Kilerci, B.; Hollebecque, A.; Sun Leong, H.; Smith, N.K.; Sahoo, S.; Descamps, T.; et al. Analysis of circulating cell-free DNA identifies KRAS copy number gain and mutation as a novel prognostic marker in Pancreatic cancer. Sci. Rep. 2019, 9, 11610. [Google Scholar] [CrossRef]
- Sefrioui, D.; Blanchard, F.; Toure, E.; Basile, P.; Beaussire, L.; Dolfus, C.; Perdrix, A.; Paresy, M.; Antonietti, M.; Iwanicki-Caron, I.; et al. Diagnostic value of CA19.9, circulating tumour DNA and circulating tumour cells in patients with solid pancreatic tumours. Br. J. Cancer 2017, 117, 1017–1025. [Google Scholar] [CrossRef]
- Sugimori, M.; Sugimori, K.; Tsuchiya, H.; Suzuki, Y.; Tsuyuki, S.; Kaneta, Y.; Hirotani, A.; Sanga, K.; Tozuka, Y.; Komiyama, S.; et al. Quantitative monitoring of circulating tumor DNA in patients with advanced pancreatic cancer undergoing chemotherapy. Cancer Sci. 2020, 111, 266–278. [Google Scholar] [CrossRef]
- Watanabe, F.; Suzuki, K.; Tamaki, S.; Abe, I.; Endo, Y.; Takayama, Y.; Ishikawa, H.; Kakizawa, N.; Saito, M.; Futsuhara, K.; et al. Longitudinal monitoring of KRAS-mutated circulating tumor DNA enables the prediction of prognosis and therapeutic responses in patients with pancreatic cancer. PLoS ONE 2019, 14, e0227366. [Google Scholar] [CrossRef] [PubMed]
- Woo, S.M.; Kim, M.K.; Joo, J.; Yoon, K.A.; Park, B.; Park, S.J.; Han, S.S.; Lee, J.H.; Hong, E.K.; Kim, Y.H.; et al. Induction Chemotherapy with Gemcitabine and Cisplatin Followed by Simultaneous Integrated Boost-Intensity Modulated Radiotherapy with Concurrent Gemcitabine for Locally Advanced Unresectable Pancreatic Cancer: Results from a Feasibility Study. Cancer Res. Treat. 2017, 49, 1022–1032. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra224. [Google Scholar] [CrossRef]
- Lee, B.; Cohen, J.; Lipton, R.L.; Tie, J.; Javed, A.A.; Li, L.; Goldstein, D.; Cooray, P.; Nagrial, A.; Burge, M.E.; et al. Potential role of circulating tumor DNA (ctDNA) in the early diagnosis and post-operative management of localised pancreatic cancer. J. Clin. Oncol. 2017, 35, 4101. [Google Scholar] [CrossRef]
- Jen, J.; Wu, L.; Sidransky, D. An overview on the isolation and analysis of circulating tumor DNA in plasma and serum. Ann. N. Y. Acad. Sci. 2000, 906, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.H.; Montalvo, L.; Chrebtow, V.; Busch, M.P. Quantitation of genomic DNA in plasma and serum samples: Higher concentrations of genomic DNA found in serum than in plasma. Transfusion 2001, 41, 276–282. [Google Scholar] [CrossRef]
- Taback, B.; O’Day, S.J.; Hoon, D.S. Quantification of circulating DNA in the plasma and serum of cancer patients. Ann. N. Y. Acad. Sci. 2004, 1022, 17–24. [Google Scholar] [CrossRef] [PubMed]
- Vallee, A.; Marcq, M.; Bizieux, A.; Kouri, C.E.; Lacroix, H.; Bennouna, J.; Douillard, J.Y.; Denis, M.G. Plasma is a better source of tumor-derived circulating cell-free DNA than serum for the detection of EGFR alterations in lung tumor patients. Lung Cancer 2013, 82, 373–374. [Google Scholar] [CrossRef] [PubMed]
- Andersen, R.F.; Jakobsen, A. Screening for circulating RAS/RAF mutations by multiplex digital PCR. Clin. Chim. Acta. 2016, 458, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Seufferlein, T.; Berger, A.W.; Schwerdel, D.; Ettrich, T.J.; Schmidt, S.A.; Kleger, A.; Marienfeld, R. Non-invasive diagnosis and tracking of tumor evolution by targeted sequencing of circulating tumor DNA in metastatic pancreatic cancer patients. J. Clin. Oncol. 2017, 35, e15769. [Google Scholar] [CrossRef]
- Wahl, S.G.F.; Dai, H.Y.; Emdal, E.F.; Ottestad, A.L.; Dale, V.G.; Richardsen, E.; Halvorsen, T.O.; Gronberg, B.H. Prognostic value of absolute quantification of mutated KRAS in circulating tumour DNA in lung adenocarcinoma patients prior to therapy. J. Pathol. Clin. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Michaelidou, K.; Koutoulaki, C.; Mavridis, K.; Vorrias, E.; Papadaki, M.A.; Koutsopoulos, A.V.; Mavroudis, D.; Agelaki, S. Detection of KRAS G12/G13 Mutations in Cell Free-DNA by Droplet Digital PCR, Offers Prognostic Information for Patients with Advanced Non-Small Cell Lung Cancer. Cells 2020, 9, 2514. [Google Scholar] [CrossRef]
- Guibert, N.; Pradines, A.; Farella, M.; Casanova, A.; Gouin, S.; Keller, L.; Favre, G.; Mazieres, J. Monitoring KRAS mutations in circulating DNA and tumor cells using digital droplet PCR during treatment of KRAS-mutated lung adenocarcinoma. Lung Cancer 2016, 100, 1–4. [Google Scholar] [CrossRef]
- Zhong, Q.; Bhattacharya, S.; Kotsopoulos, S.; Olson, J.; Taly, V.; Griffiths, A.D.; Link, D.R.; Larson, J.W. Multiplex digital PCR: Breaking the one target per color barrier of quantitative PCR. Lab Chip 2011, 11, 2167–2174. [Google Scholar] [CrossRef] [PubMed]
- McDermott, G.P.; Do, D.; Litterst, C.M.; Maar, D.; Hindson, C.M.; Steenblock, E.R.; Legler, T.C.; Jouvenot, Y.; Marrs, S.H.; Bemis, A.; et al. Multiplexed target detection using DNA-binding dye chemistry in droplet digital PCR. Anal. Chem. 2013, 85, 11619–11627. [Google Scholar] [CrossRef] [PubMed]
- Rice, A.; Del Rio Hernandez, A. The Mutational Landscape of Pancreatic and Liver Cancers, as Represented by Circulating Tumor DNA. Front. Oncol. 2019, 9, 952. [Google Scholar] [CrossRef]
- Adamo, P.; Cowley, C.M.; Neal, C.P.; Mistry, V.; Page, K.; Dennison, A.R.; Isherwood, J.; Hastings, R.; Luo, J.; Moore, D.A.; et al. Profiling tumour heterogeneity through circulating tumour DNA in patients with pancreatic cancer. Oncotarget 2017, 8, 87221–87233. [Google Scholar] [CrossRef] [PubMed]
- Berger, A.W.; Schwerdel, D.; Ettrich, T.J.; Hann, A.; Schmidt, S.A.; Kleger, A.; Marienfeld, R.; Seufferlein, T. Targeted deep sequencing of circulating tumor DNA in metastatic pancreatic cancer. Oncotarget 2018, 9, 2076–2085. [Google Scholar] [CrossRef] [PubMed]
- Takai, E.; Totoki, Y.; Nakamura, H.; Morizane, C.; Nara, S.; Hama, N.; Suzuki, M.; Furukawa, E.; Kato, M.; Hayashi, H.; et al. Clinical utility of circulating tumor DNA for molecular assessment in pancreatic cancer. Sci. Rep. 2015, 5, 18425. [Google Scholar] [CrossRef] [PubMed]
- Buscail, E.; Maulat, C.; Muscari, F.; Chiche, L.; Cordelier, P.; Dabernat, S.; Alix-Panabieres, C.; Buscail, L. Liquid Biopsy Approach for Pancreatic Ductal Adenocarcinoma. Cancers (Basel) 2019, 11, 852. [Google Scholar] [CrossRef]
- Fang, Z.; Meng, Q.; Zhang, B.; Shi, S.; Liu, J.; Liang, C.; Hua, J.; Yu, X.; Xu, J.; Wang, W. Prognostic value of circulating tumor DNA in pancreatic cancer: A systematic review and meta-analysis. Aging (Albany NY) 2020, 12. [Google Scholar] [CrossRef]
- Wang, Z.Y.; Ding, X.Q.; Zhu, H.; Wang, R.X.; Pan, X.R.; Tong, J.H. KRAS Mutant Allele Fraction in Circulating Cell-Free DNA Correlates With Clinical Stage in Pancreatic Cancer Patients. Front. Oncol. 2019, 9, 1295. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.S.; Fakih, M.G.; Strickler, J.H.; Desai, J.; Durm, G.A.; Shapiro, G.I.; Falchook, G.S.; Price, T.J.; Sacher, A.; Denlinger, C.S.; et al. KRAS(G12C) Inhibition with Sotorasib in Advanced Solid Tumors. N. Engl. J. Med. 2020, 383, 1207–1217. [Google Scholar] [CrossRef]
- Silveira, A.B.; Bidard, F.C.; Kasperek, A.; Melaabi, S.; Tanguy, M.L.; Rodrigues, M.; Bataillon, G.; Cabel, L.; Buecher, B.; Pierga, J.Y.; et al. High-Accuracy Determination of Microsatellite Instability Compatible with Liquid Biopsies. Clin. Chem. 2020, 66, 606–613. [Google Scholar] [CrossRef] [PubMed]
- Hussung, S.; Follo, M.; Klar, R.F.U.; Michalczyk, S.; Fritsch, K.; Nollmann, F.; Hipp, J.; Duyster, J.; Scherer, F.; von Bubnoff, N.; et al. Development and Clinical Validation of Discriminatory Multitarget Digital Droplet PCR Assays for the Detection of Hot Spot KRAS and NRAS Mutations in Cell-Free DNA. J. Mol. Diagn. 2020, 22, 943–956. [Google Scholar] [CrossRef] [PubMed]
- Peitz, C.; Sprussel, A.; Linke, R.B.; Astrahantseff, K.; Grimaldi, M.; Schmelz, K.; Toedling, J.; Schulte, J.H.; Fischer, M.; Messerschmidt, C.; et al. Multiplexed Quantification of Four Neuroblastoma DNA Targets in a Single Droplet Digital PCR Reaction. J. Mol. Diagn. 2020. [Google Scholar] [CrossRef] [PubMed]

| First author (Year) [Ref.] | Stage (N) | Sample | Method (Target) | ddPCR Results |
|---|---|---|---|---|
| Ako S. et al. (2017) [47] | All stages (40) | T, P, S | ddPCR (KRAS G12D/R/V) |
|
| Berger A.W. et al. (2016) [48] | Stage IV (24) | P, S | ddPCR (KRAS G12D/V and GNAS codon 201) |
|
| Bernard V. et al. (2019) [49] | All stages (194) | T, P | ddPCR (KRAS G12A/C/D/R/S/V) |
|
| Cheng H. et al. (2017) [50] | Stage IV (188) | P | NGS (Panel of 60 genes) ddPCR (KRAS G12D/R/V, BRCA2, KDR, EGFR, ERBB2) |
|
| Cheng H. et al. (2020) [51] | Stage III-IV (210) | P | ddPCR (KRAS G12D/V) |
|
| Del Re M. et al. (2017) [34] | Stage III-IV (27) | P | ddPCR (KRAS G12D/R/V, G13D) |
|
| Earl J. et al. (2015) [52] | All stages (31) | P | ddPCR (KRAS G12D/R/V) |
|
| Guo S. et al. (2020) [53] | Stage I-II-III (113) | P | NGS (Panel of 50 genes) ddPCR (KRAS G12A/C/D/R/SV) |
|
| Hadano N. et al. (2016) [10] | All stages (105) | T, P | ddPCR (KRAS G12D/R/V) |
|
| Kim M.K. et al. (2018) [54] | All stages (106) | P | ddPCR (KRAS G12A/C/D/R/S/V, G13D) |
|
| Kinugasa H. et al. (2015) [55] | All stage (75 and 66) | T, S | ddPCR (KRAS G12D/R/V) |
|
| Lin M. et al. (2018) [56] | All stages (65) | P | ddPCR (KRAS G12D/R/V) |
|
| Mohan S. et al. (2019) [57] | Stage III-IV (55) | P | NGS (Panel of 641 genes) ddPCR (KRAS G12A/C/D/R/S/V, G13D) |
|
| Sausen M. et al. (2015) [9] | Stage II (51) | T, P | NGS (WES) ddPCR (KRAS, BRAF and PIK3CA) |
|
| Sefrioui D. et al. (2017) [58] | All stages (56) | P | ddPCR (KRAS G12A/C/D/R/S/V, G13D) |
|
| Sugimori M. et al. (2020) [59] | All stages (47) | T, S | ddPCR (KRAS G12A/C/D/F/G/L/R/S/V G13A/C/D/G/R/S/V) |
|
| Watanabe F. et al. (2019) [60] | All stages (78) | T, P | ddPCR (KRAS G12D/R/V, Q61H) |
|
| Woo S.M. et al. (2017) [61] | Stage III (locally advanced) (44) | S | ddPCR (KRAS G12A/C/D/R/S/V, G13D) |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huerta, M.; Roselló, S.; Sabater, L.; Ferrer, A.; Tarazona, N.; Roda, D.; Gambardella, V.; Alfaro-Cervelló, C.; Garcés-Albir, M.; Cervantes, A.; et al. Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review. Cancers 2021, 13, 994. https://doi.org/10.3390/cancers13050994
Huerta M, Roselló S, Sabater L, Ferrer A, Tarazona N, Roda D, Gambardella V, Alfaro-Cervelló C, Garcés-Albir M, Cervantes A, et al. Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review. Cancers. 2021; 13(5):994. https://doi.org/10.3390/cancers13050994
Chicago/Turabian StyleHuerta, Marisol, Susana Roselló, Luis Sabater, Ana Ferrer, Noelia Tarazona, Desamparados Roda, Valentina Gambardella, Clara Alfaro-Cervelló, Marina Garcés-Albir, Andrés Cervantes, and et al. 2021. "Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review" Cancers 13, no. 5: 994. https://doi.org/10.3390/cancers13050994
APA StyleHuerta, M., Roselló, S., Sabater, L., Ferrer, A., Tarazona, N., Roda, D., Gambardella, V., Alfaro-Cervelló, C., Garcés-Albir, M., Cervantes, A., & Ibarrola-Villava, M. (2021). Circulating Tumor DNA Detection by Digital-Droplet PCR in Pancreatic Ductal Adenocarcinoma: A Systematic Review. Cancers, 13(5), 994. https://doi.org/10.3390/cancers13050994








