Simple Summary
The efficacy of cancer therapeutics depends on several factors, including the tumor genome, epigenome, and transcriptome. In addition, the tumor microenvironment, which consists mainly of immune cells, can influence cancer treatment outcomes. Hence, effectively leveraging host immunity is an important aspect of cancer treatment strategies. The human gut microbiome is involved in the regulation of the immune responses and affects the efficacy of chemotherapeutic and immunotherapeutic agents, including oxaliplatin, cyclophosphamide, and immune checkpoint inhibitors. This study reveals an additional dimension to the Bifidobacterium strain-specific determination of anti-cancer therapeutic efficacy using flow cytometry and transcriptome analysis of bacterial strain-fed mice and bacterial whole transcriptome analysis. We hope that our work will contribute to leveraging the gut microbiome to improve anti-cancer therapies.
Abstract
Colorectal cancer (CRC) is among the leading causes of cancer-related death in the world. The development of CRC is associated with smoking, diet, and microbial exposure. Previous studies have shown that dysbiosis of the gut microbiome affects cancer development, because it leads to inflammation and genotoxicity. Supplementation with specific microbiota induces anti-tumor effects by enhancing of anti-tumor immunity. Here, we observed that supplementation with either of two B. breve strains reduces tumor growth in MC38 colon carcinoma-bearing mice. Interestingly, only one B. breve strain boosted the efficacy of cancer therapeutics, including oxaliplatin and PD-1 blockade. Extensive immune profiling and transcriptomic analysis revealed that the boosting B. breve strain augments lymphocyte-mediated anti-cancer immunity. Our results suggest that supplementation with B. breve strains could potentially be used as a strategy to enhance the efficacy of CRC therapeutics.
1. Introduction
It is increasingly reported that the gut microbiota plays a critical role in host metabolism and immunity [1], and that the gut microbiome composition changes with age, diet, and diseases [2,3]. Colorectal cancer (CRC) is among the leading causes of cancer death in men and women in the United States of America (USA). The development of CRC is related to genetic and epigenetic alterations, inflammation, and microbial exposure [4,5,6]. Specific gut microbiota, such as Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis, and Escherichia coli are correlated with the development of intestinal adenomas and carcinomas, because they damage the DNA and affect immune and intestinal barrier functions [7,8,9]. Several studies have demonstrated that supplementation with gut microbiota species, including Enterococcus hirae, Lactobacillus johnsonii, and Bifidobacterium species reinforces the efficacy of platinum-based chemotherapy, cyclophosphamide, and immunotherapy by enhancing anti-tumor immune responses [10,11,12].
Bifidobacterium is the dominant member of the early life gut microbiome, mainly consisting of B. infantis, B. longum, and B. breve [13,14,15]. As an important part of the gut microbiome in infants, Bifidobacterium spp. reduce the risk of intestinal infections, including neonatal necrotizing enterocolitis, by regulating immune responses and protecting against intestinal barrier dysfunction [16,17,18]. Bifidobacterium spp. also affect immunotherapy responses, and supplementation with Bifidobacterium spp. in a mouse cancer model significantly improved anti-tumor immunity by CD8+ T cells [12]. In this study, we identified B. breve strains that influence cancer growth and the efficacy of cancer therapeutics through immune activation, and further investigated the underlying genetic mechanisms. Our results provide important insights regarding the gut microbiome and its applications in cancer patients.
2. Materials and Methods
2.1. Mice
All animal experiments were performed with approval from the Institutional Animal Care and Use Committee of CHA University (permission number IACIC180010). Mice used in this study were maintained and handled according to policies approved by CHA University. Five-week-old female C57B6/N mice were purchased from the Orient Bio (Gapyeong, Gyeonggi, Korea). For the tumor mice model, mice were orally administered Bifidobacterium strains for 14 days (−14 day). Then, mice were implanted with 2 × 105 MC38 colon adenocarcinoma cells subcutaneously and treated with 3 mg/kg oxaliplatin (S1224, Selleckchem, Houston, TX, USA) or 2 mg/kg anti-PD-1 mAb (clone RMP1-14, BioXCell, USA) intraperitoneally. Syngeneic tumor mice were treated 6 times with anti-PD-1 mAb or oxaliplatin on days 3, 7, 10, 14, 17, and 21. Tumor size was monitored three times a week until the study endpoint. Tumor volume was calculated as length × width2 × 0.5.
2.2. Cell Culture
MC38 cells were purchased from Kerafast (Cat# EZH204, Winston-Salem, NC, USA) and authenticated using DNA fingerprint analysis. All cells were free of mycoplasma contamination. Cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM, GIBCO) with 10% fetal bovine serum (FBS, GIBCO) and 100 units/mL penicillin (GIBCO) and streptomycin (GIBCO). All cells were maintained in a humidified incubator at 37 °C in a 5% CO2 atmosphere.
2.3. Bacteria
B. breve JCM92 (B. bre JCM92) were purchased from the Japan Culture Collection of Microorganisms (JCM). B. breve Bb03 (B. bre Bb03) were purchased from DuPont (Wilmington, NC USA). To verify bacterial viability, we cultured aliquoted cells on BL (Blood Liver) agar plates at 37 °C in anaerobic conditions (O2 < 1 ppm, 5% CO2, 5% N2) for 48 h and counted viable colonies (viable cells). For in vivo experiments, each B. breve strain was cultured in BL medium (Kisanbio, Seoul, Korea) at 37 °C in an anaerobic condition (O2 < 1 ppm, 5% CO2, 5% N2) for 48 h. After culture, the bacteria were lyophilized for preservation. Next, we measured colony forming unit (CFU) counts using a PBS serial dilution method (CORNING, 21-040-CVR). For each sampling point, an aliquot of culture was diluted to 10−6, 10−7, and 10−8, and 100 μL was spread on BL agar plates. After counting CFUs, each B. breve strain was divided into daily administration doses and stored at 4 °C. Each B. breve strain was treated at 1 × 109 CFU orally once daily. For bacterial whole transcriptome analysis, each B. breve strain was harvested in the exponential phase. Then, we cultured two B. breve strains in BL agar media at 37 °C in anaerobic conditions (O2 < 1 ppm, 5% CO2, 5% N2) for 48 h. After colony isolation from the BL agar plate, the colony was cultured in BL broth media at 37 °C for 48 h under anaerobic conditions (O2 < 1 ppm, 5% CO2, 5% N2).
2.4. Flow Cytometry Analysis
Tissues from experimental mice were obtained at day 15 after tumor cell implantation. Tumors were dissected into small pieces and transferred into RPMI 1640 media (GIBCO) supplemented with 2.5 mg/mL collagenase I, 1.5 mg/mL collagenase II, 1 mg/mL collagenase IV, 50 μg/mL DNase 1, and 0.25 mg/mL hyaluronidase Type IV-S. The tissues were incubated at 37 °C for 50 min and filtered using a 70 μm cell strainer (BD Bioscience, San Jose, CA, USA). Spleens were homogenized in RPMI 1640 media, incubated in red blood cell (RBC) lysis buffer (eBioscience, San Diego, CA, USA) and filtered using a 70 μm cell strainer. Splenocytes and tumor cells were incubated with anti-mouse CD16/CD32 (BD Bioscience) for 10 min at 4 °C to block the Fc receptor. After evaluating cell viability and surface staining, fixation/permeabilization buffer solution (BioLegend, San Diego, CA) was added. Splenocytes and tumor cells were stained with the following mouse antibodies: anti-CD45 (Biolegend, Cat# 103116), CD3 (Biolegend, Cat# 100218), NK1.1 (Biolegend, Cat# 108708), CD49b (Biolegend, Cat# 108910), CD4 (Biolegend, Cat#100422), CD25 (Biolegend, Cat#101904), Foxp3 (Invitrogen, Waltham, MA, Cat#17-5773-82), CD44 (Biolegend, Cat# 103008), CD62L (Biolegend, Cat#104412), and CD8a (Biolegend, Cat#100706). Cell acquisition was performed on a CANTO II flow cytometer (BD Bioscience, Franklin Lakes, NJ, USA) and data were analyzed using FlowJo software (TreeStar, San Carlos, CA, USA).
2.5. Quantitative PCR (qPCR) Analysis of Tumor Tissue
Total RNA from mice tumors was extracted using an RNeasy Plus Mini Kit (Qiagen, Germantown, MD) and reverse transcription was conducted using a PrimeScript First Strand cDNA Synthesis Kit (Takara, Mountain View, CA). qPCR analysis was performed using SYBR Premix Ex Taq (Tli RNase H Plus) (Takara) and a CFX384 Touch instrument (Bio-Rad, Hercules, CA, USA). The primers used in this study: IFN-γ, 5′- GAAAGCCTAGAAAGTCTGAATAACT-3′ and 5′- ATCAGCAGCGACTCCTTTTCCGCTT-3′; IL-2, 5′-ATGTACAGCATGCAGCTCGCATC-3′ and 5′-GGCTTGTTGAGATGATGCTTTGACA-3′; IL-10, 5′-TGAAGACCCTCAGGATGCGG-3′ and 5′-AGAGCTCTGTCTAGGTCCTGG-3′; and β-actin, 5′- CGTGCGTGACATCAAAGAGAA-3′ and 5′-TGGATGCCACAGGATTCCAT-3′. PCR conditions were as follows. For IFN-γ, IL-2, and β-actin: initial denaturation at 95 °C for 1 min; 35 cycles of denaturation (95 °C for 30 s), annealing (60 °C for 30 s), and extension (72 °C for 1 min), and a final extension at 72 °C for 1 min. For IL-10: initial denaturation at 95 °C for 1 min, 40 cycles of denaturation (95 °C for 30 s), annealing (60 °C for 30 s), and extension (72 °C for 1 min); and a final extension at 72 °C for 1 min.
2.6. Mice RNA Sequencing and Data Analysis
Intestines were harvested at day 22 after MC38 tumor cell inoculation, and an RNeasy Mini Kit (Qiagen) was used to extract RNA. After RNA extraction, 151-bp paired-end libraries were constructed from 1 μg RNA using a TruSeq RNA Sample Prep Kit v2 (Illumina, San Diego, CA). Whole-RNA sequencing was performed on an Illumina HiSeq instrument. RNA-seq reads were aligned to the mouse reference genome (GRCm38) using the STAR aligner [19]. Quantification of gene expression and differential expression analysis were performed using RSEM [20] and the edgeR package [21]. The ClueGO plug-in (v2.5.4, http://www.ici.upmc.fr/cluego/, 1 January 2009) in Cytoscape software (v3.3.0, http://cytoscape.org/, 1 January 2003) was used to analyze gene ontology (GO) [22]. Functionally-related GO terms for biological processes in Mus musculus (version: 27 February 2019) were grouped based on a kappa score > 0.4 and a network specificity of 5–10 using GO term fusion. Statistical significance was calculated using a two-sided hypergeometric test, and the false discovery rate was corrected using the Bonferroni step-down method.
2.7. Bacterial RNA Sequencing and Data Analysis
Bacterial RNA was extracted using the ZymoBIOMICS RNA Miniprep Kit (Zymo Research, Irvine, CA). Sequencing and library construction were performed on the Illumina Hiseq 2500 with 101 bp paired-end. Ribosomal RNA was removed using the Ribo-Zero™ rRNA Removal Kit (Bacteria) (Epicentre, Madison, WI). Libraries were prepared with the TruSeq RNA Sample Prep kit v2 (Illumina). RNA-sequenced reads were mapped on the reference genome of Bifidobacterium breve (NZ_AP012324.1) using STAR with alignIntron MAX 1 [19]. Then, the mapped reads were used to calculate read counts of genes using cufflinks [23], and the gene list was inputted into Cytoscape plug-in ClueGO v2.5.4 [22] to annotate functionally grouped networks. Functionally related GO terms for biological processes in Escherichia coli (version: November 18, 2016) were grouped based on a kappa score greater than 0.4 with a network specificity of 4–10. Statistical significance was calculated using two-sided hypergeometric tests, and the false discovery rate was corrected using the Bonferroni step down method.
2.8. Statistical Analysis
Statistical analyses were performed using GraphPad Prism 8.4.3. Differences between groups were analyzed using one-way or two-way ANOVA with Tukey’s post-test. Differences between groups were considered significant if p < 0.05. Statistical details are provided in the figure legends.
3. Results
3.1. Mice Supplemented with B. breve Strains Exhibit Anti-Tumor Effects
To examine the anti-tumor effects of Bifidobacterium spp. on colon cancer, we administered Bifidobacterium strains alone (B. breve JCM1192, B. bre JCM92; B. breve Bb03, B. bre Bb03) with oxaliplatin to MC38 colon carcinoma-bearing mice (Figure 1A and Table S1). Oral administration of B. breve in tumor-bearing mice significantly decreased tumor growth, compared to mice treated with the vehicle (phosphate-buffered saline; PBS) (Figure 1B). To determine the relationship between B. breve strain supplementation and cancer therapeutics, we treated colon carcinoma-bearing mice with each B. breve strain alone and in combination with oxaliplatin (Figure 1A). Oxaliplatin is a third-generation platinum-derivative chemotherapeutic agent and is an important metastatic CRC medication [24,25]. Interestingly, when mice were treated with oxaliplatin and B. breve strains, only mice treated with B. bre JCM92 showed significantly decreased tumor growth compared to mice treated with oxaliplatin alone (Figure 1B). To understand why only B. bre JCM92 enhanced the effect of oxaliplatin, we performed RNA sequencing of B. bre JCM92 (boosting strain) and B. bre Bb03 (non-boosting strain). We observed that 88 genes were upregulated in B. bre JCM92 compared to B. bre Bb03 (Table S2, Fold Change > 2). ClueGO analysis showed that 27 terms, including protein processing, intracellular protein transport, cellular amino acid metabolic process, ribonucleoside metabolic process, and peptide metabolic process were significantly enriched in B. bre JCM92 (Figure 1C and Table S3).
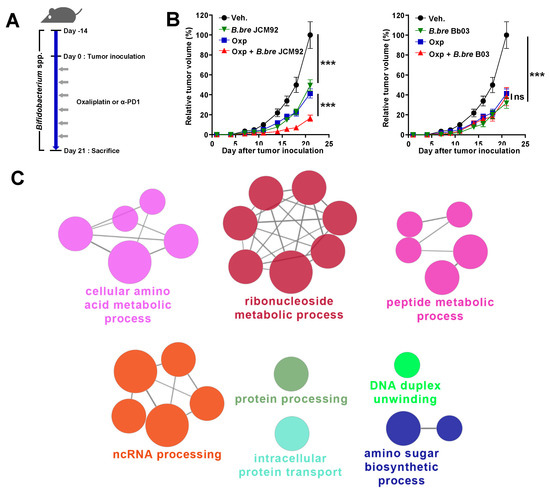
Figure 1.
Supplementation of Bifidobacterium strains exhibits anti-tumor effects. (A) Experimental protocol: each Bifidobacterium strain was administered orally 14 days before MC38 inoculation and continued until the end of the experiment. After tumor inoculation, oxaliplatin or PD-1 blockade was injected intraperitoneally. (B) Tumor growth curves after administering Bifidobacterium strains, with or without oxaliplatin. Oxp, oxaliplatin; B. bre JCM92, B. breve JCM92; B. bre Bb03, B. breve Bb03. Data are expressed as the means ± standard error of means (SEM). p-values were determined by two-way ANOVA using Tukey’s post-test. (C) Representation of the ClueGO functional network analysis of the upregulated genes in B. bre JCM92 compared to B. bre Bb03 (Fold change > 2). *** p < 0.001, and ns, not significant.
3.2. B. bre JCM92 Boosts the Efficacy of Oxaliplatin by Enhancing Anti-Tumor Immunity
To investigate the mechanism by which Bifidobacterium strains induce anti-tumor effects, we performed immune cell profiling of spleen and tumor tissue 15 days after tumor transplantation. Mice treated with B. bre JCM92 and B. bre Bb03 had increased CD4+ T, CD8+ T, and effector CD8+ T, and NK cells. These mice also had an increased ratio of CD4+/regulatory T cells (Tregs), CD8+/Tregs, and effector CD8+/Tregs. However, the mice had decreased Treg counts in splenocytes (Figure 2A,B). When we compared immune cell profiling between oxaliplatin alone and oxaliplatin + B. breve-treated groups, B. bre JCM92 enhanced the effect of oxaliplatin by increasing CD4+ T, CD8+ T, NK cells and the CD4+/Treg, CD8+/Treg, and effector CD8+/Treg ratios (Figure 2A). However, B. bre Bb03 did not boost the anti-tumor effect of oxaliplatin (Figure 2B).
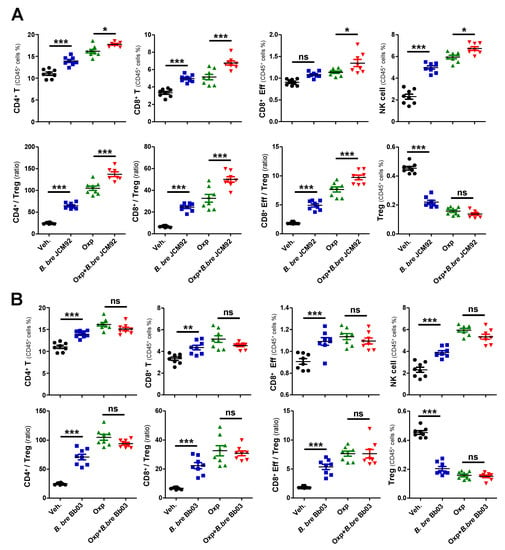
Figure 2.
B. bre JCM92 boosts oxaliplatin efficacy by enhancing anti-tumor immunity in the spleen. (A,B) Immune cell profiling of the spleen in mice treated with oxaliplatin in combination with B. bre JCM92 (A) or B. bre Bb03 (B) using flow cytometry analysis. Data are expressed as the means ± SEM. p-values were determined by one-way ANOVA using Tukey’s post-test. * p < 0.05, ** p < 0.001, *** p < 0.001, and ns, not significant.
Immune cell profiling of tumor-infiltrating lymphocytes (TILs) revealed that B. bre JCM92 and B. bre Bb03 significantly increased the CD4+/Treg, CD8+/Treg, and the effector CD8+/Treg ratio (Figure 3A,B). Furthermore, oxaliplatin combined with B. bre JCM92 significantly increased CD8+ T cells and the CD4+/Treg and CD8+/Treg ratios (Figure 3A,B). Next, we examined intra-tumor cytokine expression using qPCR. Interferon-gamma (IFN-γ) and interleukin-2 (IL-2) expression in tumor tissue was much higher in mice treated with oxaliplatin with or without B. breve strains (B. bre JCM92 and B. bre Bb03) compared to IFN-γ and IL-2 expression in vehicle-treated mice. Notably, mice treated with combined oxaliplatin and B. bre JCM92 exhibited significantly higher IFN-γ and IL-2 expression than mice treated with oxaliplatin alone (Figure 3C). In contrast, interleukin 10 (IL-10) expression was much lower in the mice treated with oxaliplatin with or without B. breve strains than in mice treated with the vehicle (Figure 3C). These results demonstrate that B. breve monotherapy induces anti-tumor effects by augmenting anti-tumor immunity, but only B. bre JCM92 enhances the anti-tumor effects of oxaliplatin.
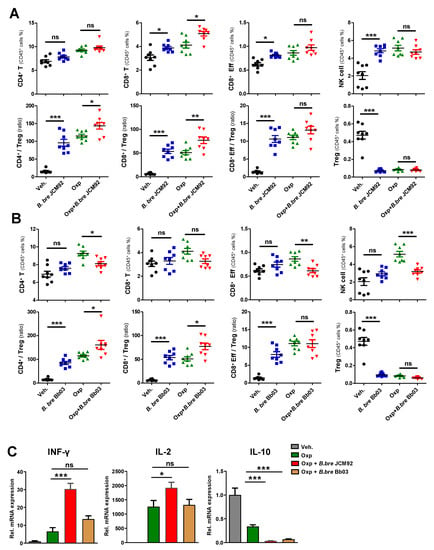
Figure 3.
B. bre JCM92 boosts oxaliplatin efficacy by enhancing anti-tumor immunity. (A,B) Immune cell profiling of tumor tissue in mice treated with oxaliplatin in combination with B. bre JCM92 (A) or B. bre Bb03 (B) using flow cytometry analysis. Data are expressed as the means ± SEM. p-values were determined by one-way ANOVA using Tukey’s post-test. (C) Expression of intratumoral cytokines in mice treated with oxaliplatin combined with B. bre JCM92 or B. bre Bb03 were measured using qPCR. Data are expressed as the means ± SEM. p-values were determined by one-way ANOVA using Tukey’s post-test within oxaliplatin treated groups. For all graphs, * p < 0.05, ** p < 0.01, *** p < 0.001, and ns, not significant.
3.3. B. bre JCM92 Boosts the Efficacy of PD-1 Blockade by Enhancing Anti-Tumor Immunity
Several studies have revealed that the gut microbiota enhances the response of immune checkpoint inhibitors that target cytotoxic T lymphocyte antigen-4 (CTLA-4) and programmed cell death protein-1 (PD-1) [12,26]. Therefore, we tested whether Bifidobacterium strains boost the efficacy of PD-1 blockade (Figure 4A). Similar to previous results, mice treated with a PD-1 blockade and B. bre JCM92 exhibited significantly decreased tumor growth compared to mice treated with PD-1 blockade alone (Figure 4A).
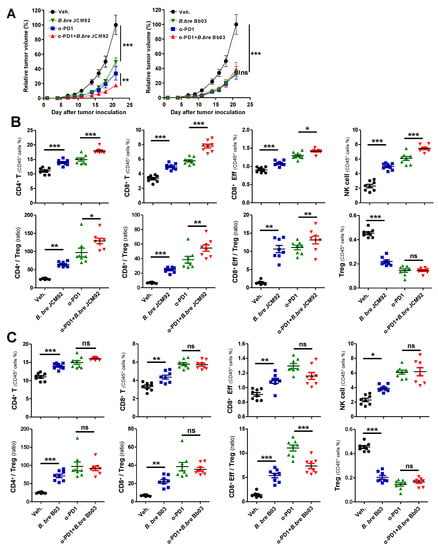
Figure 4.
B. bre JCM92 boosted PD-1 blockade efficacy. (A) Tumor growth curves after administering B. breve strains, with or without PD-1 blockade. Data are expressed as the means ± SEM. p-values were determined by two-way ANOVA using Tukey’s post-test. (B,C) Immune cell profiling of spleen tissue in mice treated with PD-1 blockade in combination with B. bre JCM92 (B) or B. bre Bb03 (C) using flow cytometry analysis. Data are expressed as the means ± SEM. p-values were determined by one-way ANOVA using Tukey’s post-test. For all graphs, * p < 0.05, ** p < 0.01, *** p < 0.001, and ns, not significant.
Splenic immune cell profiling showed that combined PD-1 blockade and B. bre JCM92 significantly enhanced anti-tumor immunity by increasing CD8+ T, effector CD8+ T, and NK cells. Additionally, combined PD-1 blockade and B. bre JCM92 increased the CD8+/Treg and effector CD8+/Treg ratios compared to PD-1 blockade alone (Figure 4B). In tumor tissue, PD-1 blockade combined with B. bre JCM92 significantly increased the CD8+ T cell level and the CD8+/Treg ratio compared to PD-1 blockade alone (Figure 5A). However, immune cell profiling of the spleen and tumor tissue revealed that B. bre Bb03 did not enhance the anti-tumor immunity of PD-1 blockade (Figure 4C and Figure 5B). Intra-tumoral cytokine expression revealed that mice treated with PD-1 blockade and B. bre JCM92 had much higher IFN-γ and IL-2 expression compared to mice treated with PD-1 blockade alone (Figure 5C). Although IFN-γ expression was significantly enhanced in mice treated with PD-1 blockade combined with B. bre Bb03, the combination of PD-1 blockade and B. bre JCM92 resulted in higher IFN-γ expression (Figure 5C). IL-10 expression was significantly decreased in mice treated with PD-1 blockade and either B. breve strain (Figure 5C).
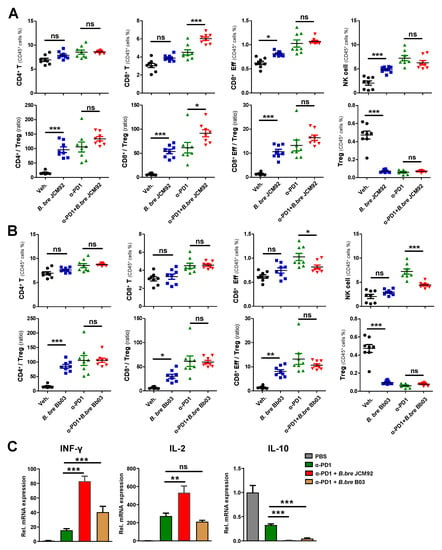
Figure 5.
B. bre JCM92 boosts PD-1 blockade efficacy by enhancing anti-tumor immunity in tumor tissue. (A,B) Immune cell profiling of tumor in mice treated with PD-1 blockade in combination with B. bre JCM92 (A) or B. bre Bb03 (B) using flow cytometry analysis. (C) Expression of intra-tumoral cytokines in mice treated with PD-1 blockade in combination with B. bre JCM92 or B. bre Bb03 were measured using qPCR. Data are expressed as the means ± SEM. p-values were determined by one-way ANOVA using Tukey’s post-test within the PD-1 blockade-treated groups. * p < 0.05, ** p < 0.01, *** p < 0.001, and ns, not significant.
3.4. Transcriptome Analysis of Intestinal Tissue Reveals That B. bre JCM92 Has the Ability to Boost the Effects of Cancer Therapeutics
To understand the mechanism by which the administration of the gut microbiome influences the intestine and anti-tumor immune responses, we performed whole transcriptome sequencing on the intestine of mice treated with cancer therapeutics and B. bre JCM92 and B. bre Bb03. Hierarchical clustering analysis revealed that intestinal gene-expression patterns were distinct between mice treated with B. bre JCM92 and B. bre Bb03 (Figure 6A). We analyzed differentially expressed genes (DEGs) between the enhanced (oxaliplatin + B. bre JCM92 and PD-1 blockade + B. bre JCM92) and unenhanced groups (oxaliplatin + B. bre Bb03 and PD-1 blockade + B. bre Bb03; p < 0.001; Figure 6B). Cytoscape network visualization exhibited that DEGs from mice treated with cancer therapeutics and B. breve strains (B. bre JCM92 and B. bre Bb03) are biologically related (Figure 6C). ClueGO analysis revealed that the term “Regulation of cytokines” was significantly enriched in mice treated with cancer therapeutics combined with B. bre JCM92 compared to mice treated with cancer therapeutics and B. bre Bb03 (Figure 6D and Table S4). Next, we performed gene set enrichment analysis (GSEA) between the boosted group and non-boosted group. The significantly enriched gene signatures in mice treated with the cancer therapeutics (oxaliplatin and PD-1 blockade) and B. bre JCM92 compared to mice treated with cancer therapeutics and B. bre Bb03 included gene sets involving E2F targets, tumor necrosis factor-α (TNF-α) signaling via nuclear factor kappa B (NFκB), allograft rejection, and inflammatory response (Figure 6E and Figure S1). Consistent with tumor cytokine expression, mice treated with cancer therapeutics and B. bre JCM92 exhibited higher IL-2, STAT5 signaling, and IFN-γ response compared to mice treated with oxaliplatin or PD-1 blockade and B. bre Bb03 (Figure 6F and Figure S2). Transcription factors regulate transcription initiation by recognizing specific binding sites, and transcription factor dysregulation is associated with human cancers [27,28,29]. To understand the regulatory mechanism of the gut microbiome, motif analysis of the intestinal transcriptome was performed. Nuclear transcription factor Y subunit beta and nuclear transcription factor Y subunit alpha genes were significantly associated with the upregulated genes in mice treated with the combination of cancer therapeutics and B. bre JCM92 (boosted group; Figure 6G). Nuclear factor Y binds to the interleukin-4 (IL-4) promoter and regulates IL-4 gene expression [30,31]. IL-4 regulates lymphocyte expansion and function [32] and exhibits anti-tumor activity by promoting Gr-1+ granulocyte and CD8+ T cell maturation [33]. These results reveal that supplementation of B. breve affects intestinal gene expression and that the changes are associated with anti-tumor immune responses.
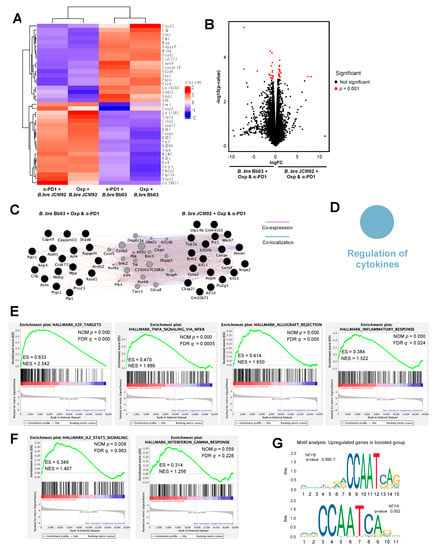
Figure 6.
Effect of cancer therapeutics and B. breve strains on the intestinal transcriptome. (A) Heat map showing hierarchical clustering of differentially expressed genes (DEGs) between mice treated with cancer therapeutics (oxaliplatin or PD-1 blockade) and B. bre JCM92 or B. bre Bb03. (B) Volcano plots illustrating genes between mice treated with cancer therapeutics and B. bre JCM92 and mice treated with cancer therapeutics and B. bre Bb03. Red dots represent significantly different genes (p < 0.001). (C) Network analysis of DEGs from mice treated with cancer therapeutics and B. bre JCM92, and mice treated with cancer therapeutics and B. bre Bb03 using Cytoscape. Co-expressed and co-localized genes are indicated by purple and blue lines, respectively. (D) Representation of the ClueGO functional network analysis shows upregulated genes in mice treated with cancer therapeutics and B. bre JCM92 compared to mice treated with cancer therapeutics and B. bre Bb03. (E,F) GSEA of intestinal transcriptome data exhibited that mice treated with cancer therapeutics and B. bre JCM92 were significantly enriched in hallmark pathways compared to mice treated with cancer therapeutics and B. bre Bb03. (G) Motifs within upregulated genes of mice treated with cancer therapeutics and B. bre JCM92. NFYB, nuclear transcription factor Y subunit beta; NFYA, nuclear transcription factor Y subunit alpha.
4. Discussion
The gut microbiome affects immune system development and differentiation by regulating innate immune responses, including innate lymphoid cells and gene expression [34]. Several diseases, such as cancer, obesity, and inflammatory bowel diseases are correlated with intestinal dysbiosis [34,35]. Previous studies suggest that supplementation with probiotics influences host homeostasis and has a therapeutic effect on immune-mediated diseases [36,37]. Bifidobacterium spp. are among the most frequently used as probiotics and have favorable effects in several diseases, including ulcerative colitis and enteropathogenic E. coli O157 infection [38,39].
Here, we found that Bifidobacterium strains confer anti-tumor properties. Although two B. breve strains increased anti-tumor immunity, only one B. breve strain boosted cancer therapeutic efficacy. Immune cell profiling of spleen and tumor tissue revealed that B. breve strains alone augmented anti-tumor immunity by increasing CD8+ T and effector CD8+ T cell numbers, and by increasing the CD8+/Treg and effector CD8+/Treg ratio. However, only B. bre JCM92 boosted the efficacy of cancer therapeutics.
Interestingly, immune cell profiling exhibited that spleens from mice treated with B. bre JCM92 and PD-1 blockade had significantly increased CD4+ and CD8+ T cells compared to PD-1 monotherapy (Figure 4B), whereas tumors from mice treated with B. bre JCM92 and PD-1 blockade only showed enhanced CD8+ T cells compared to PD-1 monotherapy (Figure 5A). We hypothesize that the CD4+ T cells activate CD8+ cells rather than directly killing tumor cells. Most CD4+ T cells could not recognize cancer cells directly due to a lack of MHCII (Major Histocompatibility complex II) in most solid cancer cells (except melanoma and breast cancer [40]), whereas CD4+ and CD8+ T cells are activated in spleens under immune activation and inflammatory conditions [41]. For those reasons, increased CD4+ and CD8+ T were observed from splenocytes analysis, while only CD8+ T cells were increased in the tumor environment.
Using bacterial transcriptome analysis, we observed that the boosting B. bre JCM92 expressed genes were significantly enriched in the ribonucleoside metabolic process, cellular amino acid metabolic process, and amino sugar biosynthetic process pathways compared to non-boosting B. bre Bb03. Previous studies showed that amino acid degrading enzymes have anti-tumor effects in preclinical and clinical trials [36,37]. The difference in gene expression between the two B. breve strains may be related to the boosting effects of B. bre JCM92. However, the exact anti-cancer molecular mechanisms and metabolic processes in B. bre JCM92 need to be further investigated. When bacteria are orally administered, the intestine is the primary colonization site [42,43]. Previous studies also showed that the gut microbiome composition is an important factor in determining the response to cancer therapeutics, especially immunotherapy. For instance, Bifidobacterium spp., Ruminococcaceae and Faecalibacterium are significantly enriched in melanoma patients who respond to anti-PD-1 treatment [44,45]. In addition, Akkermansia muciniphila abundance is associated with anti-PD-1 treatment response in epithelial tumors [46]. Therefore, we hypothesize that the boosting effects of B. breve appear by altering intestinal gene expression. Intestinal transcriptome analysis showed that mice treated with cancer therapeutics and B. bre JCM92 showed significantly enriched E2F targets, TNF-α signaling via NFκB, inflammatory responses, and IFN-γ responses. The E2F family of transcription factors plays an important role as transcriptional regulators of cell cycle-dependent gene expression [47], and also modulate apoptosis, metabolism, and angiogenesis in cancer [48,49]. E2F expression in cancer shows conflicting results in both tumor suppression and progression [50,51]. However, the relationship between E2F expression in the intestine and tumors has not yet been investigated. TNF-α is produced by various cells, including macrophages, NK cells, and T cells, and is released in response to peptidoglycan, lipopolysaccharide, and bacterial components [52,53]. Furthermore, TNF-α is involved in apoptosis, cell survival, and inflammation, and is a potential target for cancer therapy [52,54]. Previous studies have shown that TNF-α enhances antitumor effects when used in combination with other cancer therapeutics and cytokines, including adriamycin, actinomycin D, and IFN-γ [55,56]. Similarly to the intestinal transcriptome analysis, intra-tumoral IFN-γ expression was significantly enhanced in mice treated with B. bre JCM92 (boosting) and cancer therapeutics compared to mice treated with B. bre Bb03 (non-boosting) and cancer therapeutics. IFN-γ is a major player in anti-tumor immunity, and exerts anti-proliferative and pro-apoptotic effects in tumor cells [57,58,59,60], recruits immune cells into the tumor microenvironment [61,62], and enhances the tumoricidal activities of innate and adaptive immune cells [63,64,65]. This cytokine also exhibits anti-angiogenic activity via its anti-proliferative and apoptotic effects on endothelial cells [66,67]. Mutations in IFN-γ signaling genes were identified in tumors resistant to cancer immunotherapy, highlighting the essential role for this protein in chemotherapy responses [68,69]. These results demonstrate that the administration of B. bre JCM92 increases anti-tumor immunity by modulating cytokines, including TNF-α and IFN-γ, through the gut–systemic–tumor pathway. However, IL-10 expression was different between intestine and tumors. This difference demonstrates that the anti-tumor effects of B. breve are not same in all tissues. We hypothesize that other molecules, including metabolites, underlie these differences. Investigating the underlying mechanisms distinguishing intestine and tumor tissue after B. breve treatment needs to be further explored.
5. Conclusions
In summary, we identified two B. breve strains that show anti-tumor effects by enhancing anti-tumor immunity. Notably, only specific Bifidobacterium strains boosted the efficacy of cancer therapeutics. Further study is needed to examine whether supplementation with Bifidobacterium strains can be used as supportive anti-tumor drugs to boost the efficacy of cancer therapeutics in clinical trials.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/13/5/957/s1, Figure S1: GSEA of intestine RNA sequencing data; Figure S2: GSEA of intestine RNA sequencing data; Table S1: Bifidobacterium strains used in this study; Table S2: Upregulated genes in B. bre JCM92 compared to B. bre Bb03; Table S3: Gene ontology of DEGs in B. bre JCM92 compared to B. bre Bb03; Table S4: Gene ontology of DEGs in mice treated with cancer therapeutics and B. bre JCM92 compared to mice treated with cancer therapeutics and B. bre Bb03.
Author Contributions
Y.Y. and G.K. wrote the manuscript. S.F. and H.P. revised the manuscript. Y.Y. and G.K. performed the mouse work. G.K. analyzed transcriptomic data. Y.Y., G.K., and B.-N.J. performed the cell biology and molecular experiments. H.P. designed and supervised all experiments and analyses. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Institutional Review Board Statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Animal Care and Use Committee of CHA University (permission number IACIC180010).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, H.P., upon reasonable request.
Acknowledgments
We thank CHA Bio Complex for excellent guidance and assistance with the performed mouse experiments. This research was supported by the Bio & Medical Technology Development Program of the National Research Foundation (NRF), funded by the Ministry of Science & ICT (grant number: NRF-2017M3A9F3046536), a GIST Research Institute (GRI) grant, funded by the GIST in 2020, and the grants from National Cancer Centre, Korea (NCC-1911267) to H.P.; as well as the Bio and Medical Technology Development Program (NRF-2017M3A9F3046538), the Korea Health Technology R&D Project (HR18C0012) from the Ministry of Health & Welfare and the faculty research grant from the Yonsei University College of Medicine (6-2018-0098) to S.F. B.-N.J. is supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (MEST) (2018R1C1B6005768).
Conflicts of Interest
The authors declare no competing interests.
References
- O’Keefe, S.J. Nutrition and colonic health: The critical role of the microbiota. Curr. Opin. Gastroenterol. 2008, 24, 51–58. [Google Scholar] [CrossRef]
- Kundu, P.; Blacher, E.; Elinav, E.; Pettersson, S. Our gut microbiome: The evolving inner self. Cell 2017, 171, 1481–1493. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Dejea, C.M.; Wick, E.C.; Hechenbleikner, E.M.; White, J.R.; Welch, J.L.M.; Rossetti, B.J.; Peterson, S.N.; Snesrud, E.C.; Borisy, G.G.; Lazarev, M. Microbiota organization is a distinct feature of proximal colorectal cancers. Proc. Natl. Acad. Sci. USA 2014, 111, 18321–18326. [Google Scholar] [CrossRef]
- Mima, K.; Sukawa, Y.; Nishihara, R.; Qian, Z.R.; Yamauchi, M.; Inamura, K.; Kim, S.A.; Masuda, A.; Nowak, J.A.; Nosho, K. Fusobacterium nucleatum and T cells in colorectal carcinoma. Jama Oncol. 2015, 1, 653–661. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Goding Sauer, A.; Fedewa, S.A.; Butterly, L.F.; Anderson, J.C.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2020. CA A Cancer J. Clin. 2020, 70, 145–164. [Google Scholar] [CrossRef] [PubMed]
- Dejea, C.M.; Fathi, P.; Craig, J.M.; Boleij, A.; Taddese, R.; Geis, A.L.; Wu, X.; Shields, C.E.D.; Hechenbleikner, E.M.; Huso, D.L. Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 2018, 359, 592–597. [Google Scholar] [CrossRef]
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; Gerner, R.R.; Moschen, A.R. The intestinal microbiota in colorectal cancer. Cancer Cell 2018, 33, 954–964. [Google Scholar] [CrossRef]
- Daillère, R.; Vétizou, M.; Waldschmitt, N.; Yamazaki, T.; Isnard, C.; Poirier-Colame, V.; Duong, C.P.; Flament, C.; Lepage, P.; Roberti, M.P. Enterococcus hirae and Barnesiella intestinihominis facilitate cyclophosphamide-induced therapeutic immunomodulatory effects. Immunity 2016, 45, 931–943. [Google Scholar] [CrossRef]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef] [PubMed]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.-L. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti–PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.-C.; Stiemsma, L.T.; Amenyogbe, N.; Brown, E.M.; Finlay, B. The intestinal microbiome in early life: Health and disease. Front. Immunol. 2014, 5, 427. [Google Scholar] [CrossRef] [PubMed]
- Matsuki, T.; Watanabe, K.; Tanaka, R.; Fukuda, M.; Oyaizu, H. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 1999, 65, 4506–4512. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Ouwehand, A.C.; Isolauri, E.; Hashimoto, H.; Benno, Y.; Salminen, S. Comparison of mucosal adhesion and species identification of bifidobacteria isolated from healthy and allergic infants. Fems Immunol. Med Microbiol. 2001, 30, 43–47. [Google Scholar] [CrossRef]
- Patole, S.K.; Rao, S.C.; Keil, A.D.; Nathan, E.A.; Doherty, D.A.; Simmer, K.N. Benefits of Bifidobacterium breve M-16V supplementation in preterm neonates-a retrospective cohort study. PLoS ONE 2016, 11, e0150775. [Google Scholar] [CrossRef]
- Jacobs, S.E.; Tobin, J.M.; Opie, G.F.; Donath, S.; Tabrizi, S.N.; Pirotta, M.; Morley, C.J.; Garland, S.M. Probiotic effects on late-onset sepsis in very preterm infants: A randomized controlled trial. Pediatrics 2013, 132, 1055–1062. [Google Scholar] [CrossRef]
- Ling, X.; Linglong, P.; Weixia, D.; Hong, W. Protective effects of bifidobacterium on intestinal barrier function in LPS-induced enterocyte barrier injury of Caco-2 monolayers and in a rat NEC model. PLoS ONE 2016, 11, e0161635. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef]
- André, T.; Boni, C.; Mounedji-Boudiaf, L.; Navarro, M.; Tabernero, J.; Hickish, T.; Topham, C.; Zaninelli, M.; Clingan, P.; Bridgewater, J. Oxaliplatin, fluorouracil, and leucovorin as adjuvant treatment for colon cancer. N. Engl. J. Med. 2004, 350, 2343–2351. [Google Scholar] [CrossRef] [PubMed]
- De Gramont, A.d.; Figer, A.; Seymour, M.; Homerin, M.; Hmissi, A.; Cassidy, J.; Boni, C.; Cortes-Funes, H.; Cervantes, A.; Freyer, G. Leucovorin and fluorouracil with or without oxaliplatin as first-line treatment in advanced colorectal cancer. J. Clin. Oncol. 2000, 18, 2938–2947. [Google Scholar] [CrossRef] [PubMed]
- Vétizou, M.; Pitt, J.M.; Daillère, R.; Lepage, P.; Waldschmitt, N.; Flament, C.; Rusakiewicz, S.; Routy, B.; Roberti, M.P.; Duong, C.P. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 2015, 350, 1079–1084. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.E. Transcription factors as targets for cancer therapy. Nat. Rev. Cancer 2002, 2, 740–749. [Google Scholar] [CrossRef]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef]
- Habib, N.; Kaplan, T.; Margalit, H.; Friedman, N. A novel Bayesian DNA motif comparison method for clustering and retrieval. PLoS Comput. Biol. 2008, 4, e1000010. [Google Scholar] [CrossRef][Green Version]
- Li-Weber, M.; Davydov, I.V.; Krafft, H.; Krammer, P.H. The role of NF-Y and IRF-2 in the regulation of human IL-4 gene expression. J. Immunol. 1994, 153, 4122–4133. [Google Scholar]
- Szabo, S.J.; Gold, J.S.; Murphy, T.L.; Murphy, K.M. Identification of cis-acting regulatory elements controlling interleukin-4 gene expression in T cells: Roles for NF-Y and NF-ATc. Mol. Cell. Biol. 1993, 13, 4793–4805. [Google Scholar] [CrossRef] [PubMed]
- Wills-Karp, M.; Finkelman, F.D. Untangling the complex web of IL-4–and IL-13–mediated signaling pathways. Sci. Signal. 2008, 1, pe55. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Chen, L.; Qin, Z. Paradoxical roles of IL-4 in tumor immunity. Cell. Mol. Immunol. 2009, 6, 415–422. [Google Scholar] [CrossRef]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Xuan, C.; Shamonki, J.M.; Chung, A.; DiNome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef] [PubMed]
- Lavasani, S.; Dzhambazov, B.; Nouri, M.; Fåk, F.; Buske, S.; Molin, G.; Thorlacius, H.; Alenfall, J.; Jeppsson, B.; Weström, B. A novel probiotic mixture exerts a therapeutic effect on experimental autoimmune encephalomyelitis mediated by IL-10 producing regulatory T cells. PLoS ONE 2010, 5, e9009. [Google Scholar] [CrossRef]
- De Oliveira, G.L.V.; Leite, A.Z.; Higuchi, B.S.; Gonzaga, M.I.; Mariano, V.S. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology 2017, 152, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Duranti, S.; Gaiani, F.; Mancabelli, L.; Milani, C.; Grandi, A.; Bolchi, A.; Santoni, A.; Lugli, G.A.; Ferrario, C.; Mangifesta, M. Elucidating the gut microbiome of ulcerative colitis: Bifidobacteria as novel microbial biomarkers. Fems Microbiol. Ecol. 2016, 92, fiw191. [Google Scholar] [CrossRef]
- Hadrup, S.; Donia, M.; Thor Straten, P. Effector CD4 and CD8 T cells and their role in the tumor microenvironment. Cancer Microenviron. 2013, 6, 123–133. [Google Scholar] [CrossRef]
- Bronte, V.; Pittet, M.J. The spleen in local and systemic regulation of immunity. Immunity 2013, 39, 806–818. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.N.; Walker, A. Bacterial colonization and the development of intestinal defences. Can. J. Gastroenterol. 2004, 18, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Walker, W.A. The importance of appropriate initial bacterial colonization of the intestine in newborn, child, and adult health. Pediatric Res. 2017, 82, 387–395. [Google Scholar] [CrossRef] [PubMed]
- Matson, V.; Fessler, J.; Bao, R.; Chongsuwat, T.; Zha, Y.; Alegre, M.-L.; Luke, J.J.; Gajewski, T.F. The commensal microbiome is associated with anti–PD-1 efficacy in metastatic melanoma patients. Science 2018, 359, 104–108. [Google Scholar] [CrossRef]
- Gopalakrishnan, V.; Spencer, C.; Nezi, L.; Reuben, A.; Andrews, M.; Karpinets, T.; Prieto, P.; Vicente, D.; Hoffman, K.; Wei, S. Gut microbiome modulates response to anti–PD-1 immunotherapy in melanoma patients. Science 2018, 359, 97–103. [Google Scholar] [CrossRef]
- Routy, B.; Le Chatelier, E.; Derosa, L.; Duong, C.P.; Alou, M.T.; Daillère, R.; Fluckiger, A.; Messaoudene, M.; Rauber, C.; Roberti, M.P. Gut microbiome influences efficacy of PD-1–based immunotherapy against epithelial tumors. Science 2018, 359, 91–97. [Google Scholar] [CrossRef]
- Kent, L.N.; Leone, G. The broken cycle: E2F dysfunction in cancer. Nat. Rev. Cancer 2019, 19, 326–338. [Google Scholar] [CrossRef]
- Chen, H.-Z.; Tsai, S.-Y.; Leone, G. Emerging roles of E2Fs in cancer: An exit from cell cycle control. Nat. Rev. Cancer 2009, 9, 785–797. [Google Scholar] [CrossRef]
- Dyson, N.J. RB1: A prototype tumor suppressor and an enigma. Genes Dev. 2016, 30, 1492–1502. [Google Scholar] [CrossRef]
- Müller, H.; Helin, K. The E2F transcription factors: Key regulators of cell proliferation. Biochim. Et Biophys. Acta (Bba)-Rev. Cancer 2000, 1470, M1–M12. [Google Scholar]
- Kent, L.N.; Rakijas, J.B.; Pandit, S.K.; Westendorp, B.; Chen, H.-Z.; Huntington, J.T.; Tang, X.; Bae, S.; Srivastava, A.; Senapati, S. E2f8 mediates tumor suppression in postnatal liver development. J. Clin. Investig. 2016, 126, 2955–2969. [Google Scholar] [CrossRef]
- Van Horssen, R.; Ten Hagen, T.L.; Eggermont, A.M. TNF-α in cancer treatment: Molecular insights, antitumor effects, and clinical utility. Oncologist 2006, 11, 397–408. [Google Scholar] [CrossRef] [PubMed]
- Locksley, R.M.; Killeen, N.; Lenardo, M.J. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell 2001, 104, 487–501. [Google Scholar] [CrossRef]
- Roberts, N.J.; Zhou, S.; Diaz Jr, L.A.; Holdhoff, M. Systemic use of tumor necrosis factor alpha as an anticancer agent. Oncotarget 2011, 2, 739. [Google Scholar] [CrossRef] [PubMed]
- Alexander, R.B.; Isaacs, J.T.; Coffey, D.S. Tumor necrosis factor enhances the in vitro and in vivo efficacy of chemotherapeutic drugs targeted at DNA topoisomerase II in the treatment of murine bladder cancer. J. Urol. 1987, 138, 427–429. [Google Scholar] [CrossRef]
- Aggarwal, B.B.; Eessalu, T.E.; Hass, P.E. Characterization of receptors for human tumour necrosis factor and their regulation by γ-interferon. Nature 1985, 318, 665–667. [Google Scholar] [CrossRef] [PubMed]
- Chin, Y.E.; Kitagawa, M.; Su, W.-C.S.; You, Z.-H.; Iwamoto, Y.; Fu, X.-Y. Cell growth arrest and induction of cyclin-dependent kinase inhibitor p21WAF1/CIP1 mediated by STAT1. Science 1996, 272, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Bromberg, J.F.; Horvath, C.M.; Wen, Z.; Schreiber, R.D.; Darnell, J.E. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. USA 1996, 93, 7673–7678. [Google Scholar] [CrossRef]
- Chin, Y.E.; Kitagawa, M.; Kuida, K.; Flavell, R.A.; Fu, X.-Y. Activation of the STAT signaling pathway can cause expression of caspase 1 and apoptosis. Mol. Cell. Biol. 1997, 17, 5328–5337. [Google Scholar] [CrossRef]
- Xu, X.; Fu, X.-Y.; Plate, J.; Chong, A.S. IFN-γ induces cell growth inhibition by Fas-mediated apoptosis: Requirement of STAT1 protein for up-regulation of Fas and FasL expression. Cancer Res. 1998, 58, 2832–2837. [Google Scholar]
- Groom, J.R.; Luster, A.D. CXCR3 ligands: Redundant, collaborative and antagonistic functions. Immunol. Cell Biol. 2011, 89, 207–215. [Google Scholar] [CrossRef]
- Melero, I.; Rouzaut, A.; Motz, G.T.; Coukos, G. T-cell and NK-cell infiltration into solid tumors: A key limiting factor for efficacious cancer immunotherapy. Cancer Discov. 2014, 4, 522–526. [Google Scholar] [CrossRef] [PubMed]
- Celada, A.; Gray, P.W.; Rinderknecht, E.; Schreiber, R. Evidence for a gamma-interferon receptor that regulates macrophage tumoricidal activity. J. Exp. Med. 1984, 160, 55–74. [Google Scholar] [CrossRef] [PubMed]
- Kursunel, M.A.; Esendagli, G. The untold story of IFN-γ in cancer biology. Cytokine Growth Factor Rev. 2016, 31, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Ni, L.; Lu, J. Interferon gamma in cancer immunotherapy. Cancer Med. 2018, 7, 4509–4516. [Google Scholar] [CrossRef]
- Campanella, G.S.; Colvin, R.A.; Luster, A.D. CXCL10 can inhibit endothelial cell proliferation independently of CXCR3. PLoS ONE 2010, 5, e12700. [Google Scholar] [CrossRef]
- Feldman, E.D.; Weinreich, D.M.; Carroll, N.M.; Burness, M.L.; Feldman, A.L.; Turner, E.; Xu, H.; Alexander, H.R. Interferon γ–inducible protein 10 selectively inhibits proliferation and induces apoptosis in endothelial cells. Ann. Surg. Oncol. 2006, 13, 125–133. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Shi, L.Z.; Zhao, H.; Chen, J.; Xiong, L.; He, Q.; Chen, T.; Roszik, J.; Bernatchez, C.; Woodman, S.E. Loss of IFN-γ pathway genes in tumor cells as a mechanism of resistance to anti-CTLA-4 therapy. Cell 2016, 167, 397–404. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.J.; Sanjana, N.E.; Kishton, R.J.; Eidizadeh, A.; Vodnala, S.K.; Cam, M.; Gartner, J.J.; Jia, L.; Steinberg, S.M.; Yamamoto, T.N. Identification of essential genes for cancer immunotherapy. Nature 2017, 548, 537. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).