Evaluation of Fast Molecular Detection of Lymph Node Metastases in Prostate Cancer Patients Using One-Step Nucleic Acid Amplification (OSNA)
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Population
2.2. Study Design
2.3. OSNA
2.4. Histopathology
2.5. RNA-Quality/Quantity
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.; Kneib, T.H.; Wasylow, C.; Reinhardt, L.; Henke, R.-P.; Engels, S.; Gerullis, H.; Wawroschek, F. Updated nomogram incorporating percentage of positive cores to predict probability of lymph node invasion in prostate cancer patients undergoing sentinel lymph node dissection. J. Cancer 2017, 8, 2692–2698. [Google Scholar] [CrossRef]
- Touijer, K.A.; Karnes, R.J.; Passoni, N.; Sjoberg, D.D.; Assel, M.; Fossati, N.; Gandaglia, G.; Eastham, J.A.; Scardino, P.T.; Vickers, A.; et al. Survival outcomes of men with lymph node-positive prostate cancer after radical prostatectomy: A comparative analysis of different postoperative management strategies. Eur. Urol. 2018, 73, 890–896. [Google Scholar] [CrossRef] [PubMed]
- Mottet, N.; Cornford, P.; van den Bergh, R.C.N.; Briers, E.; De Santis, M.; Fanti, S.; Gillessen, S.; Grummet, J.; Henry, A.M.; Lam, T.B.; et al. EAU-EANM-ESTRO-ESUR-SIOG Guidelines on Prostate Cancer. Available online: https://uroweb.org/guideline/prostate-cancer/ (accessed on 26 November 2020).
- Hofman, M.S.; Lawrentschuk, N.; Francis, R.J.; Tang, C.; Vela, I.; Thomas, P.; Rutherford, N.; Martin, J.M.; Frydenberg, M.; Shakher, R.; et al. Prostate-specific membrane antigen PET-CT in patients with high-risk prostate cancer before curative-intent surgery or radiotherapy (proPSMA): A prospective, randomized, multicenter study. Lancet 2020, 395, 1208–1216. [Google Scholar] [CrossRef]
- Hope, T.A.; Armstrong, W.R.; Murthy, V.; Lawhn Heath, C.; Behr, S.; Barbato, F.; Ceci, F.; Farolfi, A.; Schwarzenboeck, S.; Unterrainer, M.; et al. Accuracy of 68Ga-PSMA-11 for pelvic nodal metastasis detection prior to radical prostatectomy and pelvic lymph node dissection: A multicenter prospective phase III imaging study. J. Clin. Oncol. 2020, 38, 5502. [Google Scholar] [CrossRef]
- Wawroschek, F.; Wagner, T.; Hamm, M.; Weckermann, D.; Vogt, H.; Märkl, B.; Gordijn, R.; Harzmann, R. The influence of serial sections, immunohistochemistry, and extension of pelvic lymph node dissection on the lymph node status in clinically localized prostate cancer. Eur. Urol. 2003, 43, 132–136. [Google Scholar] [CrossRef]
- Heck, M.M.; Retz, M.; Bandur, M.; Souchay, M.; Vitzthum, E.; Weirich, G.; Mollenhauer, M.; Schuster, T.; Autenrieth, M.; Kübler, H.; et al. Topography of lymph node metastases in prostate cancer patients undergoing radical prostatectomy and extended lymphadenectomy: Results of a combined molecular and histopathologic mapping study. Eur. Urol. 2014, 66, 222–229. [Google Scholar] [CrossRef]
- Lawrence, W.D.; Association of Directors of Anatomic and Surgical Pathology. ADASP recommendations for processing and reporting of lymph node specimens submitted for evaluation of metastatic disease. Virchows Arch. 2001, 439, 601–603. [Google Scholar] [CrossRef]
- Tsujimoto, M.; Nakabayashi, K.; Yoshidome, K.; Kaneko, T.; Iwase, T.; Akiyama, F.; Kato, Y.; Tsuda, H.; Ueda, S.; Sato, K.; et al. One-step nucleic acid amplification for intraoperative detection of lymph node metastasis in breast cancer patients. Clin. Cancer Res. 2007, 13, 4807–4816. [Google Scholar] [CrossRef]
- Tamaki, Y.; Sato, N.; Homma, K.; Takabatake, D.; Nishimura, R.; Tsujimoto, M.; Yoshidome, K.; Tsuda, H.; Kinoshita, T.; Kato, H.; et al. Routine clinical use of the one-step nucleic acid amplification assay for detection of sentinel lymph node metastases in breast cancer patients. Cancer 2012, 118, 3477–3483. [Google Scholar] [CrossRef]
- Babar, M.; Madani, R.; Thwaites, L.; Jackson, P.A.; Devalia, H.L.; Chakravorty, A.; Irvine, T.E.; Layera, G.T.; Kissin, M.W. A differential intra-operative molecular biological test for the detection of sentinel lymph node metastases in breast carcinoma. An extended experience from the first UK centre routinely offering the service in clinical practice. ESJO 2014, 40, 282–288. [Google Scholar] [CrossRef]
- Terrenato, I.; D’Alicandro, V.; Casini, B.; Perracchio, L.; Rollo, F.; De Salvo, L.; Di Filippo, S.; Di Filippo, F.; Pescarmona, E.; Maugeri-Saccà, M.; et al. A cut-off of 2150 cytokeratin 19 mRNA copy number in sentinel lymph node may be a powerful predictor of non-sentinel lymph node status in breast cancer patients. PLoS ONE 2017, 12, e0171517. [Google Scholar] [CrossRef]
- Nakagawa, K.; Asamura, H.; Tsuta, K.; Nagai, K.; Yamada, E.; Ishii, G.; Mitsudomi, T.; Ito, A.; Higashiyama, A.; Tomita, Y.; et al. The novel one-step nucleic acid amplification (OSNA) assay for the diagnosis of lymph node metastasis in patients with non-small cell lung cancer (NSCLC): Results of a multi-center prospective study. Lung Cancer 2016, 97, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Kumagai, K.; Yamamoto, N.; Miyashiro, I.; Tomita, Y.; Katai, H.; Kushima, R.; Tsuda, H.; Kitagawa, Y.; Takeuchi, H.; Mukai, M.; et al. Multicenter study evaluating the clinical performance of the OSNA assay for the molecular detection of lymph node metastases in gastric cancer patients. Gastric Cancer 2014, 17, 273–280. [Google Scholar] [CrossRef]
- Croner, R.S.; Geppert, C.I.; Bader, F.G.; Nitsche, U.; Späth, C.; Rosenberg, R.; Zettl, A.; Matias-Guiu, X.; Tarragona, J.; Güller, U.; et al. Molecular staging of lymph node-negative colon carcinomas by one-step nucleic acid amplification (OSNA) results in upstaging of a quarter of patients in a prospective, European, multicenter study. Br. J. Cancer 2014, 110, 2544–2550. [Google Scholar] [CrossRef] [PubMed]
- Fanfani, F.; Monterossi, G.; Ghizzoni, V.; Rossi, E.D.; Dinoi, G.; Inzani, F.; Fagotti, A.; Aletti, S.G.; Scarpellini, F.; Nero, C.; et al. One-step nucleic acid amplification (OSNA): A fast molecular test based on CK19 mRNA concentration for assessment of lymph-nodes metastases in early stage endometrial cancer. PLoS ONE 2018, 13, e0195877. [Google Scholar] [CrossRef]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Peehl, D.M.; Sellers, R.G.; McNeal, J.E. Keratin 19 in the adult human prostate: Tissue and cell culture studies. Cell Tissue Res. 1996, 285, 171–176. [Google Scholar] [CrossRef] [PubMed]
- Winter, A.; Engels, S.; Goos, P.; Süykers, M.-C.; Henke, R.-P.; Gerullis, H.; Wawroschek, F. Detection of CK19 mRNA using one-step nucleic acid amplification (OSNA) in prostate cancer: Preliminary results. J. Cancer 2018, 9, 4611–4617. [Google Scholar] [CrossRef] [PubMed]
- Sedrakyan, A.; Campbell, B.; Merino, J.G.; Kuntz, R.; Hirst, A.; McCulloch, P. IDEAL-D: A rational framework for evaluating and regulating the use of medical devices. BMJ 2016, 353, i2372. [Google Scholar] [CrossRef]
- Marra, G.; Valerio, M.; Heidegger, I.; Tsaur, I.; Mathieu, R.; Ceci, F.; Ploussard, G.; van den Bergh, R.C.N.; Kretschmer, A.; Thibault, C.; et al. Management of patients with node-positive prostate cancer at radical prostatectomy and pelvic lymph node dissection: A systematic review. Eur. Urol. Oncol. 2020, 3, 565–581. [Google Scholar] [CrossRef]
- Prendeville, S.; van der Kwast, T.H. Lymph node staging in prostate cancer: Perspective for the pathologist. Clin. Pathol. 2016, 69, 1039–1045. [Google Scholar] [CrossRef]
- Schiavina, R.; Capizzi, E.; Borghesi, M.; Vagnoni, V.; Romagnoli, D.; Rocca, G.C.; Giunchi, F.; D’Errico, A.; De Giovanni, A.; Rizzi, S.; et al. Nodal occult metastases in intermediate- and high-risk prostate cancer patients detected using serial section, immunohistochemistry, and real-time reverse transcriptase polymerase chain reaction: Prospective evaluation with matched-pair analysis. Clin. Genitourin. Cancer 2015, 13, e55–e64. [Google Scholar] [CrossRef]
- Lunger, L.; Retz, M.; Bandur, M.; Souchay, M.; Vitzthum, E.; Jäger, M.; Weirich, G.; Schuster, T.; Autenrieth, M.; Kübler, H.; et al. KLK3 and TMPRSS2 for molecular lymph-node staging in prostate cancer patients undergoing radical prostatectomy. Prostate Cancer Prostatic Dis. 2020. advanced online publication. [Google Scholar] [CrossRef]
- Haas, C.J.; Wagner, T.; Wawroschek, F.; Arnholdt, H. Combined application of RT-PCR and immunohistochemistry on paraffin embedded sentinel lymph nodes of prostate cancer patients. Pathol. Res. Pract. 2005, 200, 763–770. [Google Scholar] [CrossRef]
- Yaguchi, Y.; Sugasawa, H.; Tsujimoto, H.; Takata, H.; Nakabayashi, K.; Ichikura, T.; Ono, S.; Hiraki, S.; Sakamoto, N.; Horio, T.; et al. One-Step Nucleic Acid Amplification (OSNA) for the application of sentinel node concept in gastric cancer. Ann. Surg. Oncol. 2011, 18, 2289–2296. [Google Scholar] [CrossRef]
- National Institute for Health and Care Excellence. Intraoperative Tests (RD 100i OSNA System and Metasin Test) for Detecting Sentinel Lymph Node Metastases in Breast Cancer. Diagnostics Guidance [DG8]. Available online: https://www.nice.org.uk/guidance/dg8 (accessed on 26 November 2020).
- Bernet, L.; Piñero, A.; Vidal-Sicart, S.; Peg, V.; Giménez, J.; Algara, M.; Dueñas, B.; Tresserra, F.; Cano, R.; Cordero, J.M. Consenso sobre la biopsia selectiva del ganglio centinela en el cáncer de mama. Revisión 2013 de la Sociedad Española de Senología y Patología Mamaria. Rev. De Senol. Y Patol. Mamar. 2014, 27, 43–53. [Google Scholar] [CrossRef]
- Gligorov, J.; Penault-Llorca, F.; Aapro, M.; Aimard, L.; Alfonsi, J.-P.; André, F.; Antoine, M.; Antoine, E.C.; Azria, D.; Azuar, P.; et al. Sixièmes Recommandations Pour la Pratique Clinique de la Prise en Charge des Cancers du Sein de Nice—St Paul de Vence 2015. Available online: https://www.cours-rpc-nice-saintpaul.fr/wp-content/uploads/2017/01/RPC-2015-RESUME-SITE.pdf (accessed on 15 February 2021).
- Tamaki, Y. One-step nucleic acid amplification (OSNA)- where do we go with it? Int. J. Clin. Oncol. 2017, 22, 3–10. [Google Scholar] [CrossRef]
- Wit, E.M.K.; Acar, C.; Grivas, N.; Yuan, C.; Horenblas, S.; Liedberg, F.; Valdes Olmos, R.A.; van Leeuwen, F.W.B.; van den Berg, N.S.; Winter, A.; et al. Sentinel node procedure in prostate cancer: A systematic review to assess diagnostic accuracy. Eur. Urol. 2017, 71, 596–605. [Google Scholar] [CrossRef]
- Van der Poel, H.G.; Wit, E.M.; Acar, C.; van den Berg, N.S.; van Leeuwen, F.W.B.; Olmos, R.A.V.; Winter, A.; Wawroschek, F.; Liedberg, F.; MacLennan, S.; et al. Sentinel node biopsy for prostate cancer: Report from a consensus panel meeting. BJU Int. 2017, 120, 204–211. [Google Scholar] [CrossRef]
- Rakislova, N.; Montironi, C.; Aldecoa, I.; Fernandez, E.; Bombi, J.A.; Jimeno, M.; Balaguer, F.; Pellise, M.; Castells, A.; Cuatrecasas, M. Lymph node pooling: A feasible and efficient method of lymph node molecular staging in colorectal carcinoma. J. Transl. Med. 2017, 15, 14. [Google Scholar] [CrossRef]
- Castellano, I.; Macrì, L.; Deambrogio, C.; Balmativola, D.; Bussone, R.; Ala, A.; Coluccia, C.; Sapino, A. Reliability of whole sentinel lymph node analysis by one-step nucleic acid amplification for intraoperative diagnosis of breast cancer metastases. Ann. Surg. 2012, 255, 334–342. [Google Scholar] [CrossRef]
- Fougo, J.L.; Amendoeira, I.; Brito, M.J.; Correia, A.P.; Gonçalves, A.; Honavar, M.; Machado, A.; Magalhães, A.; Marta, S.; Nogueira, M.; et al. Sentinel node total tumour load as a predictive factor for non-sentinel node status in early breast cancer patients—The porttle study. Surgic. Oncol. 2020, 32, 108–114. [Google Scholar] [CrossRef]
- Peña, K.B.; Kepa, A.; Cochs, A.; Riu, F.; Parada, D.; Gumà, J. Total tumor load of mRNA cytokeratin 19 in the sentinel lymph node as a predictive value of axillary lymphadenectomy in patients with neoadjuvant breast cancer. Genes 2021, 12, 77. [Google Scholar] [CrossRef]
- Vieites, B.; López-García, M.Á.; Martín-Salvago, M.D.; Ramirez-Tortosa, C.L.; Rezola, R.; Sancho, M.; López-Vilaró, L.; Villardell, F.; Burgués, O.; Fernández-Rodriguez, B.; et al. Predictive and prognostic value of total tumor load in sentinel lymph nodes in breast cancer patients after neoadjuvant treatment using one-step nucleic acid amplification: The NEOVATTL study. Clin. Transl. Oncol. 2021. advanced online publication. [Google Scholar] [CrossRef] [PubMed]
- Vodicka, J.; Mukensnabl, P.; Vejvodova, S.; Spidlen, V.; Kulda, V.; Topolcan, O.; Pesta, M. A more sensitive detection of micrometastases of NSCLC in lymph nodes using the one-step nucleic acid amplification (OSNA) method. J. Surg. Oncol. 2018, 117, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Daniele, L.; Annaratone, L.; Allia, E.; Mariani, S.; Armando, E.; Bosco, M.; Macrì, L.; Cassoni, P.; D’Armento, G.; Bussolati, G.; et al. Technical limits of comparison of step-sectioning, immunohistochemistry and RT-PCR on breast cancer sentinel nodes: A study on methacarn-fixed tissue. J. Cell Mol. Med. 2009, 13, 4042–4050. [Google Scholar] [CrossRef]
- Di Filippo, F.; Giannarelli, D.; Bouteille, C.; Bernet, L.; Cano, R.; Cunnick, G.; Sapino, A. Elaboration of a nomogram to predict non sentinel node status in breast cancer patients with positive sentinel node, intra-operatively assessed with one step nucleic acid amplification method. J. Exp. Clin. Cancer Res. 2015, 34, 136. [Google Scholar] [CrossRef]
- Osako, T.; Iwase, T.; Ushijima, M.; Yonekura, R.; Ohno, S.; Akiyama, F. A new molecular-based lymph node staging classification determines the prognosis of breast cancer patients. Br. J. Cancer 2017, 117, 1470–1477. [Google Scholar] [CrossRef] [PubMed][Green Version]
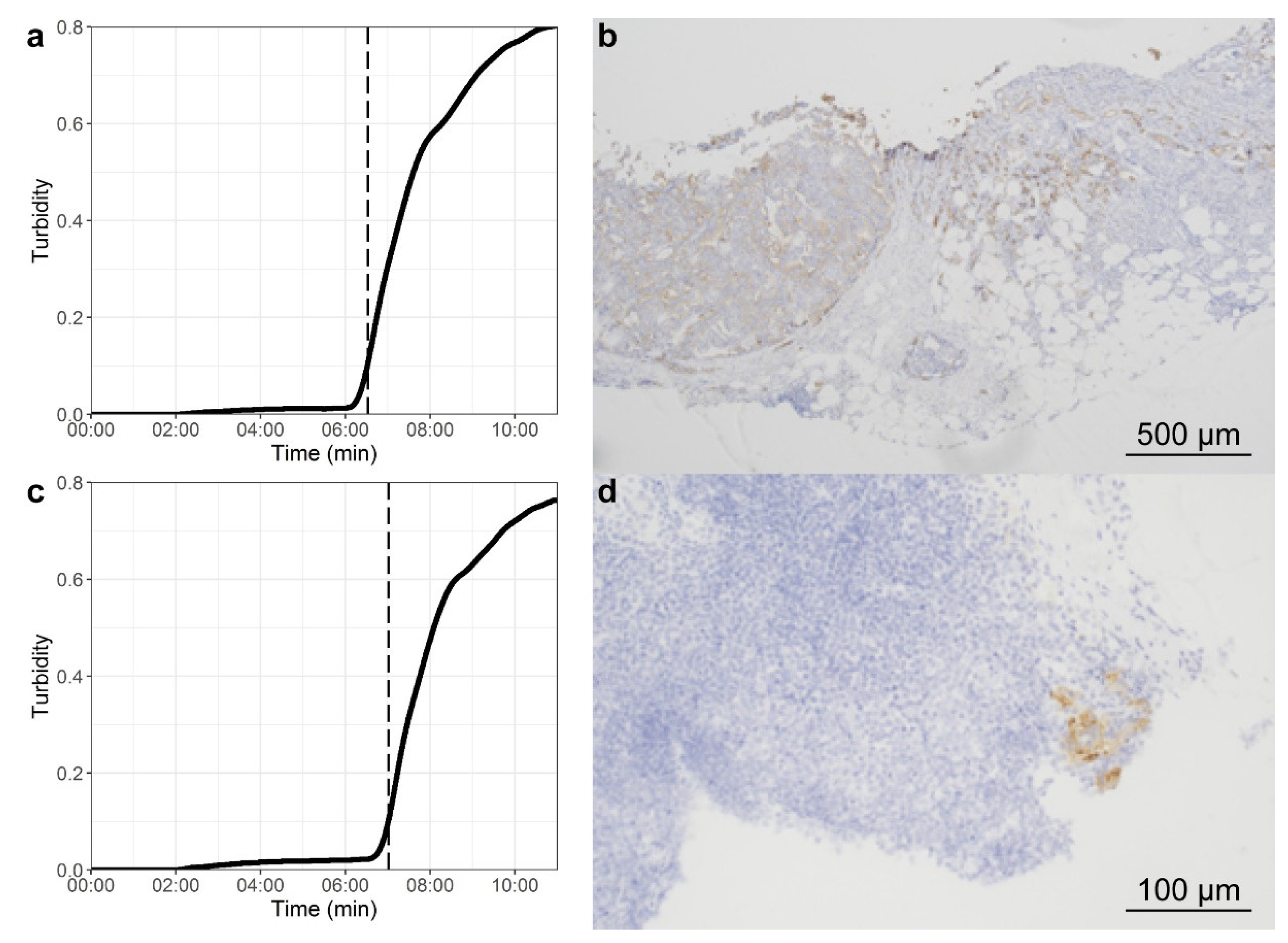
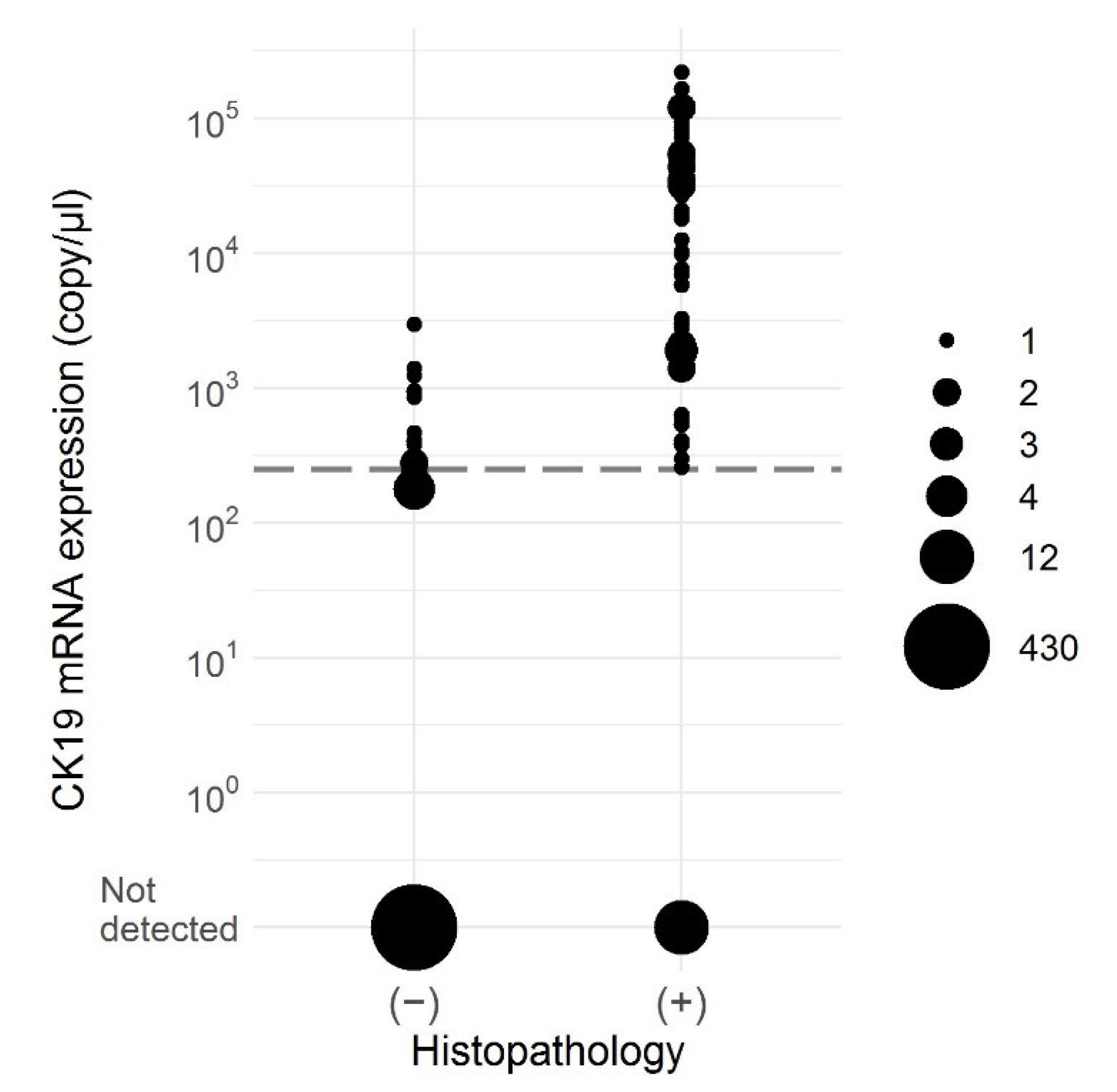
| Characteristic | Overall n = 64 | Patients with Pathological Negative LNs n = 41 (64%) | Patients with Pathological Positive LNs n = 23 (36%) |
|---|---|---|---|
| Age | 69 (65.5–73) | 69 (63.5–73.5) | 69 (67–73) |
| Total PSA, ng/ mL | 11.3 (9.3–17.8) | 10.7 (9.0–14.2) | 14.9 (9.9–22.3) |
| No. of LN removed | 17 (13–22) | 17 (13–22) | 15 (13–20) |
| No. of SLN removed | 8 (5–12) | 9 (5–13) | 8 (5–11) |
| No. of positive LN | 3 (1–8) | ||
| Tumor stage (%) | |||
| cT1c | 22 (34.4) | 21 (51.2) | 1 (4.4) |
| cT2a | 18 (28.1) | 10 (24.4) | 8 (34.8) |
| cT2b | 3 (4.7) | 2 (4.9) | 1 (4.4) |
| cT2c | 16 (25.0) | 7 (17.1) | 9 (39.1) |
| cT3 | 5 (7.8) | 1 (2.4) | 4 (17.4) |
| Biopsy Gleason score (%) | |||
| 6 (3 + 3) | 1 (1.6) | 1 (2.4) | 0 |
| 7 (3 + 4) | 30 (46.9) | 25 (61.0) | 5 (21.7) |
| 7 (4 + 3) | 10 (15.6) | 8 (19.5) | 2 (8.7) |
| ≥8 | 23 (35.9) | 7 (17.1) | 16 (69.6) |
| Postoperative Gleason score (%) | |||
| 6 (3 + 3) | 0 | 0 | 0 |
| 7 (3 + 4) | 25 (39.1) | 24 (58.5) | 1 (4.4) |
| 7 (4 + 3) | 19 (29.7) | 11 (26.8) | 8 (34.8) |
| ≥8 | 20 (31.3) | 6 (14.6) | 14 (60.9) |
| Pathologic stage (%) | |||
| pT2 | 23 (35.9) | 23 (56.1) | 0 |
| pT3a | 12 (18.8) | 11 (26.8) | 1 (4.4) |
| pT3b | 29 (45.3) | 7 (17.1) | 22 (95.7) |
| pT4 | 0 | 0 | 0 |
| LN No. | OSNA * | Micrometastasis ** | Histopathology | CK19 IHC |
|---|---|---|---|---|
| 3-1 | + | + | − | − |
| 3-2 | + | + | − | − |
| 3-4 | + | + | − | − |
| 11-7 | + | + | − | − |
| 14-1 | + | + | − | − |
| 14-3 | + | + | − | − |
| 15-5 | + | + | − | − |
| 15-21 | + | + | − | − |
| 17-10 | + | + | − | − |
| 28-8 | + | + | − | − |
| 37-2 | + | + | − | − |
| 42-4 | + | + | − | − |
| 45-11 | + | + | − | − |
| 45-19 | + | + | − | − |
| 52-17 | + | + | − | − |
| 54-4 | + | + | − | − |
| 62-3 | + | + | − | − |
| 63-2 | + | + | − | − |
| LN No. | OSNA * | Histopathology | CK19 IHC | Comments |
|---|---|---|---|---|
| 1-1 | − | macrometastasis | − | |
| 1-3 | − | macrometastasis | − | |
| 15-15 | − | − | + | macrometastasis only found in LN slices for IHC |
| 44-1 | − | macrometastasis | + | only single CK19 positive cells |
| 64-6 | − | macrometastasis | + | only single CK19 positive cells |
| 64-11 | − | macrometastasis | + | in parts faint CK19 positive cells |
| 19-22 | − | micrometastasis | + | |
| 28-2 | − | micrometastasis | + | only single CK19 positive cells |
| 31-13 | − | micrometastasis | + | |
| 43-1 | − | micrometastasis | + | |
| 62-9 | − | − | + | micrometastasis only found in LN slices for IHC |
| 64-1 | − | micrometastasis | − |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Engels, S.; Brautmeier, L.; Reinhardt, L.; Wasylow, C.; Hasselmann, F.; Henke, R.P.; Wawroschek, F.; Winter, A. Evaluation of Fast Molecular Detection of Lymph Node Metastases in Prostate Cancer Patients Using One-Step Nucleic Acid Amplification (OSNA). Cancers 2021, 13, 1117. https://doi.org/10.3390/cancers13051117
Engels S, Brautmeier L, Reinhardt L, Wasylow C, Hasselmann F, Henke RP, Wawroschek F, Winter A. Evaluation of Fast Molecular Detection of Lymph Node Metastases in Prostate Cancer Patients Using One-Step Nucleic Acid Amplification (OSNA). Cancers. 2021; 13(5):1117. https://doi.org/10.3390/cancers13051117
Chicago/Turabian StyleEngels, Svenja, Lutz Brautmeier, Lena Reinhardt, Clara Wasylow, Friederike Hasselmann, Rolf P. Henke, Friedhelm Wawroschek, and Alexander Winter. 2021. "Evaluation of Fast Molecular Detection of Lymph Node Metastases in Prostate Cancer Patients Using One-Step Nucleic Acid Amplification (OSNA)" Cancers 13, no. 5: 1117. https://doi.org/10.3390/cancers13051117
APA StyleEngels, S., Brautmeier, L., Reinhardt, L., Wasylow, C., Hasselmann, F., Henke, R. P., Wawroschek, F., & Winter, A. (2021). Evaluation of Fast Molecular Detection of Lymph Node Metastases in Prostate Cancer Patients Using One-Step Nucleic Acid Amplification (OSNA). Cancers, 13(5), 1117. https://doi.org/10.3390/cancers13051117






