Therapeutic Efficacy of Variable Biological Effectiveness of Proton Therapy in U-CH2 and MUG-Chor1 Human Chordoma Cell Death
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
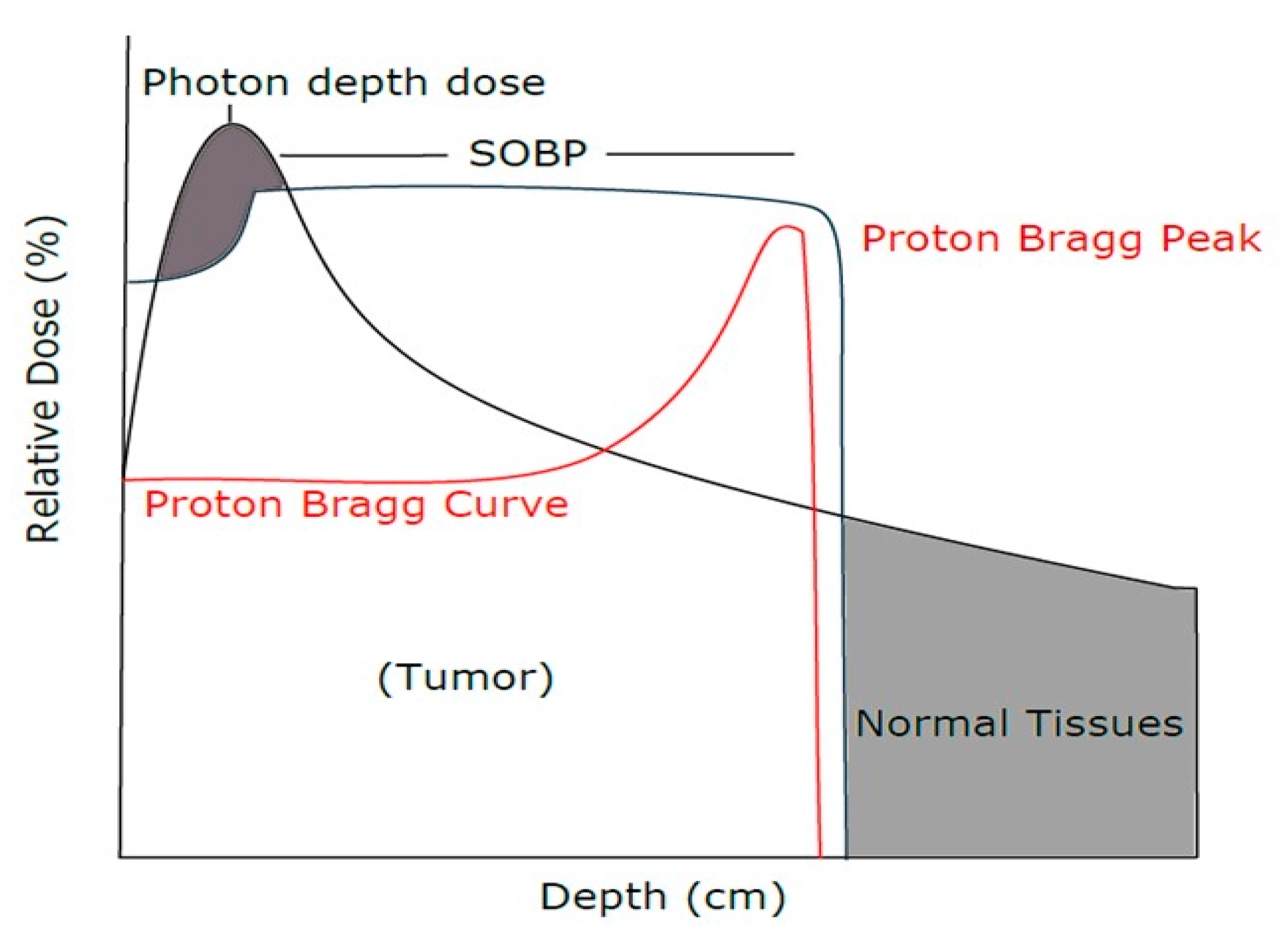
2.2. Proton Beam Radiation Treatment
2.3. Colony Forming Assay
2.4. Proton Radiation Treatment
2.5. RNA Extraction and Purification
2.6. Microarray Analysis
2.7. Statistical Analysis
3. Results
3.1. E-SOBP Treatment Enhanced Significant Killing of Chordoma Cells Compared to M-SOBP Treatment
3.2. Comparison of Relative Biological Effect in M-SOBP and E-SOBP
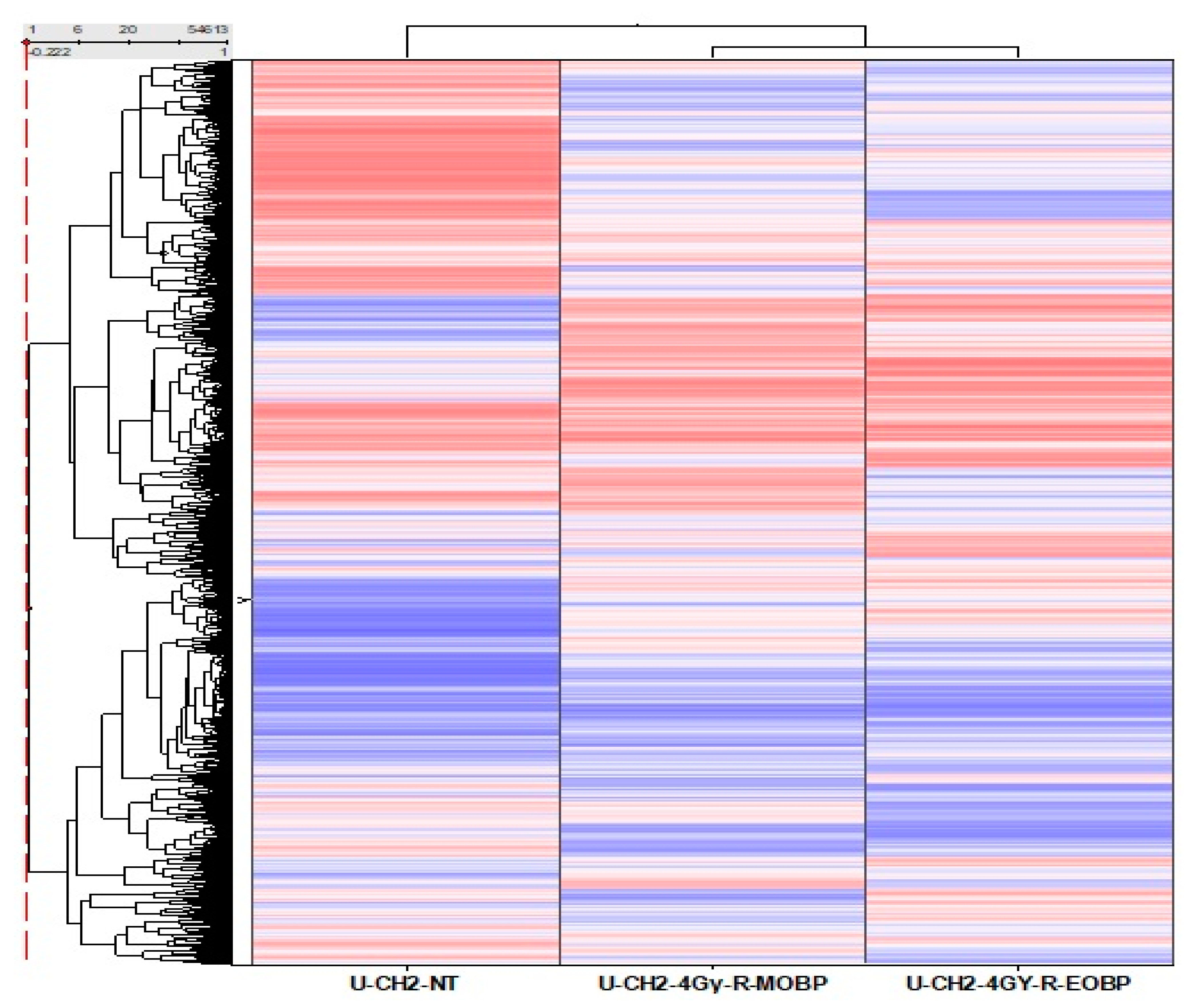
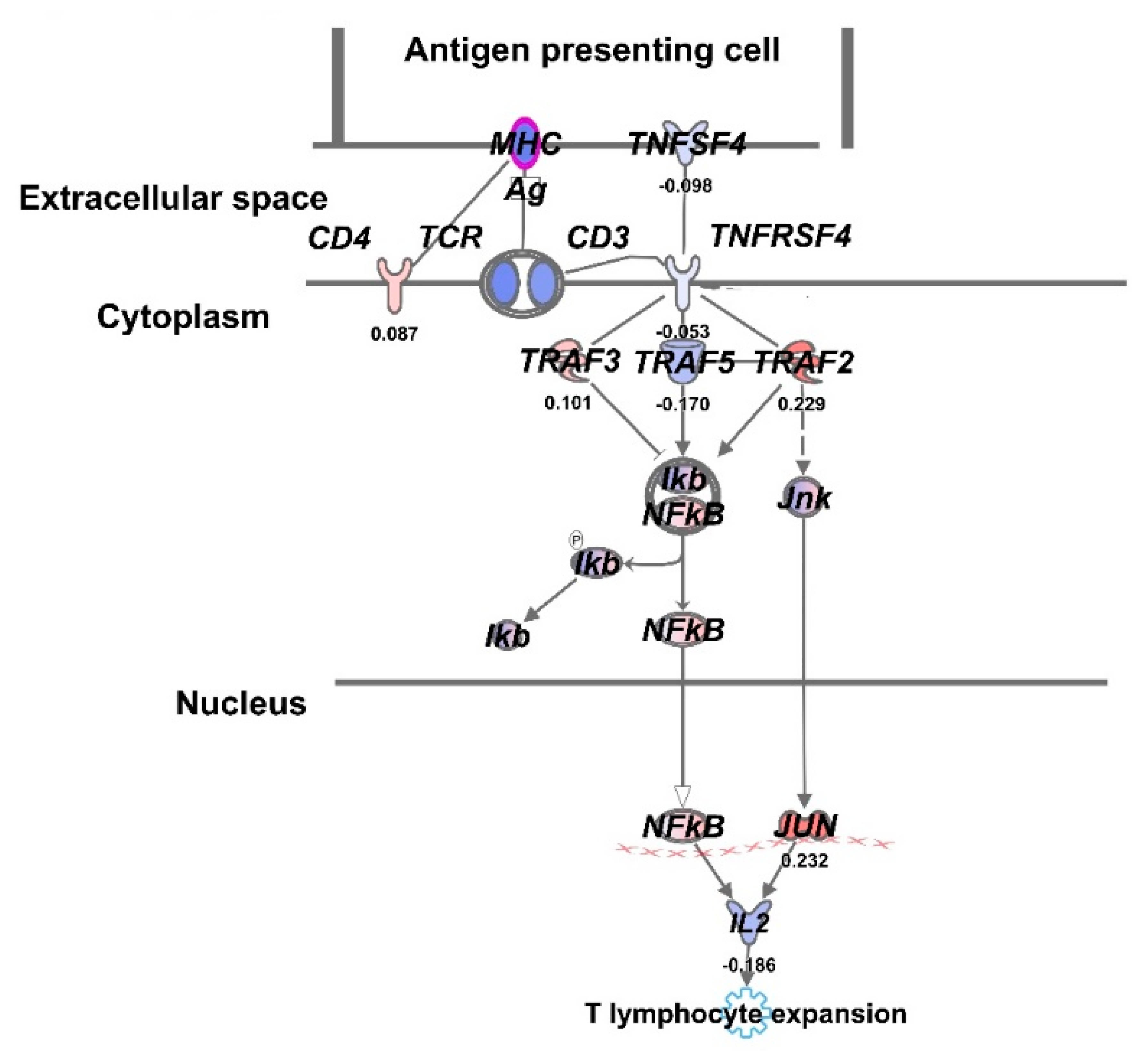
3.3. Comparative Gene Expression Analysis of UCH2 Cells Treated at E-SOBP and M-SOBP of Proton Radiation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Barker, H.E.; Paget, J.T.; Khan, A.A.; Harrington, K.J. The tumour microenvironment after radiotherapy: Mechanisms of resistance and recurrence. Nat. Rev. Cancer 2015, 15, 409–425. [Google Scholar] [CrossRef] [PubMed]
- Durante, M.; Paganetti, H. Nuclear physics in particle therapy: A review. Rep. Prog. Phys. 2016, 79, 096702. [Google Scholar]
- Fossati, P.; Vavassori, A.; Deantonio, L.; Ferrara, E.; Krengli, M.; Orecchia, R. Review of photon and proton radiotherapy for skull base tumours. Rep. Pract. Oncol. Radiother. 2016, 21, 336–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noël, G.; Gondi, V. Proton therapy for tumors of the base of the skull. Chin. Clin. Oncol. 2016, 5, 51. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.; Marshall, T.; Perozziello, F.; Manti, L.; Currell, F.J.; Hanton, F.; McMahon, S.J.; Kavanagh, J.N.; Cirrone, G.A.P.; Romano, F. Relative Biological Effectiveness Variation Along Monoenergetic and Modulated Bragg Peaks of a 62-MeV Therapeutic Proton Beam: A Preclinical Assessment. Int. J. Radiat. Oncol. Biol. Phys. 2014, 90, 27–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lomax, A. What will the medical physics of proton therapy look like 10 yr from now? A personal view. Med. Phys. 2018, 45, e984–e993. [Google Scholar] [PubMed] [Green Version]
- Oeck, S.; Szymonowicz, K.; Wiel, G.; Krysztofiak, A.; Lambert, J.; Koska, B.; Iliakis, G.; Timmermann, B.; Jendrossek, V. Relating Linear Energy Transfer to the Formation and Resolution of DNA Repair Foci After Irradiation with Equal Doses of X-ray Photons, Plateau, or Bragg-Peak Protons. Int. J. Mol. Sci. 2018, 19, 3779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Indelicato, D.; Rotondo, R.; Begosh-Mayne, D.; Scarborough, M.; Gibbs, C.; Morris, C.; Mendenhall, W.M. A Prospective Outcomes Study of Proton Therapy for Chordomas and Chondrosarcomas of the Spine. Int. J. Radiat. Oncol. 2016, 95, 297–303. [Google Scholar] [CrossRef]
- Franken, N.A.; Hovingh, S.; Ten Cate, R.; Krawczyk, P.; Stap, J.; Hoebe, R.; Aten, J.; Barendsen, G.W. Relative biological effectiveness of high linear energy transfer α-particles for the induction of DNA-double-strand breaks, chromosome aberrations and reproductive cell death in SW-1573 lung tumor cells. Oncol. Rep. 2012, 27, 769–774. [Google Scholar] [CrossRef] [Green Version]
- Girdhani, S.; Sachs, R.; Hlatky, L. Biological effects of proton radiation: What we know and don’t know. Radiat. Res. 2013, 179, 257–272. [Google Scholar]
- Paganetti, H. Relative biological effectiveness (RBE) values for proton beam therapy. Variations as a function of biological endpoint, dose, and linear energy transfer. Phys. Med. Biol. 2014, 59, R419–R472. [Google Scholar] [CrossRef]
- Mitteer, R.A.; Wang, Y.; Shah, J.; Gordon, S.; Fager, M.; Butter, P.P.; Kim, H.J.; Guardiola-Salmeron, C.; Carabe-Fernandez, A.; Fan, Y. Proton beam radiation induces DNA damage and cell apoptosis in glioma stem cells through reactive oxygen species. Sci. Rep. 2015, 5, 13961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cuaron, J.J.; Chang, C.; Lovelock, M.; Higginson, D.S.; Mah, D.; Cahlon, O.; Powell, S. Exponential Increase in Relative Biological Effectiveness Along Distal Edge of a Proton Bragg Peak as Measured by Deoxyribonucleic Acid Double-Strand Breaks. Int. J. Radiat. Oncol. Biol. Phys. 2016, 95, 62–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitti, E.T.; Parsons, J.L. The Radiobiological Effects of Proton Beam Therapy: Impact on DNA Damage and Repair. Cancers 2019, 11, 946. [Google Scholar] [CrossRef] [Green Version]
- Elizadeh, E.; Sanz, A.G.; Madugundu, G.S.; García, G.; Wagner, J.R.; Sanche, L. Thymidine decomposition induced by low-energy electrons and soft X rays under N2 and O2 atmospheres. Radiat. Res. 2014, 181, 629–640. [Google Scholar] [CrossRef] [Green Version]
- de Vera, P.; Surdutovich, E.; Solov’yov, A.V. The role of shock waves on the biodamage induced by ion beam radiation. Cancer Nanotechnol. 2019, 10, 5. [Google Scholar] [CrossRef] [Green Version]
- Brenner, D.J.; Hlatky, L.R.; Hahnfeldt, P.J.; Huang, Y.; Sachs, R.K. The linear-quadratic model and most other common radiobiological models result in similar predictions of time-dose relationships. Radiat. Res. 1998, 150, 83–91. [Google Scholar] [CrossRef]
- Britten, R.A.; Nazaryan, V.; Davis, L.K.; Klein, S.B.; Nichiporov, D.; Mendonca, M.S.; Wolanski, M.; Nie, X.; George, J.; Keppel, C. Variations in the RBE for cell killing along the depth-dose profile of a modulated proton therapy beam. Radiat. Res. 2013, 179, 21–28. [Google Scholar] [CrossRef]
- Frese, M.C.; Wilkens, J.J.; Huber, P.E.; Jensen, A.D.; Oelfke, U.; Taheri-Kadkhoda, Z. Application of constant vs. variable relative biological effectiveness in treatment planning of intensity-modulated proton therapy. Int. J. Radiat. Oncol. Biol. Phys. 2011, 79, 80–88. [Google Scholar] [CrossRef]
- Dasu, A.; Toma-Dasu, I. Prostate alpha/beta revisited—An analysis of clinical results from 14 168 patients. Acta Oncol. 2012, 51, 963–974. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.; Scholz, M. Treatment planning for heavy-ion radiotherapy: Calculation and optimization of biologically effective dose. Phys. Med. Biol. 2000, 45, 3319–3330. [Google Scholar] [CrossRef] [PubMed]
- Kramer, M.; Scholz, M. Rapid calculation of biological effects in ion radiotherapy. Phys. Med. Biol. 2006, 51, 1959–1970. [Google Scholar] [CrossRef]
- Wouters, B.G.; Lam, G.K.; Oelfke, U.; Gardey, K.; Durand, R.E.; Skarsgard, L.D. Measurements of relative biological effectiveness of the 70 MeV proton beam at TRIUMF using Chinese hamster V79 cells and the high-precision cell sorter assay. Radiat. Res. 1996, 146, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Ward, J.F. The complexity of DNA damage: Relevance to biological consequences. Int. J. Radiat. Biol. 1994, 66, 427–432. [Google Scholar] [CrossRef]
- Goodhead, D.T. Mechanisms for the biological effectiveness of high-LET radiations. J. Radiat. Res. 1999, 40, S1–S13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.G.; Lee, K.S.; Nam, K.S. The association of changes in RAD51 and survivin expression levels with the proton beam sensitivity of Capan-1 and Panc-1 human pancreatic cancer cell. Int. J. Oncol. 2019, 54, 744–752. [Google Scholar] [CrossRef] [Green Version]
- Migliorini, D.; Mach, N.; Aguiar, D.; Vernet, R.; Landis, B.N.; Becker, M.; McKee, T.; Dutoit, V.; Dietrich, P.-Y. First report of clinical responses to immunotherapy in 3 relapsing cases of chordoma after failure of standard therapies. Oncoimmunology 2017, 6, e1338235. [Google Scholar] [CrossRef] [Green Version]



| UCH2 | α (Gy-1) (95% CI) | β (Gy-2) (95% CI) | RBE (D = 2 Gy) at 95% CI |
|---|---|---|---|
| M-SOBP | 0.155 (0.114, 0.195) | 0.007 (0.004, 0.010) | 1 |
| E-SOBP | 0.298 (0.260, 0.337) | 0.000 (−0.003, 0.003) | 1.67 (−0.49–3.84) |
| MUG-Chor1 | α (Gy-1) (95% CI) | β (Gy-2) (95% CI) | RBE (D = 2Gy) 95% CI |
|---|---|---|---|
| M-SOBP | 0.243 (0.177, 0.310) | 0.043 (0.033, 0.052) | 1 |
| E-SOBP | 0.419 (0.352, 0.485) | 0.025 (0.015, 0.034) | 1.32 (0.88, 1.75) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singh, P.; Eley, J.; Mahmood, N.; Bhandary, B.; Dukic, T.; Tu, K.J.; Polf, J.; Lamichhane, N.; Mahmood, J.; Vujaskovic, Z.; et al. Therapeutic Efficacy of Variable Biological Effectiveness of Proton Therapy in U-CH2 and MUG-Chor1 Human Chordoma Cell Death. Cancers 2021, 13, 6115. https://doi.org/10.3390/cancers13236115
Singh P, Eley J, Mahmood N, Bhandary B, Dukic T, Tu KJ, Polf J, Lamichhane N, Mahmood J, Vujaskovic Z, et al. Therapeutic Efficacy of Variable Biological Effectiveness of Proton Therapy in U-CH2 and MUG-Chor1 Human Chordoma Cell Death. Cancers. 2021; 13(23):6115. https://doi.org/10.3390/cancers13236115
Chicago/Turabian StyleSingh, Prerna, John Eley, Nayab Mahmood, Binny Bhandary, Tijana Dukic, Kevin J. Tu, Jerimy Polf, Narottam Lamichhane, Javed Mahmood, Zeljko Vujaskovic, and et al. 2021. "Therapeutic Efficacy of Variable Biological Effectiveness of Proton Therapy in U-CH2 and MUG-Chor1 Human Chordoma Cell Death" Cancers 13, no. 23: 6115. https://doi.org/10.3390/cancers13236115
APA StyleSingh, P., Eley, J., Mahmood, N., Bhandary, B., Dukic, T., Tu, K. J., Polf, J., Lamichhane, N., Mahmood, J., Vujaskovic, Z., & Shukla, H. D. (2021). Therapeutic Efficacy of Variable Biological Effectiveness of Proton Therapy in U-CH2 and MUG-Chor1 Human Chordoma Cell Death. Cancers, 13(23), 6115. https://doi.org/10.3390/cancers13236115






