The lncRNA H19-Derived MicroRNA-675 Promotes Liver Necroptosis by Targeting FADD
Abstract
Simple Summary
Abstract
1. Introduction
2. Results
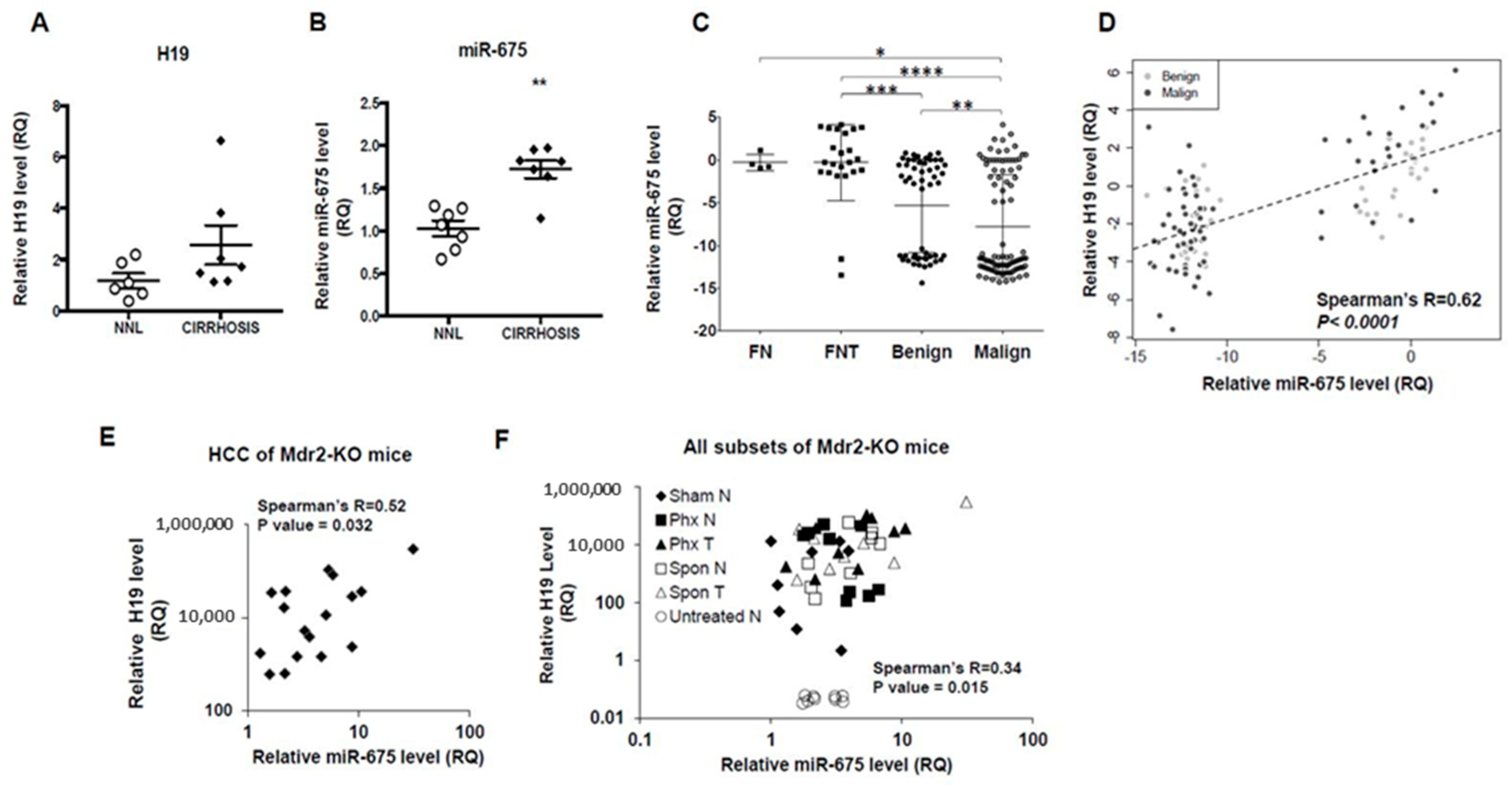
2.1. Positive Correlation between H19 and miR-675 in Human and Mouse Hepatocellular Carcinoma (HCC)
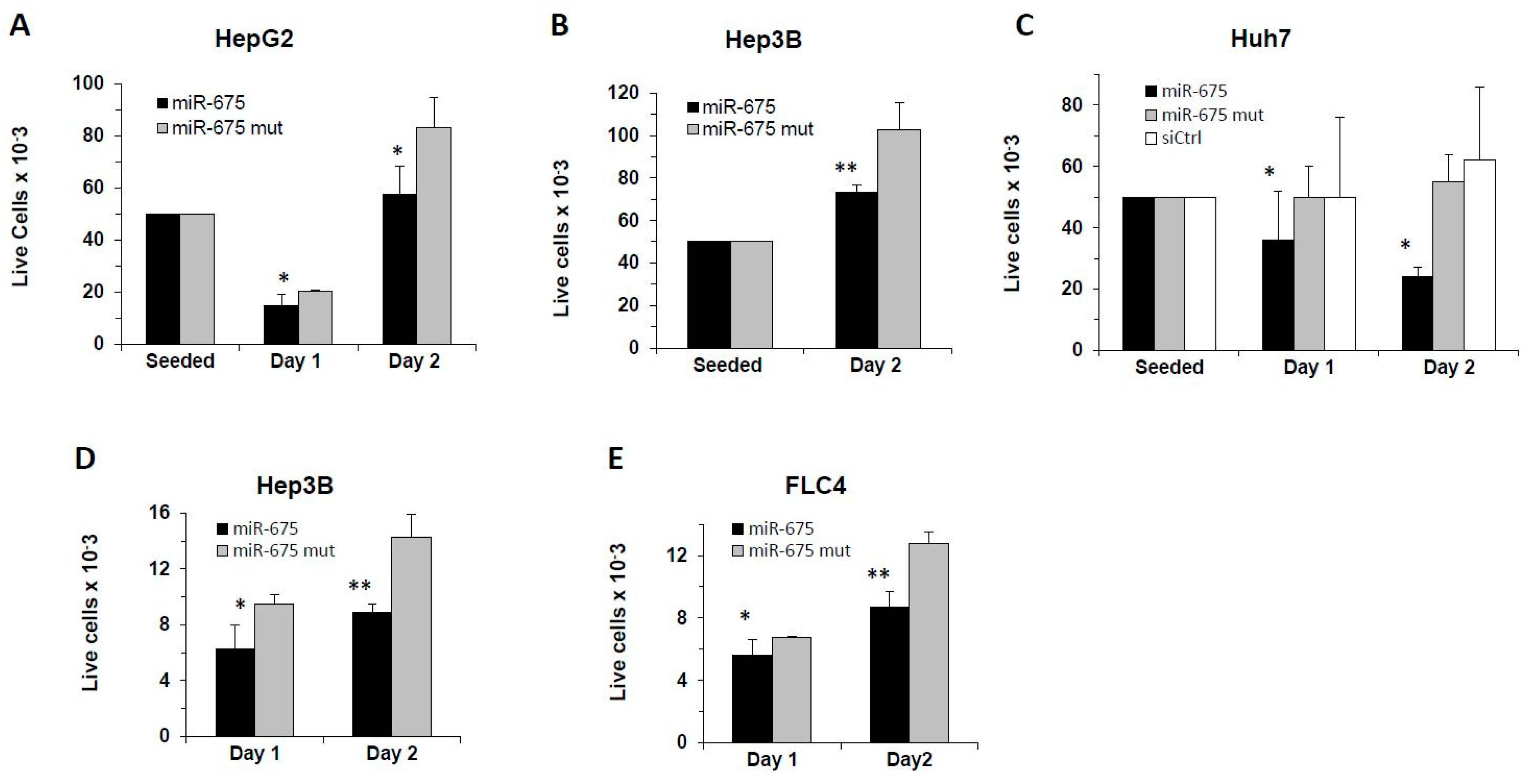
2.2. miR-675 Inhibits Cell Growth of Cultured Human HCC-Derived Cells
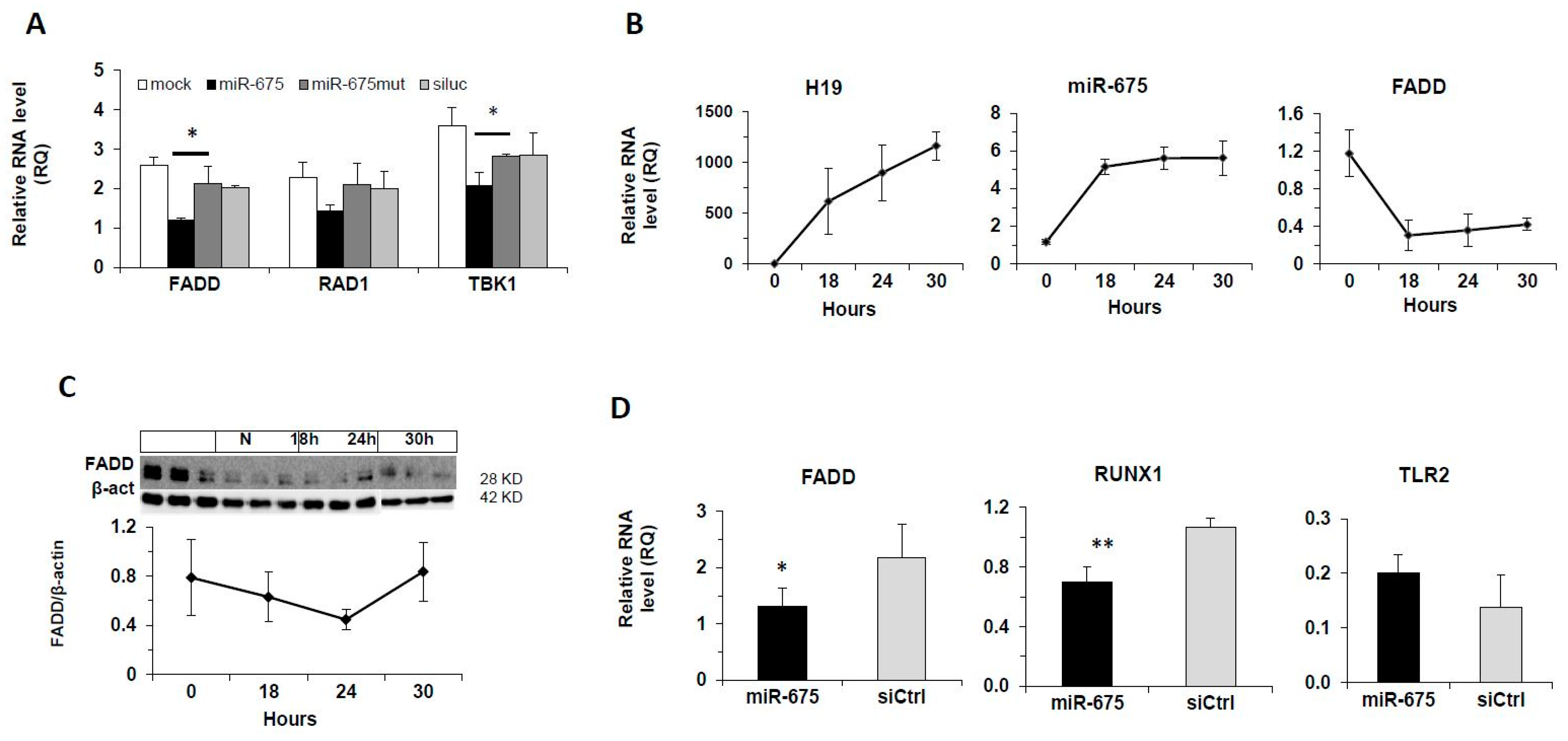
2.3. Negative Correlation between Fas-Associated Protein with Death Domain (FADD) and miR-675 in Human and Mouse HCC
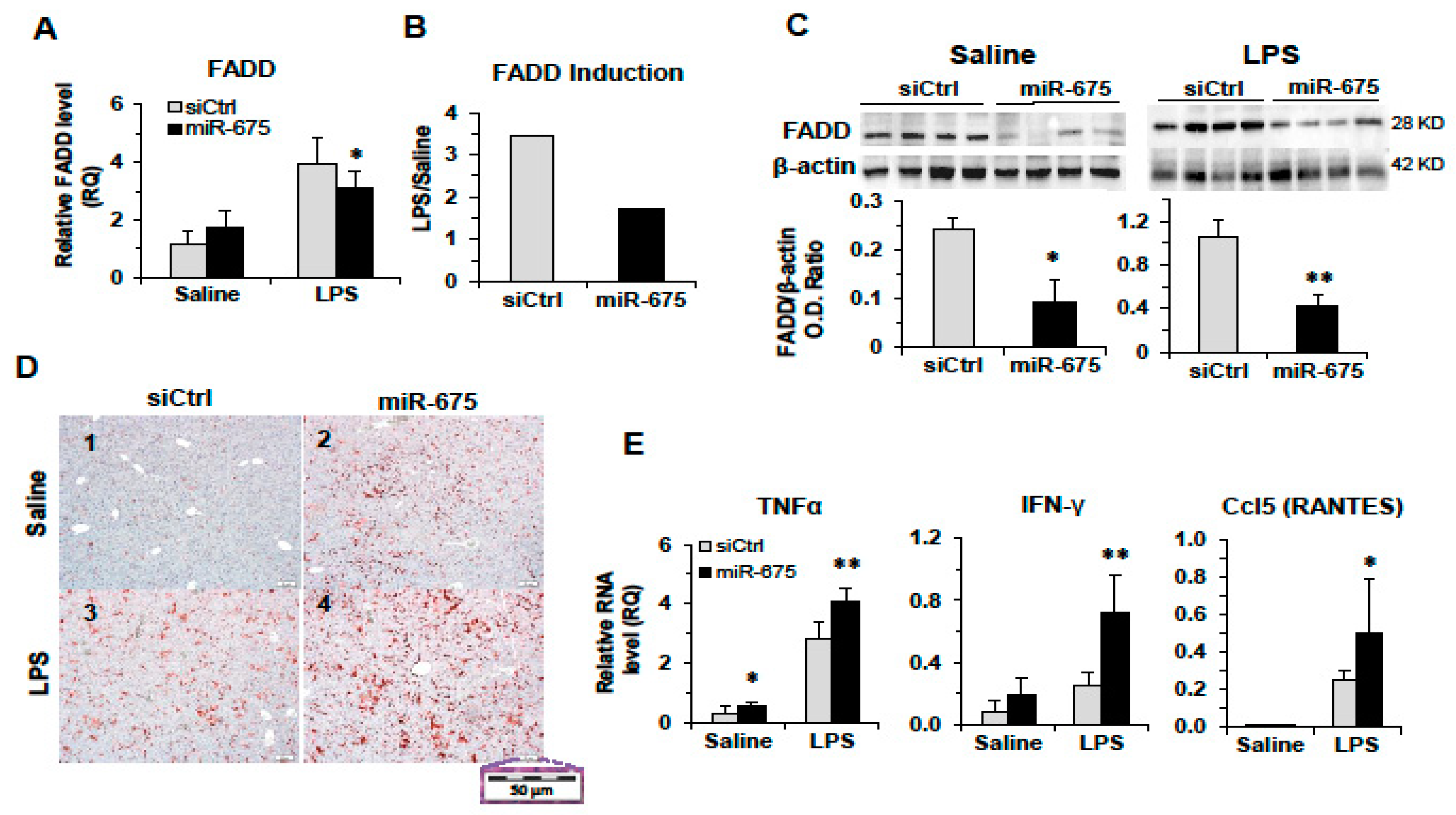
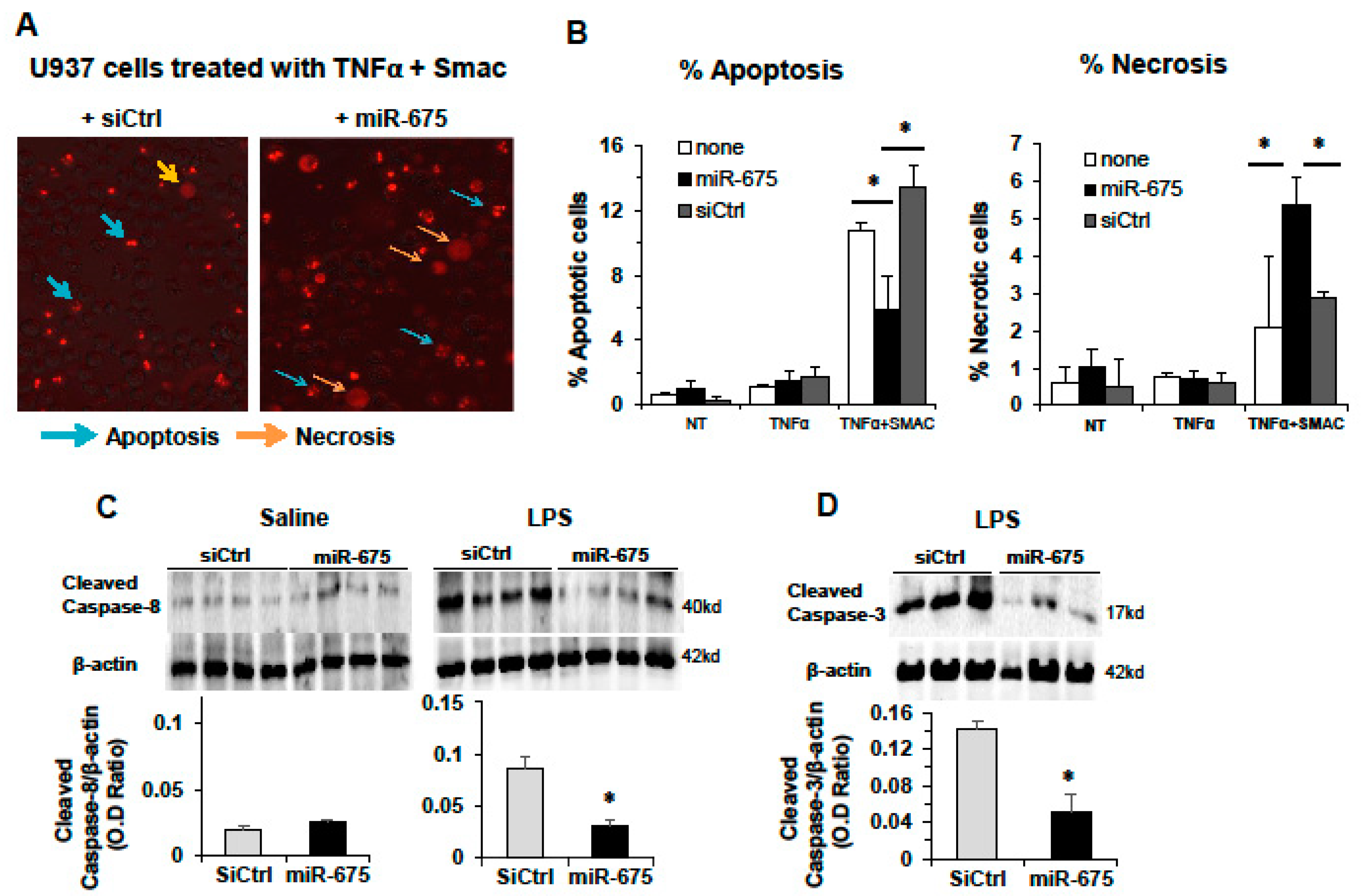
2.4. Regulation of FADD by miR-675 In Vitro and In Vivo
2.5. Overexpression of miR-675 Affects the Inflammatory Response in Mice
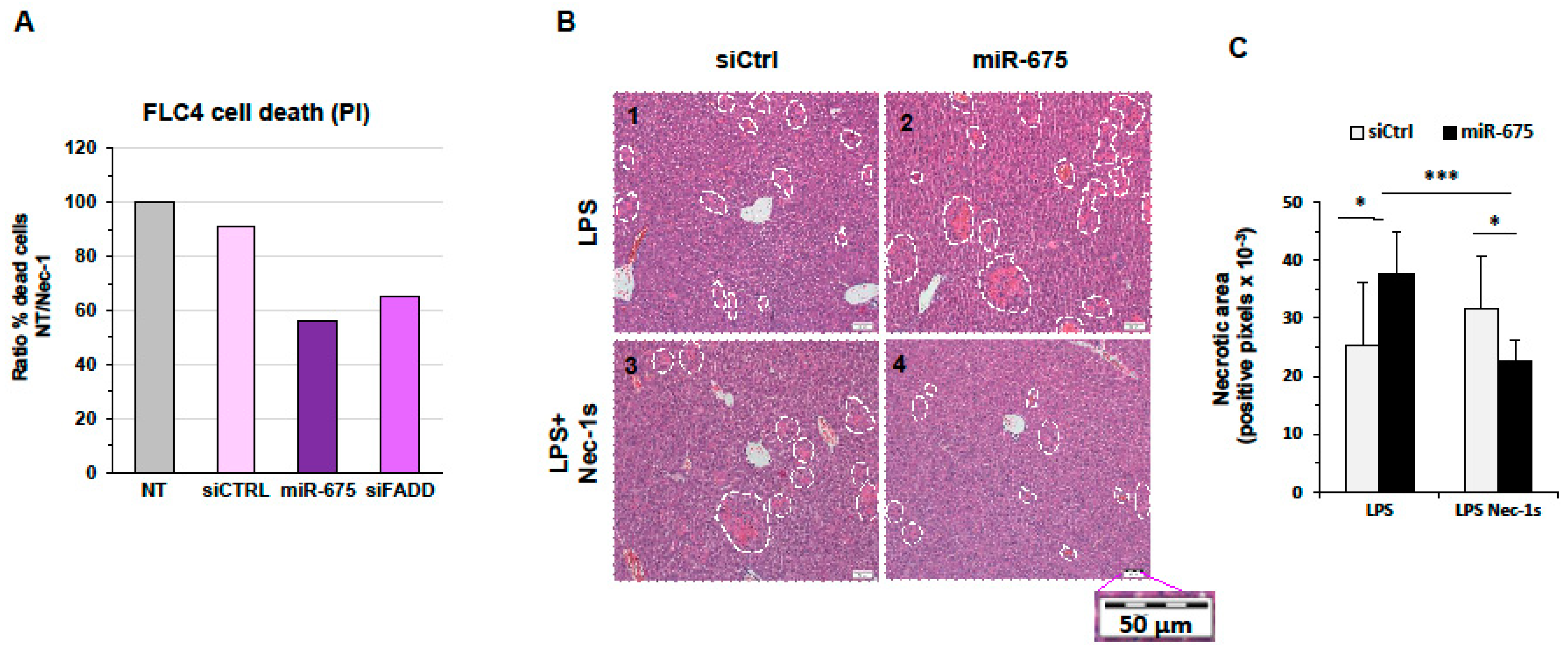
2.6. miR-675 Promotes Necrosis in Cultured Cells and Murine Livers
2.7. miR-675 Induces Necroptosis in a Human HCC Cell Line and in Mice
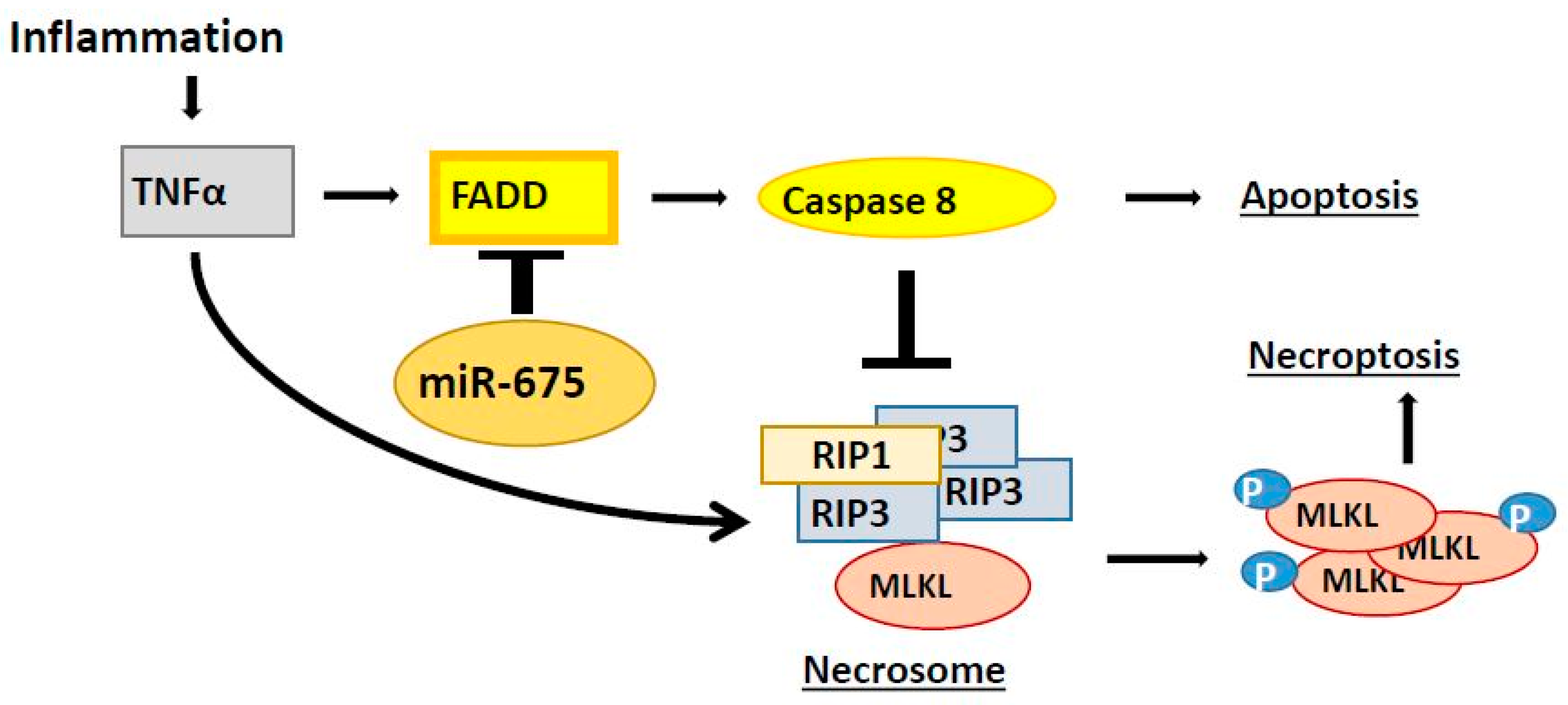
2.8. miR-675 Promotes Necroptosis by Enhancing Mixed Lineage Kinase Domain-Like Pseudokinase (MLKL) Phosphorylation
3. Discussion
4. Materials and Methods
4.1. Human Samples
4.2. Animals and Animal Procedures
4.3. Cell Lines and Cell Culture
4.4. Plasmids
4.5. Transfection Procedures and Luciferase Activity Assay
4.6. RNA Extraction and Real-Time Polymerase Chain Reaction (PCR)
4.7. Immunohistochemistry
4.8. Western Blot Analysis
4.9. Immunoprecipitation
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fischer, R.; Baumert, T.; Blum, H.E. Hepatitis C virus infection and apoptosis. World J. Gastroenterol. 2007, 13, 4865–4872. [Google Scholar] [CrossRef] [PubMed]
- Luedde, T.; Kaplowitz, N.; Schwabe, R.F. Cell death and cell death responses in liver disease: Mechanisms and clinical relevance. Gastroenterology 2014, 147, 765–783. [Google Scholar] [CrossRef] [PubMed]
- Ashkenazi, A.; Salvesen, G. Regulated cell death: Signaling and mechanisms. Annu. Rev. Cell Dev. Biol. 2014, 30, 337–356. [Google Scholar] [CrossRef] [PubMed]
- Hotchkiss, R.S.; Strasser, A.; McDunn, J.E.; Swanson, P.E. Cell death. N. Engl. J. Med. 2009, 361, 1570–1583. [Google Scholar] [CrossRef] [PubMed]
- Chan, F.K.; Baehrecke, E.H. RIP3 finds partners in crime. Cell 2012, 148, 17–18. [Google Scholar] [CrossRef] [PubMed]
- Pasparakis, M.; Vandenabeele, P. Necroptosis and its role in inflammation. Nature 2015, 517, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Guicciardi, M.E.; Malhi, H.; Mott, J.L.; Gores, G.J. Apoptosis and necrosis in the liver. Compr. Physiol. 2013, 3, 977–1010. [Google Scholar]
- Sun, L.; Wang, H.; Wang, Z.; He, S.; Chen, S.; Liao, D.; Wang, L.; Yan, J.; Liu, W.; Lei, X.; et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012, 148, 213–227. [Google Scholar] [CrossRef]
- Lin, Y.; Devin, A.; Rodriguez, Y.; Liu, Z.G. Cleavage of the death domain kinase RIP by caspase-8 prompts TNF-induced apoptosis. Genes Dev. 1999, 13, 2514–2526. [Google Scholar] [CrossRef]
- Lee, E.W.; Seo, J.; Jeong, M.; Lee, S.; Song, J. The roles of FADD in extrinsic apoptosis and necroptosis. BMB Rep. 2012, 45, 496–508. [Google Scholar] [CrossRef]
- Raveh, E.; Matouk, I.J.; Gilon, M.; Hochberg, A. The H19 Long non-coding RNA in cancer initiation, progression and metastasis—A proposed unifying theory. Mol. Cancer 2015, 14, 184. [Google Scholar] [CrossRef]
- Chen, X.; Yamamoto, M.; Fujii, K.; Nagahama, Y.; Ooshio, T.; Xin, B.; Okada, Y.; Furukawa, H.; Nishikawa, Y. Differential reactivation of fetal neonatal genes in mouse liver tumors induced in cirrhotic and non-cirrhotic conditions. Cancer Sci. 2015, 106, 972–981. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Cullen, B.R. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA 2007, 13, 313–316. [Google Scholar] [CrossRef] [PubMed]
- Schultz, N.A.; Andersen, K.K.; Roslind, A.; Willenbrock, H.; Wojdemann, M.; Johansen, J.S. Prognostic microRNAs in cancer tissue from patients operated for pancreatic cancer--five microRNAs in a prognostic index. World J. Surg. 2012, 36, 2699–2707. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, Y.C.; Lu, Y.D.; Ma, C.P. Integrated bioinformatics analyses identify dysregulated miRNAs in lung cancer. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 2270–2274. [Google Scholar] [PubMed]
- Li, H.; Li, J.; Jia, S.; Wu, M.; An, J.; Zheng, Q.; Zhang, W.; Lu, D. miR-675 upregulates long noncoding RNA H19 through activating EGR1 in human liver cancer. Oncotarget 2015, 6, 31958–31984. [Google Scholar] [CrossRef]
- Zhai, L.L.; Wang, P.; Zhou, L.Y.; Yin, J.Y.; Tang, Q.; Zhang, T.J.; Wang, Y.X.; Yang, D.Q.; Lin, J.; Deng, Z.Q. Over-expression of miR-675 in formalin-fixed paraffin-embedded (FFPE) tissues of breast cancer patients. Int. J. Clin. Exp. Med. 2015, 8, 11195–11201. [Google Scholar]
- Li, H.; Yu, B.; Li, J.; Su, L.; Yan, M.; Zhu, Z.; Liu, B. Overexpression of lncRNA H19 enhances carcinogenesis and metastasis of gastric cancer. Oncotarget 2014, 5, 2318–2329. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, Y.; Luan, W.; Wang, P.; Tao, T.; Zhang, J.; Qian, J.; Liu, N.; You, Y. Long non-coding RNA H19 promotes glioma cell invasion by deriving miR-675. PLoS ONE 2014, 9, e86295. [Google Scholar] [CrossRef]
- Tsang, W.P.; Ng, E.K.; Ng, S.S.; Jin, H.; Yu, J.; Sung, J.J.; Kwok, T.T. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis 2010, 31, 350–358. [Google Scholar] [CrossRef]
- Mauad, T.H.; van Nieuwkerk, C.M.; Dingemans, K.P.; Smit, J.J.; Schinkel, A.H.; Notenboom, R.G.; van den Bergh Weerman, M.A.; Verkruisen, R.P.; Groen, A.K.; Oude Elferink, R.P.; et al. Mice with homozygous disruption of the mdr2 P-glycoprotein gene. A novel animal model for studies of nonsuppurative inflammatory cholangitis and hepatocarcinogenesis. Am. J. Pathol. 1994, 145, 1237–1245. [Google Scholar] [PubMed]
- Ella, E.; Heim, D.; Stoyanov, E.; Harari-Steinfeld, R.; Steinfeld, I.; Pappo, O.; Perlman, T.S.; Nachmansson, N.; Rivkin, L.; Olam, D.; et al. Specific genomic and transcriptomic aberrations in tumors induced by partial hepatectomy of a chronically inflamed murine liver. Oncotarget 2014, 5, 10318–10331. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Barash, H.; Gross, E.R.; Edrei, Y.; Ella, E.; Israel, A.; Cohen, I.; Corchia, N.; Ben-Moshe, T.; Pappo, O.; Pikarsky, E.; et al. Accelerated carcinogenesis following liver regeneration is associated with chronic inflammation-induced double-strand DNA breaks. Proc. Natl. Acad. Sci. USA 2010, 107, 2207–2212. [Google Scholar] [CrossRef]
- Pikarsky, E.; Porat, R.M.; Stein, I.; Abramovitch, R.; Amit, S.; Kasem, S.; Gutkovich-Pyest, E.; Urieli-Shoval, S.; Galun, E.; Ben-Neriah, Y. NF-kappaB functions as a tumour promoter in inflammation-associated cancer. Nature 2004, 431, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Matouk, I.J.; DeGroot, N.; Mezan, S.; Ayesh, S.; Abu-lail, R.; Hochberg, A.; Galun, E. The H19 non-coding RNA is essential for human tumor growth. PLoS ONE 2007, 2, e845. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; Zhang, Y.; She, Q.; Li, X.; Peng, L.; Wang, X.; Liu, S.; Shen, X.; Zhang, W.; Dong, Y.; et al. Long Noncoding RNA H19/miR-675 Axis Promotes Gastric Cancer via FADD/Caspase 8/Caspase 3 Signaling Pathway. Cell Physiol. Biochem. 2017, 42, 2364–2376. [Google Scholar] [CrossRef]
- Matouk, I.J.; Mezan, S.; Mizrahi, A.; Ohana, P.; Abu-Lail, R.; Fellig, Y.; Degroot, N.; Galun, E.; Hochberg, A. The oncofetal H19 RNA connection: Hypoxia, p53 and cancer. Biochim. Biophys. Acta 2010, 1803, 443–451. [Google Scholar] [CrossRef]
- Keniry, A.; Oxley, D.; Monnier, P.; Kyba, M.; Dandolo, L.; Guillaume Smits, G.; Reik, W. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat. Cell Biol. 2012, 14, 659–665. [Google Scholar] [CrossRef]
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2000, 438, 685–689. [Google Scholar] [CrossRef]
- Zhuang, M.; Gao, W.; Xu, J.; Wang, P.; Shu, Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochem. Biophys. Res. Commun. 2014, 448, 315–322. [Google Scholar] [CrossRef]
- Blander, J.M. A long-awaited merger of the pathways mediating host defence and programmed cell death. Nat. Rev. Immunol. 2014, 14, 601–618. [Google Scholar] [CrossRef] [PubMed]
- Luedde, T.; Schwabe, R.F. NF-κB in the liver—linking injury, fibrosis and hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 108–118. [Google Scholar] [CrossRef] [PubMed]
- Karin, M.; Ben-Neriah, Y. Phosphorylation meets ubiquitination: The control of NF-[kappa]B activity. Annu. Rev. Immunol. 2000, 18, 621–663. [Google Scholar] [CrossRef] [PubMed]
- Genin, P.; Algarte, M.; Roof, P.; Lin, R.; Hiscott, J. Regulation of RANTES Chemokine Gene Expression Requires Cooperativity Between NF-kB and IFN-Regulatory Factor Transcription Factors. J. Immunol. 2000, 164, 5352–5361. [Google Scholar] [CrossRef] [PubMed]
- Palomba, L.; Sestili, P.; Columbaro, M.; Falcieri, E.; Cantoni, O. Apoptosis and necrosis following exposure of U937 cells to increasing concentrations of hydrogen peroxide: The effect of the poly(ADP-ribose)polymerase inhibitor 3-aminobenzamide. Biochem. Pharmacol. 1999, 58, 1743–1750. [Google Scholar] [CrossRef]
- Du, C.; Fang, M.; Li, Y.; Li, L.; Wang, X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 2000, 102, 33–42. [Google Scholar] [CrossRef]
- Degterev, A.; Huang, Z.; Boyce, M.; Li, Y.; Jagtap, P.; Mizushima, N.; Cuny, G.D.; Mitchison, T.J.; Moskowitz, M.A.; Yuan, J. Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 2005, 1, 112–119. [Google Scholar] [CrossRef]
- Hernandez, J.M.; Elahi, A.; Clark, C.W.; Wang, J.; Humphries, L.A.; Centeno, B.; Bloom, G.; Fuchs, B.C.; Yeatman, T.; Shibata, D. miR-675 mediates downregulation of Twist1 and Rb in AFP-secreting hepatocellular carcinoma. Ann. Surg. Oncol. 2013, 20, 625–635. [Google Scholar] [CrossRef]
- Gamaev, L.; Mizrahi, L.; Friehmann, T.; Rosenberg, N.; Pappo, O.; Olam, D.; Zeira, E.; Bahar Halpern, K.; Caruso, S.; Zucman-Rossi, J.; et al. The pro-oncogenic effect of the lncRNA H19 in the development of chronic inflammation-mediated hepatocellular carcinoma. Oncogene 2020. [Google Scholar] [CrossRef]
- Nath, B.; Szabo, G. Hypoxia and hypoxia inducible factors: Diverse roles in liver diseases. Hepatology 2012, 55, 622–633. [Google Scholar] [CrossRef]
- Lin, D.; Wu, J. Hypoxia inducible factor in hepatocellular carcinoma: A therapeutic target. World J. Gastroenterol. 2015, 21, 12171–12178. [Google Scholar] [CrossRef] [PubMed]
- Vanlangenakker, N.; Bertrand, M.J.; Bogaert, P.; Vandenabeele, P.; Vanden Berghe, T. TNF-induced necroptosis in L929 cells is tightly regulated by multiple TNFR1 complex I and II members. Cell Death Dis. 2011, 2, e230. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Vanden Berghe, T.; Vandenabeele, P.; Kroemer, G. The molecular machinery of regulated cell death. Cell Res. 2019, 29, 347–364. [Google Scholar] [CrossRef] [PubMed]
- Weber, K.; Roelandt, R.; Bruggeman, I.; Estornes, Y.; Vandenabeele, P. Nuclear RIPK3 and MLKL contribute to cytosolic necrosome formation and necroptosis. Commun. Biol. 2018, 1, 6. [Google Scholar] [CrossRef]
- Wang, T.; Jin, Y.; Yang, W.; Zhang, L.; Jin, X.; Liu, X.; He, Y.; Li, X. Necroptosis in cancer: An angel or a demon? Tumor Biol. 2017, 39, 1010428317711539. [Google Scholar] [CrossRef]
- Lottin, S.; Vercoutter-Edouart, A.S.; Adriaenssens, E.; Czeszak, X.; Lemoine, J.; Roudbaraki, M.; Coll, J.; Hondermarck, H.; Dugimont, T.; Curgy, J.J. Thioredoxin post-transcriptional regulation by H19 provides a new function to mRNA-like non-coding RNA. Oncogene 2002, 21, 1625–1631. [Google Scholar] [CrossRef]
- Rivkin, M.; Simerzin, A.; Zorde-Khvalevsky, E.; Chai, C.; Yuval, J.B.; Rosenberg, N. Inflammation-Induced Expression and Secretion of MicroRNA 122 Leads to Reduced Blood Levels of Kidney- Derived Erythropoietin and Anemia. Gastroenterology 2016, 151, 999–1010. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Harari-Steinfeld, R.; Gefen, M.; Simerzin, A.; Zorde-Khvalevsky, E.; Rivkin, M.; Ella, E.; Friehmann, T.; Gerlic, M.; Zucman-Rossi, J.; Caruso, S.; et al. The lncRNA H19-Derived MicroRNA-675 Promotes Liver Necroptosis by Targeting FADD. Cancers 2021, 13, 411. https://doi.org/10.3390/cancers13030411
Harari-Steinfeld R, Gefen M, Simerzin A, Zorde-Khvalevsky E, Rivkin M, Ella E, Friehmann T, Gerlic M, Zucman-Rossi J, Caruso S, et al. The lncRNA H19-Derived MicroRNA-675 Promotes Liver Necroptosis by Targeting FADD. Cancers. 2021; 13(3):411. https://doi.org/10.3390/cancers13030411
Chicago/Turabian StyleHarari-Steinfeld, Rona, Maytal Gefen, Alina Simerzin, Elina Zorde-Khvalevsky, Mila Rivkin, Ezra Ella, Tomer Friehmann, Mordechay Gerlic, Jessica Zucman-Rossi, Stefano Caruso, and et al. 2021. "The lncRNA H19-Derived MicroRNA-675 Promotes Liver Necroptosis by Targeting FADD" Cancers 13, no. 3: 411. https://doi.org/10.3390/cancers13030411
APA StyleHarari-Steinfeld, R., Gefen, M., Simerzin, A., Zorde-Khvalevsky, E., Rivkin, M., Ella, E., Friehmann, T., Gerlic, M., Zucman-Rossi, J., Caruso, S., Leveille, M., Estall, J. L., Goldenberg, D. S., Giladi, H., Galun, E., & Bromberg, Z. (2021). The lncRNA H19-Derived MicroRNA-675 Promotes Liver Necroptosis by Targeting FADD. Cancers, 13(3), 411. https://doi.org/10.3390/cancers13030411





