Immune Milieu and Genomic Alterations Set the Triple-Negative Breast Cancer Immunomodulatory Subtype Tumor Behavior
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Selection
2.2. Nucleic Acid Extraction
2.3. Gene Expression Profiling
2.4. Classification of TNBC Patients in IM and Non-IM Subtypes
2.5. Immunohistochemistry of Tumor Sections
2.6. Next-Generation Sequencing
2.7. Statistical Analyses
3. Results
3.1. Clinicopathological Features of TNBC Patients
3.2. Treatment Management of TNBC Patients
3.3. Classification of TNBC Patients into IM and Non-IM Subtypes
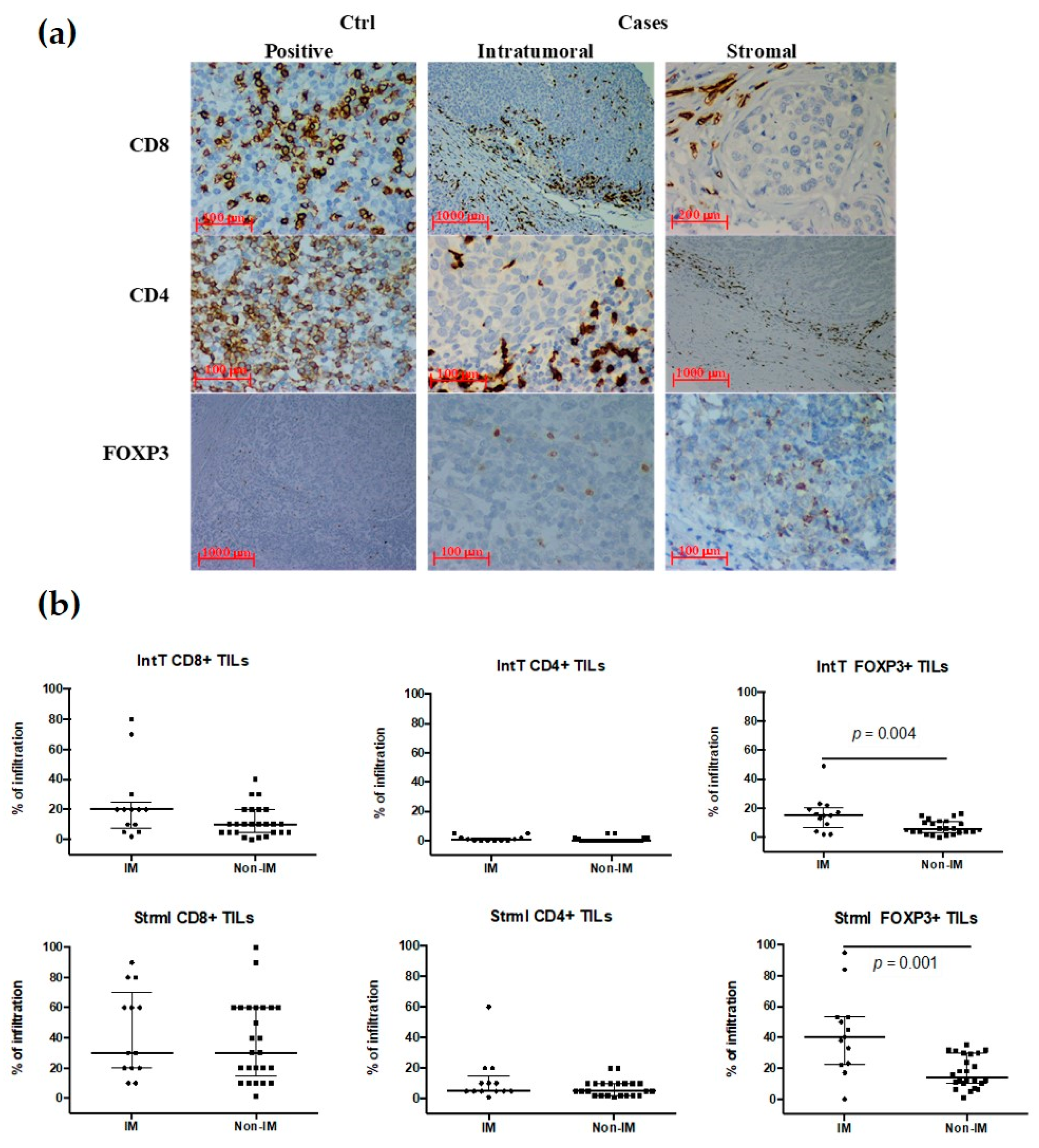
3.4. TILs Subpopulations in the IM Subtype
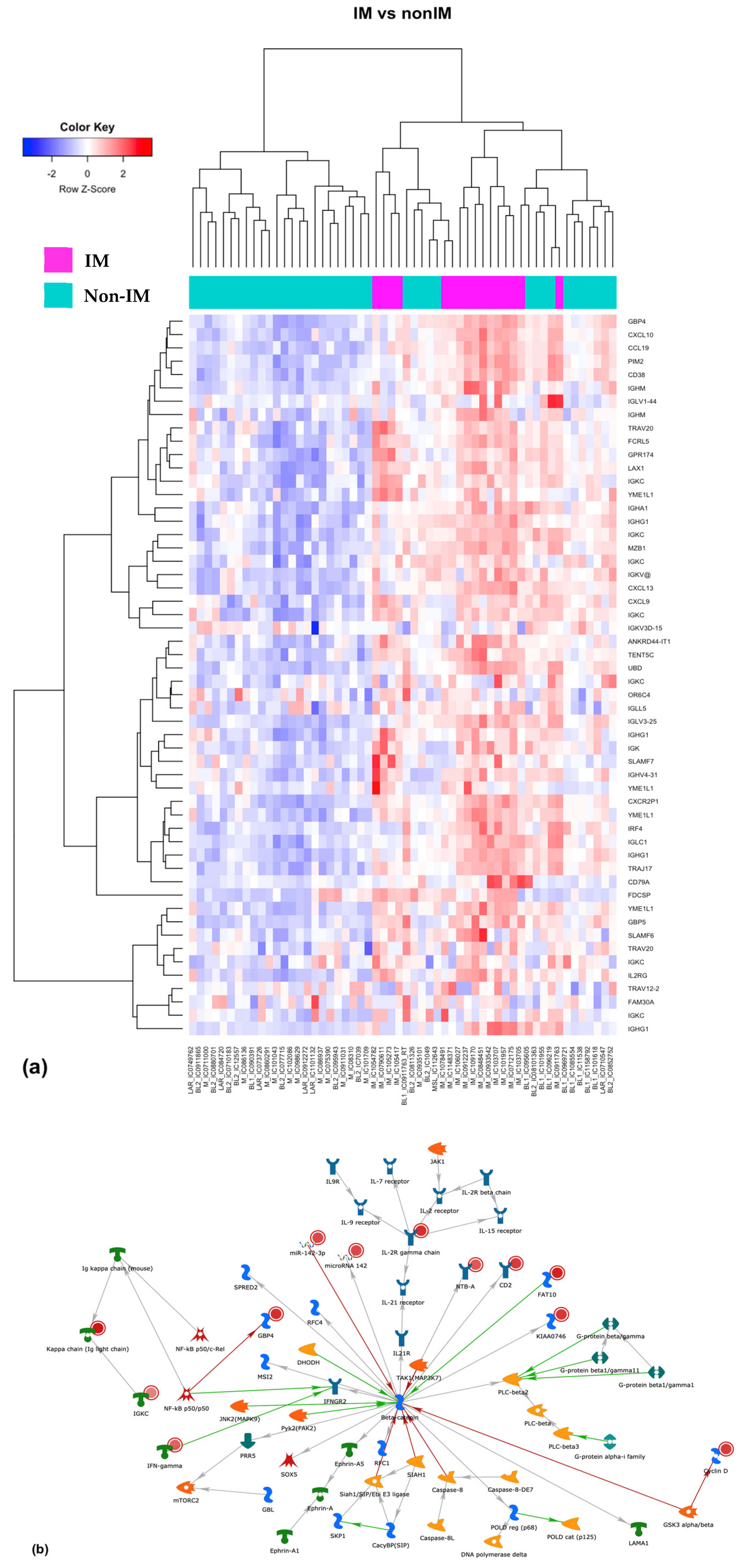
3.5. Differential Expression of Coding and Long Non-Coding RNAs
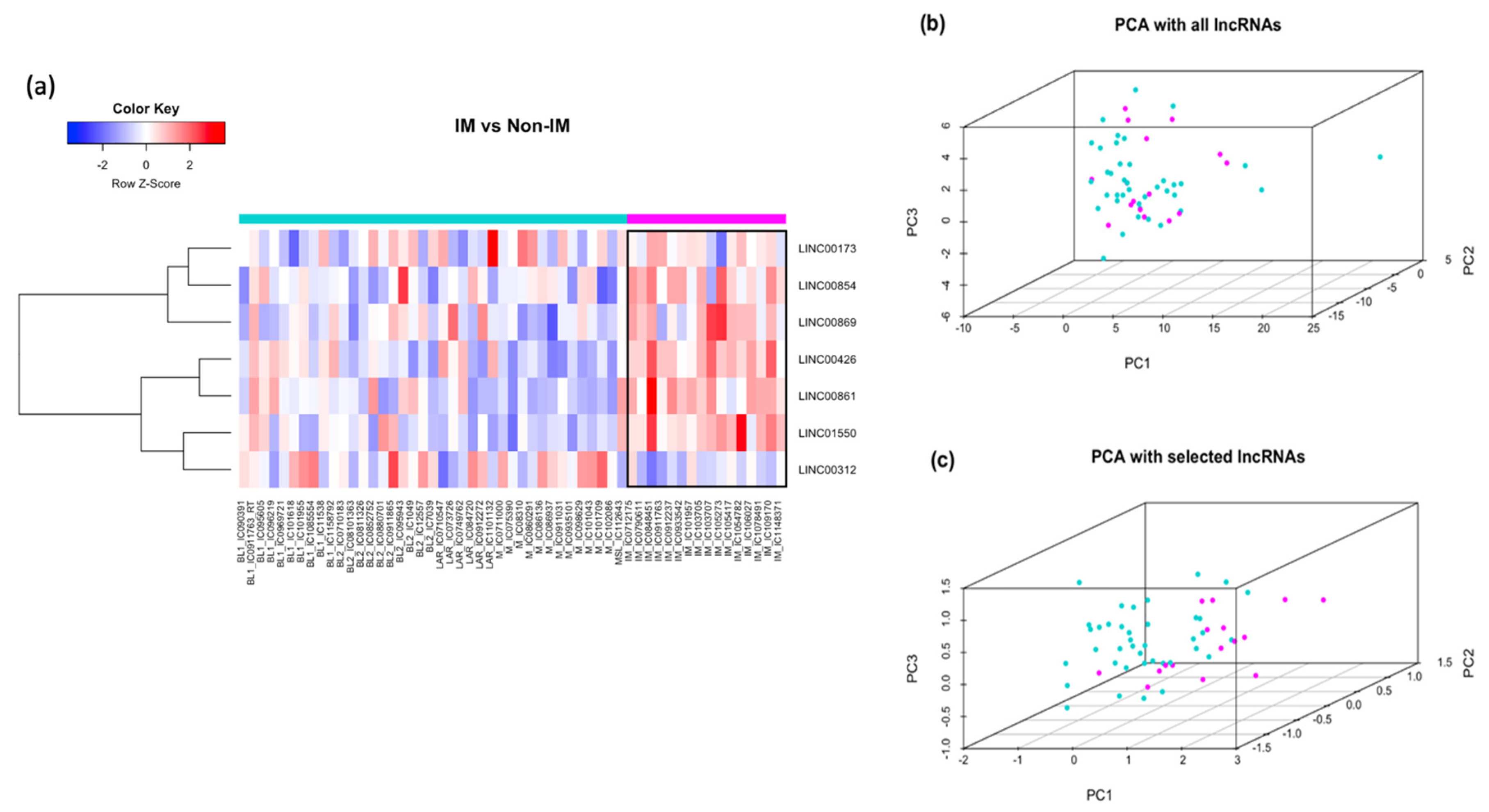
3.6. LncRNAs Associated with the IM Subtype
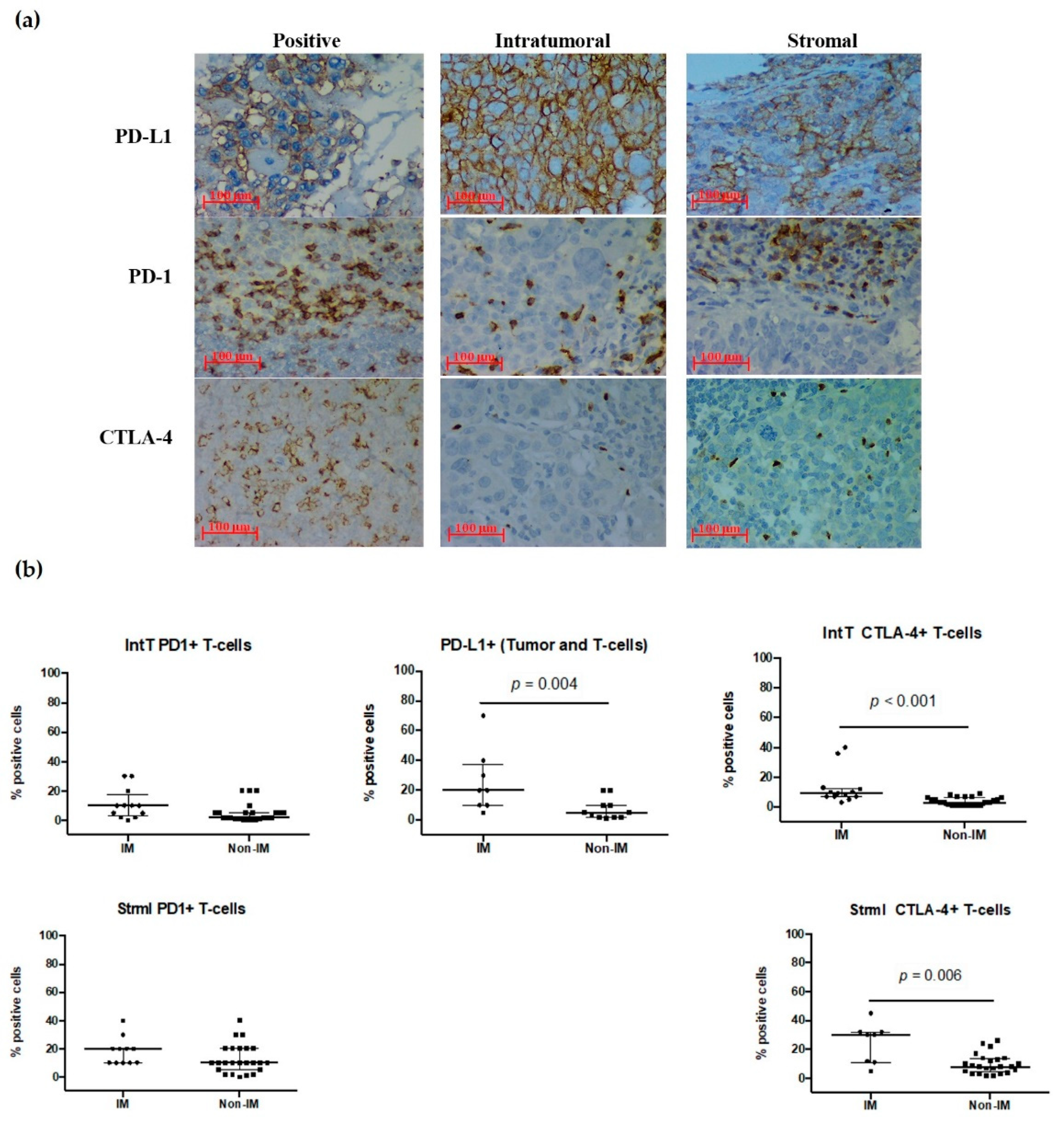
3.7. PD-1, PD-L1, and CTL-4 Expression in the IM Subtype
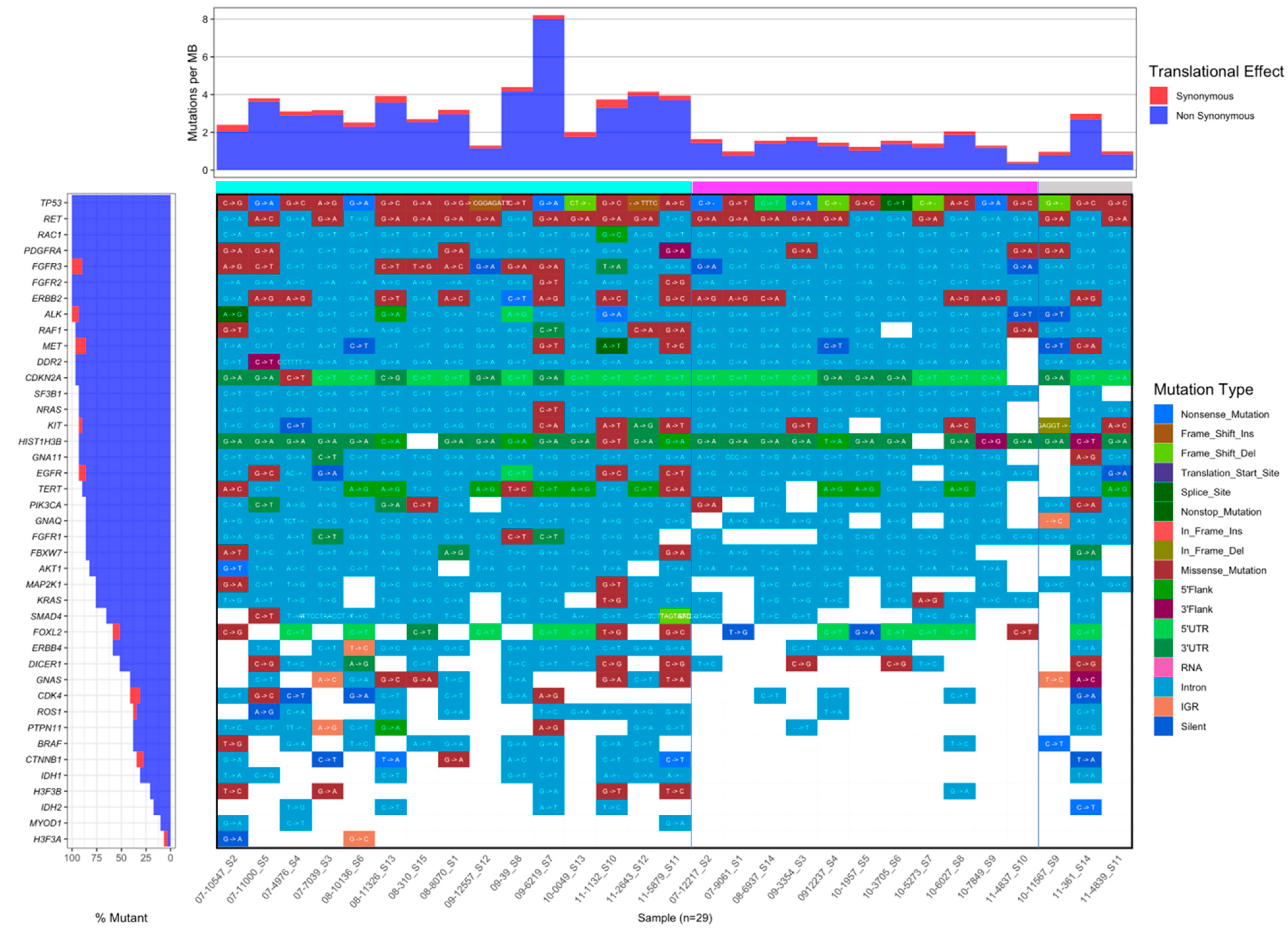
3.8. Comparison of Mutation Frequency between IM and Non-IM Subtypes
3.9. Clinical Variables and Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lara-Medina, F.; Pérez-Sánchez, V.; Saavedra-Pérez, D.; Blake-Cerda, M.; Arce, C.; Motola-Kuba, D.; Villarreal-Garza, C.; González-Angulo, A.M.; Bargalló, E.; Aguilar, J.L.; et al. Triple-Negative Breast Cancer in Hispanic Patients. Cancer 2011, 117, 3658–3669. [Google Scholar] [CrossRef] [PubMed]
- Rey-Vargas, L.; Sanabria-Salas, M.C.; Fejerman, L.; Serrano-Gomez, S.J. Risk Factors for Triple-Negative Breast Cancer among Latina Women. Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Soc. Prev. Oncol. 2019, 28, 1771–1783. [Google Scholar] [CrossRef] [PubMed]
- Burstein, M.D.; Tsimelzon, A.; Poage, G.M.; Covington, K.R.; Contreras, A.; Fuqua, S.A.W.; Savage, M.I.; Osborne, C.K.; Hilsenbeck, S.G.; Chang, J.C.; et al. Comprehensive Genomic Analysis Identifies Novel Subtypes and Targets of Triple-Negative Breast Cancer. Clin. Cancer Res. 2015, 21, 1688–1698. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of Human Triple-Negative Breast Cancer Subtypes and Preclinical Models for Selection of Targeted Therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- Wang, D.Y.; Jiang, Z.; Ben-David, Y.; Woodgett, J.R.; Zacksenhaus, E. Molecular Stratification within Triple-Negative Breast Cancer Subtypes. Sci. Rep. 2019, 9, 19107. [Google Scholar] [CrossRef]
- Yousefi, H.; Yuan, J.; Keshavarz-Fathi, M.; Murphy, J.F.; Rezaei, N. Immunotherapy of Cancers Comes of Age. Expert Rev. Clin. Immunol. 2017, 13, 1001–1015. [Google Scholar] [CrossRef]
- Disis, M.L.; Stanton, S.E. Triple-Negative Breast Cancer: Immune Modulation as the New Treatment Paradigm. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, e25–e30. [Google Scholar] [CrossRef]
- Ono, M.; Tsuda, H.; Shimizu, C.; Yamamoto, S.; Shibata, T.; Yamamoto, H.; Hirata, T.; Yonemori, K.; Ando, M.; Tamura, K.; et al. Tumor-Infiltrating Lymphocytes Are Correlated with Response to Neoadjuvant Chemotherapy in Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2012, 132, 793–805. [Google Scholar] [CrossRef]
- Dieci, M.V.; Criscitiello, C.; Goubar, A.; Viale, G.; Conte, P.; Guarneri, V.; Ficarra, G.; Mathieu, M.C.; Delaloge, S.; Curigliano, G.; et al. Prognostic Value of Tumor-Infiltrating Lymphocytes on Residual Disease after Primary Chemotherapy for Triple-Negative Breast Cancer: A Retrospective Multicenter Study. Ann. Oncol. 2014, 25, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Du, Q.; Che, J.; Jiang, X.; Li, L.; Luo, X.; Li, Q. PD-L1 Acts as a Promising Immune Marker to Predict the Response to Neoadjuvant Chemotherapy in Breast Cancer Patients. Clin. Breast Cancer 2020, 20, e99–e111. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Lachapelle, J.; Leung, S.; Gao, D.; Foulkes, W.D.; Nielsen, T.O. CD8 +lymphocyte Infiltration Is an Independent Favorable Prognostic Indicator in Basal-like Breast Cancer. Breast Cancer Res. 2012, 14, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, S.M.A.; Paish, E.C.; Powe, D.G.; Macmillan, R.D.; Grainge, M.J.; Lee, A.H.S.; Ellis, I.O.; Green, A.R. Tumor-Infiltrating CD8+ Lymphocytes Predict Clinical Outcome in Breast Cancer. J. Clin. Oncol. 2011, 29, 1949–1955. [Google Scholar] [CrossRef] [PubMed]
- Gao, G.; Wang, Z.; Qu, X.; Zhang, Z. Prognostic Value of Tumor-Infiltrating Lymphocytes in Patients with Triple-Negative Breast Cancer: A Systematic Review and Meta-Analysis. BMC Cancer 2020, 20, 179. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, M.; Sasano, H.; Tamaki, K.; Chan, M.; Hirakawa, H.; Suzuki, A.; Tada, H.; Watanabe, G.; Nemoto, N.; Nakagawa, S.; et al. Tumor-Infiltrating CD8+ and FOXP3+ Lymphocytes in Triple-Negative Breast Cancer: Its Correlation with Pathological Complete Response to Neoadjuvant Chemotherapy. Breast Cancer Res. Treat. 2014, 148, 525–534. [Google Scholar] [CrossRef] [PubMed]
- Schmid, P.; Rugo, H.S.; Adams, S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Henschel, V.; Molinero, L.; Chui, S.Y.; et al. Atezolizumab plus Nab-Paclitaxel as First-Line Treatment for Unresectable, Locally Advanced or Metastatic Triple-Negative Breast Cancer (IMpassion130): Updated Efficacy Results from a Randomised, Double-Blind, Placebo-Controlled, Phase 3 Trial. Lancet Oncol. 2020, 21, 44–59. [Google Scholar] [CrossRef]
- Schmid, P.; Adams, S.; Rugo, H.S.; Schneeweiss, A.; Barrios, C.H.; Iwata, H.; Diéras, V.; Hegg, R.; Im, S.-A.; Shaw Wright, G.; et al. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N. Engl. J. Med. 2018, 379, 2108–2121. [Google Scholar] [CrossRef] [PubMed]
- Cortes, J.; Cescon, D.W.; Rugo, H.S.; Nowecki, Z.; Im, S.A.; Yusof, M.M.; Gallardo, C.; Lipatov, O.; Barrios, C.H.; Holgado, E.; et al. Pembrolizumab plus Chemotherapy versus Placebo plus Chemotherapy for Previously Untreated Locally Recurrent Inoperable or Metastatic Triple-Negative Breast Cancer (KEYNOTE-355): A Randomised, Placebo-Controlled, Double-Blind, Phase 3 Clinical Trial. Lancet 2020, 396, 1817–1828. [Google Scholar] [CrossRef]
- Schmid, P.; Cortes, J.; Pusztai, L.; McArthur, H.; Kümmel, S.; Bergh, J.; Denkert, C.; Park, Y.H.; Hui, R.; Harbeck, N.; et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N. Engl. J. Med. 2020, 382, 810–821. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Zhang, H.; Barrios, C.H.; Saji, S.; Jung, K.H.; Hegg, R.; Koehler, A.; Sohn, J.; Iwata, H.; Telli, M.L.; et al. Neoadjuvant Atezolizumab in Combination with Sequential Nab-Paclitaxel and Anthracycline-Based Chemotherapy versus Placebo and Chemotherapy in Patients with Early-Stage Triple-Negative Breast Cancer (IMpassion031): A Randomised, Double-Blind, Phase 3 Trial. Lancet 2020, 396, 1090–1100. [Google Scholar] [CrossRef]
- Stovgaard, E.S.; Dyhl-Polk, A.; Roslind, A.; Balslev, E.; Nielsen, D. PD-L1 Expression in Breast Cancer: Expression in Subtypes and Prognostic Significance: A Systematic Review. Breast Cancer Res. Treat. 2019, 174, 571–584. [Google Scholar] [CrossRef]
- Mittendorf, E.A.; Philips, A.V.; Meric-Bernstam, F.; Qiao, N.; Wu, Y.; Harrington, S.; Su, X.; Wang, Y.; Gonzalez-Angulo, A.M.; Akcakanat, A.; et al. PD-L1 Expression in Triple-Negative Breast Cancer. Cancer Immunol. Res. 2014, 2, 361–370. [Google Scholar] [CrossRef]
- Peng, Z.; Su, P.; Yang, Y.; Yao, X.; Zhang, Y.; Jin, F.; Yang, B. Identification of CTLA-4 Associated with Tumor Microenvironment and Competing Interactions in Triple Negative Breast Cancer by Co-Expression Network Analysis. J. Cancer 2020, 11, 6365–6375. [Google Scholar] [CrossRef]
- Karn, T.; Jiang, T.; Hatzis, C.; Sänger, N.; El-Balat, A.; Rody, A.; Holtrich, U.; Becker, S.; Bianchini, G.; Pusztai, L. Association Between Genomic Metrics and Immune Infiltration in Triple-Negative Breast Cancer. JAMA Oncol. 2017, 3, 1707–1711. [Google Scholar] [CrossRef] [PubMed]
- Allison, K.H.; Hammond, M.E.H.; Dowsett, M.; McKernin, S.E.; Carey, L.A.; Fitzgibbons, P.L.; Hayes, D.F.; Lakhani, S.R.; Chavez-MacGregor, M.; Perlmutter, J.; et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Oncol. 2020, 38, 1346–1366. [Google Scholar] [CrossRef] [PubMed]
- Irizarry, R.A.; Bolstad, B.M.; Collin, F.; Cope, L.M.; Hobbs, B.; Speed, T.P. Summaries of Affymetrix GeneChip Probe Level Data. Nucleic Acids Res. 2003, 31, e15. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, J.; Gray, W.H.; Lehmann, B.D.; Bauer, J.A.; Shyr, Y.; Pietenpol, J.A. TNBCtype: A Subtyping Tool for Triple-Negative Breast Cancer. Cancer Inform. 2012, 11, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Babraham Bioinformatics—FastQC A Quality Control Tool for High Throughput Sequence Data. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 12 November 2021).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce Framework for Analyzing next-Generation DNA Sequencing Data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Skidmore, Z.L.; Wagner, A.H.; Lesurf, R.; Campbell, K.M.; Kunisaki, J.; Griffith, O.L.; Griffith, M. GenVisR: Genomic Visualizations in R. Bioinformatics 2016, 32, 3012–3014. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A.; Cruz, C.; Eder, J.P.; Braiteh, F.; Chung, C.; Tolaney, S.M.; Kuter, I.; Nanda, R.; Cassier, P.A.; Delord, J.P.; et al. Long-Term Clinical Outcomes and Biomarker Analyses of Atezolizumab Therapy for Patients with Metastatic Triple-Negative Breast Cancer: A Phase 1 Study. JAMA Oncol. 2019, 5, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Miles, D.; Gligorov, J.; André, F.; Cameron, D.; Schneeweiss, A.; Barrios, C.; Xu, B.; Wardley, A.; Kaen, D.; Andrade, L.; et al. Primary Results from IMpassion131, a Double-Blind, Placebo-Controlled, Randomised Phase III Trial of First-Line Paclitaxel with or without Atezolizumab for Unresectable Locally Advanced/Metastatic Triple-Negative Breast Cancer. Ann. Oncol. 2021, 32, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Roche—Roche Provides Update on Tecentriq US Indication for PD-L1-Positive, Metastatic Triple-Negative Breast Cancer. Available online: https://www.roche.com/media/releases/med-cor-2021-08-27.htm (accessed on 16 November 2021).
- Zhao, S.; Ma, D.; Xiao, Y.; Li, X.; Ma, J.; Zhang, H.; Xu, X.; Lv, H.; Jiang, W.; Yang, W.; et al. Molecular Subtyping of Triple-Negative Breast Cancers by Immunohistochemistry: Molecular Basis and Clinical Relevance. Oncologist 2020, 25, e1481. [Google Scholar] [CrossRef]
- Santonja, A.; Sánchez-Muñoz, A.; Lluch, A.; Chica-Parrado, M.R.; Albanell, J.; Chacón, J.I.; Antolín, S.; Jerez, J.M.; de la Haba, J.; de Luque, V.; et al. Triple Negative Breast Cancer Subtypes and Pathologic Complete Response Rate to Neoadjuvant Chemotherapy. Oncotarget 2018, 9, 26406. [Google Scholar] [CrossRef]
- Funakoshi, Y.; Wang, Y.; Semba, T.; Masuda, H.; Hout, D.; Ueno, N.T.; Wang, X. Comparison of Molecular Profile in Triple-Negative Inflammatory and Non-Inflammatory Breast Cancer Not of Mesenchymal Stem-like Subtype. PLoS ONE 2019, 14, e0222336. [Google Scholar] [CrossRef] [PubMed]
- Bareche, Y.; Venet, D.; Ignatiadis, M.; Aftimos, P.; Piccart, M.; Rothe, F.; Sotiriou, C. Unravelling Triple-Negative Breast Cancer Molecular Heterogeneity Using an Integrative Multiomic Analysis. Ann. Oncol. 2018, 29, 895–902. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Xiang, Y.; Li, F.; Yin, C.; Li, B.; Ke, X. WNT/β-Catenin Signaling Pathway Regulating T Cell-Inflammation in the Tumor Microenvironment. Front. Immunol. 2019, 10, 2293. [Google Scholar] [CrossRef]
- Wang, B.; Tian, T.; Kalland, K.H.; Ke, X.; Qu, Y. Targeting Wnt/β-Catenin Signaling for Cancer Immunotherapy. Trends Pharmacol. Sci. 2018, 39, 648–658. [Google Scholar] [CrossRef] [PubMed]
- Yu, Q.C.; Verheyen, E.M.; Zeng, Y.A. Mammary Development and Breast Cancer: A Wnt Perspective. Cancers 2016, 8, 65. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Canonical and Non-Canonical WNT Signaling in Cancer Stem Cells and Their Niches: Cellular Heterogeneity, Omics Reprogramming, Targeted Therapy and Tumor Plasticity (Review). Int. J. Oncol. 2017, 51, 1357–1369. [Google Scholar] [CrossRef] [PubMed]
- Khramtsov, A.I.; Khramtsova, G.F.; Tretiakova, M.; Huo, D.; Olopade, O.I.; Goss, K.H. Wnt/Beta-Catenin Pathway Activation Is Enriched in Basal-like Breast Cancers and Predicts Poor Outcome. Am. J. Pathol. 2010, 176, 2911–2920. [Google Scholar] [CrossRef]
- Kwee, S.A.; Tiirikainen, M. Beta-Catenin Activation and Immunotherapy Resistance in Hepatocellular Carcinoma: Mechanisms and Biomarkers. Hepatoma Res. 2021, 7, 8. [Google Scholar] [CrossRef]
- Morin, P.J.; Kinzler, K.W.; Sparks, A.B. β-Catenin Mutations: Insights into the APC Pathway and the Power of Genetics. Cancer Res. 2016, 76, 5587–5589. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez Bautista, R.; Ortega Gómez, A.; Hidalgo Miranda, A.; Zentella Dehesa, A.; Villarreal-Garza, C.; Ávila-Moreno, F.; Arrieta, O. Long Non-Coding RNAs: Implications in Targeted Diagnoses, Prognosis, and Improved Therapeutic Strategies in Human Non- and Triple-Negative Breast Cancer. Clin. Epigenet. 2018, 10, 88. [Google Scholar] [CrossRef] [PubMed]
- Beltrán-Anaya, F.O.; Romero-Córdoba, S.; Rebollar-Vega, R.; Arrieta, O.; Bautista-Piña, V.; Dominguez-Reyes, C.; Villegas-Carlos, F.; Tenorio-Torres, A.; Alfaro-Riuz, L.; Jiménez-Morales, S.; et al. Expression of Long Non-Coding RNA ENSG00000226738 (LncKLHDC7B) Is Enriched in the Immunomodulatory Triple-Negative Breast Cancer Subtype and Its Alteration Promotes Cell Migration, Invasion, and Resistance to Cell Death. Mol. Oncol. 2019, 13, 909–927. [Google Scholar] [CrossRef]
- Koliaraki, V.; Prados, A.; Armaka, M.; Kollias, G. The Mesenchymal Context in Inflammation, Immunity and Cancer. Nat. Immunol. 2020, 21, 974–982. [Google Scholar] [CrossRef]
- Li, H.; Mu, Q.; Zhang, G.; Shen, Z.; Zhang, Y.; Bai, J.; Zhang, L.; Zhou, D.; Zheng, Q.; Shi, L.; et al. Linc00426 Accelerates Lung Adenocarcinoma Progression by Regulating MiR-455-5p as a Molecular Sponge. Cell Death Dis. 2020, 11, 1051. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, P.; Chen, Z.; Wang, S.; Tang, S.; Chen, X.; Chen, Z.; Zhou, J. Elevated LINC01550 Induces the Apoptosis and Cell Cycle Arrest of Melanoma. Med Oncol. 2021, 38, 32. [Google Scholar] [CrossRef] [PubMed]
- Fan, H.; Yuan, J.; Li, X.; Ma, Y.; Wang, X.; Xu, B.; Li, X. LncRNA LINC00173 Enhances Triple-Negative Breast Cancer Progression by Suppressing MiR-490-3p Expression. Biomed. Pharmacother. 2020, 125, 109987. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Li, Y.; Wang, Q.; Zhang, X.; Wang, D.; Tang, H.C.; Meng, X.; Ding, X. Identification of an IncRNA-MiRNA-MRNA Interaction Mechanism in Breast Cancer Based on Bioinformatic Analysis. Mol. Med. Rep. 2017, 16, 5113–5120. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Qiu, F.; Huang, L.; Liu, W.; Li, L.; Ji, C.; Zeng, X.; Qiao, L.; Liu, M.; Gong, X. Long Non-Coding RNA LINC00312 Regulates Breast Cancer Progression through the MiR-9/CDH1 Axis. Mol. Med. Rep. 2020, 21, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, C.; Guo, J.; Liu, K.; Hu, Y.; Wu, D.; Fang, H.; Zou, Y.; Wei, Z.; Wang, Z.; et al. Cis- and Trans-Acting Expression Quantitative Trait Loci of Long Non-Coding RNA in 2549 Cancers With Potential Clinical and Therapeutic Implications. Front. Oncol. 2020, 10, 2279. [Google Scholar] [CrossRef]
- Yang, Q.; Tang, Y.; Tang, C.; Cong, H.; Wang, X.; Shen, X.; Ju, S. Diminished LINC00173 Expression Induced MiR-182-5p Accumulation Promotes Cell Proliferation, Migration and Apoptosis Inhibition via AGER/NF-ΚB Pathway in Non-Small-Cell Lung Cancer. Am. J. Transl. Res. 2019, 11, 4248–4262. [Google Scholar] [PubMed]
- Hu, W.; Wang, Y.; Fang, Z.; He, W.; Li, S. Integrated Characterization of LncRNA-Immune Interactions in Prostate Cancer. Front. Cell Dev. Biol. 2021, 9, 271. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Wang, J.; Shan, B.; Li, B.; Peng, W.; Dong, Y.; Shi, W.; Zhao, W.; He, D.; Duan, M.; et al. The Long Noncoding RNA LINC00312 Induces Lung Adenocarcinoma Migration and Vasculogenic Mimicry through Directly Binding YBX1. Mol. Cancer 2018, 17, 167. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-J.; Wang, Q.; Guo, B.; Liu, X.-Y.; Wang, B. Identification of Skin-Related LncRNAs as Potential Biomarkers That Involved in Wnt Pathways in Keloids. Oncotarget 2017, 8, 34236–34244. [Google Scholar] [CrossRef]
- Li, Q.; Chen, X.; Chen, L.; Yan, H.; Li, J. LINC00173 Promotes the Apoptosis of Hypertrophic Scar Fibroblasts through Increasing β-Catenin Expression. Mol. Cell. Biochem. 2020, 476, 1005–1014. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, L.; Ding, X.; Sui, X. LINC00861 Inhibits the Progression of Cervical Cancer Cells by Functioning as a CeRNA for MiR-513b-5p and Regulating the PTEN/AKT/MTOR Signaling Pathway. Mol. Med. Rep. 2021, 23, 1. [Google Scholar] [CrossRef]
- Guo, Z.; Wang, Y.H.; Xu, H.; Yuan, C.S.; Zhou, H.H.; Huang, W.H.; Wang, H.; Zhang, W. LncRNA Linc00312 Suppresses Radiotherapy Resistance by Targeting DNA-PKcs and Impairing DNA Damage Repair in Nasopharyngeal Carcinoma. Cell Death Dis. 2021, 12, 69. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wu, J.; Wang, Y.; Zhao, T.; Ma, B.; Liu, Y.; Fang, W.; Zhu, W.G.; Zhang, H. Kindlin 2 Forms a Transcriptional Complex with β-Catenin and TCF4 to Enhance Wnt Signalling. EMBO Rep. 2012, 13, 750–758. [Google Scholar] [CrossRef]
- Genovese, G.; Ghosh, P.; Li, H.; Rettino, A.; Sioletic, S.; Cittadini, A.; Sgambato, A. The Tumor Suppressor HINT1 Regulates MITF and β-Catenin Transcriptional Activity in Melanoma Cells. Cell Cycle (Georgetown, Tex.) 2012, 11, 2206–2215. [Google Scholar] [CrossRef] [PubMed]
- Satow, R.; Shitashige, M.; Jigami, T.; Fukami, K.; Honda, K.; Kitabayashi, I.; Yamada, T. β-Catenin Inhibits Promyelocytic Leukemia Protein Tumor Suppressor Function in Colorectal Cancer Cells. Gastroenterology 2012, 142, 572–581. [Google Scholar] [CrossRef]
- Fiset, A.; Xu, E.; Bergeron, S.; Marette, A.; Pelletier, G.; Siminovitch, K.A.; Olivier, M.; Beauchemin, N.; Faure, R.L. Compartmentalized CDK2 Is Connected with SHP-1 and β-Catenin and Regulates Insulin Internalization. Cell. Signal. 2011, 23, 911–919. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Ray, G.; Yoo, B.H.; Erdogan, M.; Rosen, K.V. Down-Regulation of Death-Associated Protein Kinase-2 Is Required for Beta-Catenin-Induced Anoikis Resistance of Malignant Epithelial Cells. J. Biol. Chem. 2009, 284, 2012–2022. [Google Scholar] [CrossRef] [PubMed]
- Bahmanyar, S.; Kaplan, D.D.; DeLuca, J.G.; Giddings, T.H.; O’Toole, E.T.; Winey, M.; Salmon, E.D.; Casey, P.J.; Nelson, W.J.; Barth, A.I.M. Beta-Catenin Is a Nek2 Substrate Involved in Centrosome Separation. Genes Dev. 2008, 22, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Weiske, J.; Albring, K.F.; Huber, O. The Tumor Suppressor Fhit Acts as a Repressor of Beta-Catenin Transcriptional Activity. Proc. Natl. Acad. Sci. USA 2007, 104, 20344–20349. [Google Scholar] [CrossRef]
- Lillehoj, E.P.; Lu, W.; Kiser, T.; Goldblum, S.E.; Kim, K.C. MUC1 Inhibits Cell Proliferation by a Beta-Catenin-Dependent Mechanism. Biochim. Biophys. Acta 2007, 1773, 1028–1038. [Google Scholar] [CrossRef]
- Liu, F.; Lang, R.; Zhao, J.; Zhang, X.; Pringle, G.A.; Fan, Y.; Yin, D.; Gu, F.; Yao, Z.; Fu, L. CD8+ Cytotoxic T Cell and FOXP3+ Regulatory T Cell Infiltration in Relation to Breast Cancer Survival and Molecular Subtypes. Breast Cancer Res. Treat. 2011, 130, 645–655. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.J.; Wahl, S.M. TGF-Beta: The Missing Link in CD4+CD25+ Regulatory T Cell-Mediated Immunosuppression. Cytokine Growth Factor Rev. 2003, 14, 85–89. [Google Scholar] [CrossRef]
- Liu, F.; Li, Y.; Ren, M.; Zhang, X.; Guo, X.; Lang, R.; Gu, F.; Fu, L. Peritumoral FOXP3+ Regulatory T Cell Is Sensitive to Chemotherapy While Intratumoral FOXP3+ Regulatory T Cell Is Prognostic Predictor of Breast Cancer Patients. Breast Cancer Res. Treat. 2012, 135, 459–467. [Google Scholar] [CrossRef] [PubMed]
- Yeong, J.; Thike, A.A.; Lim, J.C.T.; Lee, B.; Li, H.; Wong, S.-C.; Hue, S.S.S.; Tan, P.H.; Iqbal, J. Higher Densities of Foxp3 + Regulatory T Cells Are Associated with Better Prognosis in Triple-Negative Breast Cancer. Breast Cancer Res. Treat. 2017, 163, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Oldenhove, G.; de Heusch, M.; Urbain-Vansanten, G.; Urbain, J.; Maliszewski, C.; Leo, O.; Moser, M. CD4+ CD25+ Regulatory T Cells Control T Helper Cell Type 1 Responses to Foreign Antigens Induced by Mature Dendritic Cells In Vivo. J. Exp. Med. 2003, 198, 259. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.K.; Turner, H.; Maynard, C.L.; Oliver, J.R.; Chen, D.; Elson, C.O.; Weaver, C.T. Late Developmental Plasticity in the T Helper 17 Lineage. Immunity 2009, 30, 92. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Wu, H.; Lu, J.; Zhang, Y.; Luo, Y.; Xu, Q.; Shen, S.; Liang, Z. PD1 Protein Expression in Tumor Infiltrated Lymphocytes Rather than PDL1 in Tumor Cells Predicts Survival in Triple-Negative Breast Cancer. Cancer Biol. Ther. 2018, 19, 373–380. [Google Scholar] [CrossRef]
- Beckers, R.K.; Selinger, C.I.; Vilain, R.; Madore, J.; Wilmott, J.S.; Harvey, K.; Holliday, A.; Cooper, C.L.; Robbins, E.; Gillett, D.; et al. Programmed Death Ligand 1 Expression in Triple-Negative Breast Cancer Is Associated with Tumour-Infiltrating Lymphocytes and Improved Outcome. Histopathology 2016, 69, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Emens, L.A.; Goldstein, L.D.; Schmid, P.; Rugo, H.S.; Adams, S.; Barrios, C.H.; Schneeweiss, A.; Dieras, V.; Iwata, H.; Chang, C.-W.; et al. The Tumor Microenvironment (TME) and Atezolizumab + Nab-Paclitaxel (A+nP) Activity in Metastatic Triple-Negative Breast Cancer (MTNBC): IMpassion130. J. Clin. Oncol. 2021, 39, 1006. [Google Scholar] [CrossRef]
- Hassel, J.C.; Heinzerling, L.; Aberle, J.; Bähr, O.; Eigentler, T.K.; Grimm, M.O.; Grünwald, V.; Leipe, J.; Reinmuth, N.; Tietze, J.K.; et al. Combined Immune Checkpoint Blockade (Anti-PD-1/Anti-CTLA-4): Evaluation and Management of Adverse Drug Reactions. Cancer Treat. Rev. 2017, 57, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Sznol, M.; Ferrucci, P.F.; Hogg, D.; Atkins, M.B.; Wolter, P.; Guidoboni, M.; Lebbé, C.; Kirkwood, J.M.; Schachter, J.; Daniels, G.A.; et al. Pooled Analysis Safety Profile of Nivolumab and Ipilimumab Combination Therapy in Patients With Advanced Melanoma. J. Clin. Oncol. 2017, 35, 3815–3822. [Google Scholar] [CrossRef]

| Variables | % n/n |
|---|---|
| Age (years) median (min–max) | 49.5 (30–80) |
| ≥40 | 85.3 (58/68) |
| <40 | 14.7 (10/68) |
| Hormonal Status | |
| Premenopausal | 50 (34/68) |
| Postmenopausal | 50 (34/68) |
| Clinical Stage | |
| III | 83.8 (57/68) |
| IV | 16.2 (11/68) |
| Surgical procedure (mastectomy) | |
| Yes | 75.4 (43/57) |
| No | 24.6 (14/57) |
| Histology | |
| Ductal | 86.8 (59/68) |
| Lobular | 7.4 (5/68) |
| Other | 5.9 (4/68) |
| Nuclear Grade | |
| G3 | 88.2 (60/68) |
| G2 | 11.8 (8/68) |
| Vascular Infiltration | |
| Yes | 47.1 (32/68) |
| No | 52.9 (36/68) |
| Pathological Complete Response | |
| Yes | 24.6 (14/57) |
| No | 50.9 (29/57) |
| NE | 24.6 (14/57) |
| Systemic Treatment | |
| Neo/Adjuvant | 83.8 (57/68) |
| Palliative | 16.2 (11/68) |
| Radiotherapy | |
| No | 25 (17/68) |
| Yes | 75 (51/68) |
| Ki67(%) | |
| <14 | 1.5 (1/68) |
| 14–20 | 2.9 (2/68) |
| >20 | 55.9 (38/68) |
| NE | 39.7 (27/68) |
| CEA (ng/mL) | |
| <3.38 | 79.4 (54/68) |
| >3.38 | 19.1 (13/68) |
| NE | 1.5 (1/68) |
| CA 15-3 (U/mL) | |
| <12.32 | 20.6 (14/68) |
| >12.32 | 77.9 (53/68) |
| NE | 1.5 (1/68) |
| Chemotherapy Type | |
| A + T | 22.0 (15/68) |
| A + T + Cis | 61.8 (42/68) |
| A or T | 16.2 (11/68) |
| IM and Non-IM | Subtype | % (n/n) |
|---|---|---|
| IM | IM | 23.5 (16/68) |
| Non-IM 57.3 (39/68) | BL1 | 14.7 (10/68) |
| BL2 | 14.7 (10/68) | |
| LAR | 8.8 (6/68) | |
| M | 17.6 (12/68) | |
| MSL | 1.5 (1/68) | |
| UNS | 19.1 (13/68) |
| lncRNA | FCH | Mean Expression | t-Test | p Value | adj. p Value |
|---|---|---|---|---|---|
| LINC00861 | 1.64 | 4.800199 | 4.982293 | 5.82 × 10−6 | 0.00182405 |
| LINC00869 | 1.49 | 6.408327 | 3.61491 | 6.23 × 10−4 | 0.083770344 |
| LINC00426 | 1.38 | 4.920571 | 5.373881 | 1.38 × 10−6 | 0.000647671 |
| LINC01550 | 1.35 | 4.084667 | 5.5031 | 8.50 × 10−7 | 0.000647671 |
| LINC00854 | 1.32 | 6.853894 | 3.699587 | 4.76 × 10−4 | 0.074699771 |
| LINC00312 | −1.29 | 3.827583 | −2.835162 | 6.26 × 10−3 | 0.317551682 |
| LINC00173 | 1.26 | 4.548792 | 3.099832 | 2.97 × 10−3 | 0.199410413 |
| TNBC Molecular Subtype | Sub-Distribution Hazard Ratio (SHR) | Robust s.e. | z-Score | p-Value | 95% CI (Lower) | 95% CI (Upper) |
|---|---|---|---|---|---|---|
| BL1 | 1.0238 | 0.5039 | 0.05 | 0.962 | 0.3901 | 2.6865 |
| BL2 | 1.3908 | 0.7935 | 0.58 | 0.563 | 0.4546 | 4.2549 |
| IM | 0.2545 | 0.1511 | −2.31 | 0.021 | 0.0795 | 0.8146 |
| LAR | 1.3317 | 1.0144 | 0.38 | 0.707 | 0.2992 | 5.9265 |
| MSL | 1.047 | 0.5395 | 0.09 | 0.929 | 0.3813 | 2.8747 |
| M | 1.0000 † | — | — | — | — | — |
| UNS | 1.0000 † | — | — | — | — | — |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rodríguez-Bautista, R.; Caro-Sánchez, C.H.; Cabrera-Galeana, P.; Alanis-Funes, G.J.; Gutierrez-Millán, E.; Ávila-Ríos, S.; Matías-Florentino, M.; Reyes-Terán, G.; Díaz-Chávez, J.; Villarreal-Garza, C.; et al. Immune Milieu and Genomic Alterations Set the Triple-Negative Breast Cancer Immunomodulatory Subtype Tumor Behavior. Cancers 2021, 13, 6256. https://doi.org/10.3390/cancers13246256
Rodríguez-Bautista R, Caro-Sánchez CH, Cabrera-Galeana P, Alanis-Funes GJ, Gutierrez-Millán E, Ávila-Ríos S, Matías-Florentino M, Reyes-Terán G, Díaz-Chávez J, Villarreal-Garza C, et al. Immune Milieu and Genomic Alterations Set the Triple-Negative Breast Cancer Immunomodulatory Subtype Tumor Behavior. Cancers. 2021; 13(24):6256. https://doi.org/10.3390/cancers13246256
Chicago/Turabian StyleRodríguez-Bautista, Rubén, Claudia H. Caro-Sánchez, Paula Cabrera-Galeana, Gerardo J. Alanis-Funes, Everardo Gutierrez-Millán, Santiago Ávila-Ríos, Margarita Matías-Florentino, Gustavo Reyes-Terán, José Díaz-Chávez, Cynthia Villarreal-Garza, and et al. 2021. "Immune Milieu and Genomic Alterations Set the Triple-Negative Breast Cancer Immunomodulatory Subtype Tumor Behavior" Cancers 13, no. 24: 6256. https://doi.org/10.3390/cancers13246256
APA StyleRodríguez-Bautista, R., Caro-Sánchez, C. H., Cabrera-Galeana, P., Alanis-Funes, G. J., Gutierrez-Millán, E., Ávila-Ríos, S., Matías-Florentino, M., Reyes-Terán, G., Díaz-Chávez, J., Villarreal-Garza, C., Hernández-Pedro, N. Y., Ortega-Gómez, A., Lara-Mejía, L., Rangel-Escareño, C., & Arrieta, O. (2021). Immune Milieu and Genomic Alterations Set the Triple-Negative Breast Cancer Immunomodulatory Subtype Tumor Behavior. Cancers, 13(24), 6256. https://doi.org/10.3390/cancers13246256








