CARMA3 Promotes Colorectal Cancer Cell Motility and Cancer Stemness via YAP-Mediated NF-κB Activation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Reagents and Antibodies
2.3. Western Blotting
2.4. Cell Proliferation by MTT Assay
2.5. Transwell Migration and Invasion Assay
2.6. Sphere Formation Assay
2.7. Plasmid Constructs and Cell Transfection and Virus Infection Assay
2.8. RNA Isolation, RT-PCR and Quantitative RT-PCR (RT-qPCR)
2.9. Human Tissue Microarray (TMA) and Immunohistochemistry (IHC) Staining
2.10. Animal Xenograft Experiment
2.11. Statistical Analysis
3. Results
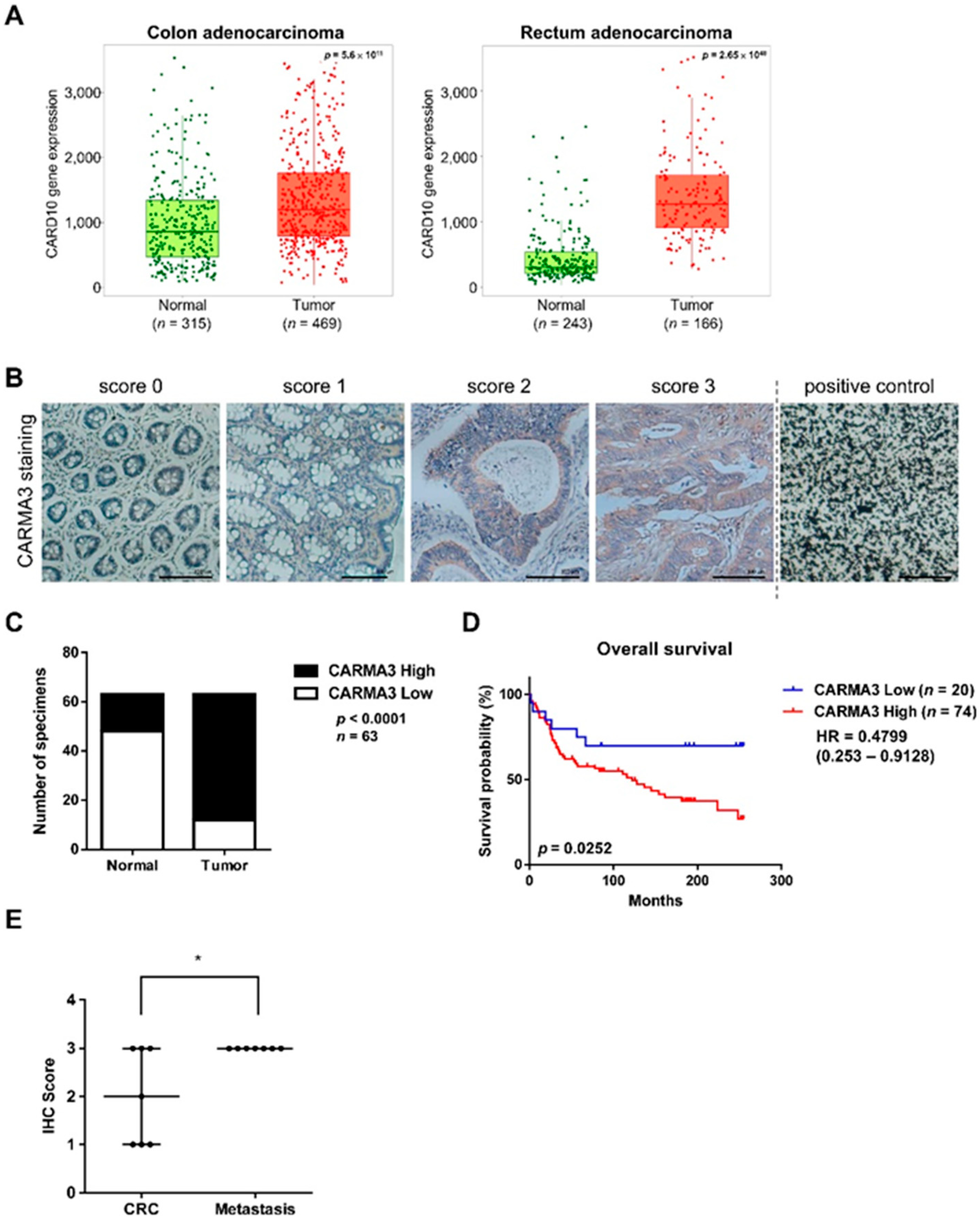
3.1. CARMA3 Overexpression Correlates with Poor Prognosis of Colorectal Cancers (CRC)
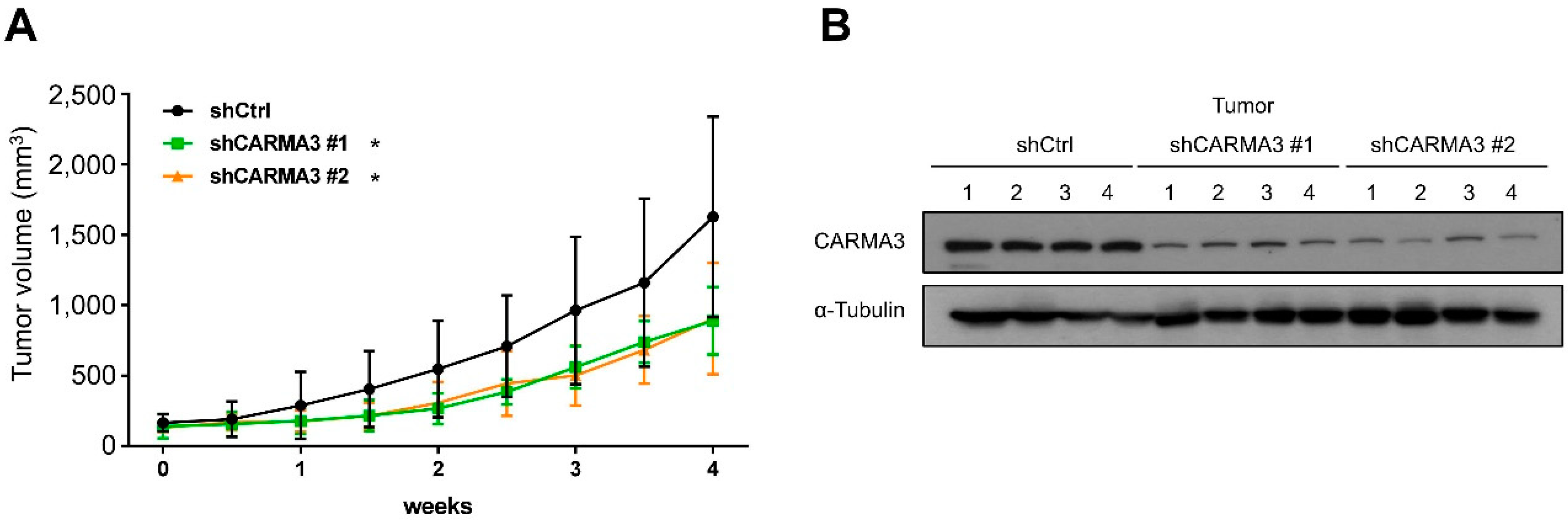
3.2. CARMA3 Expression Is Critical for CRC Aggressive Abilities
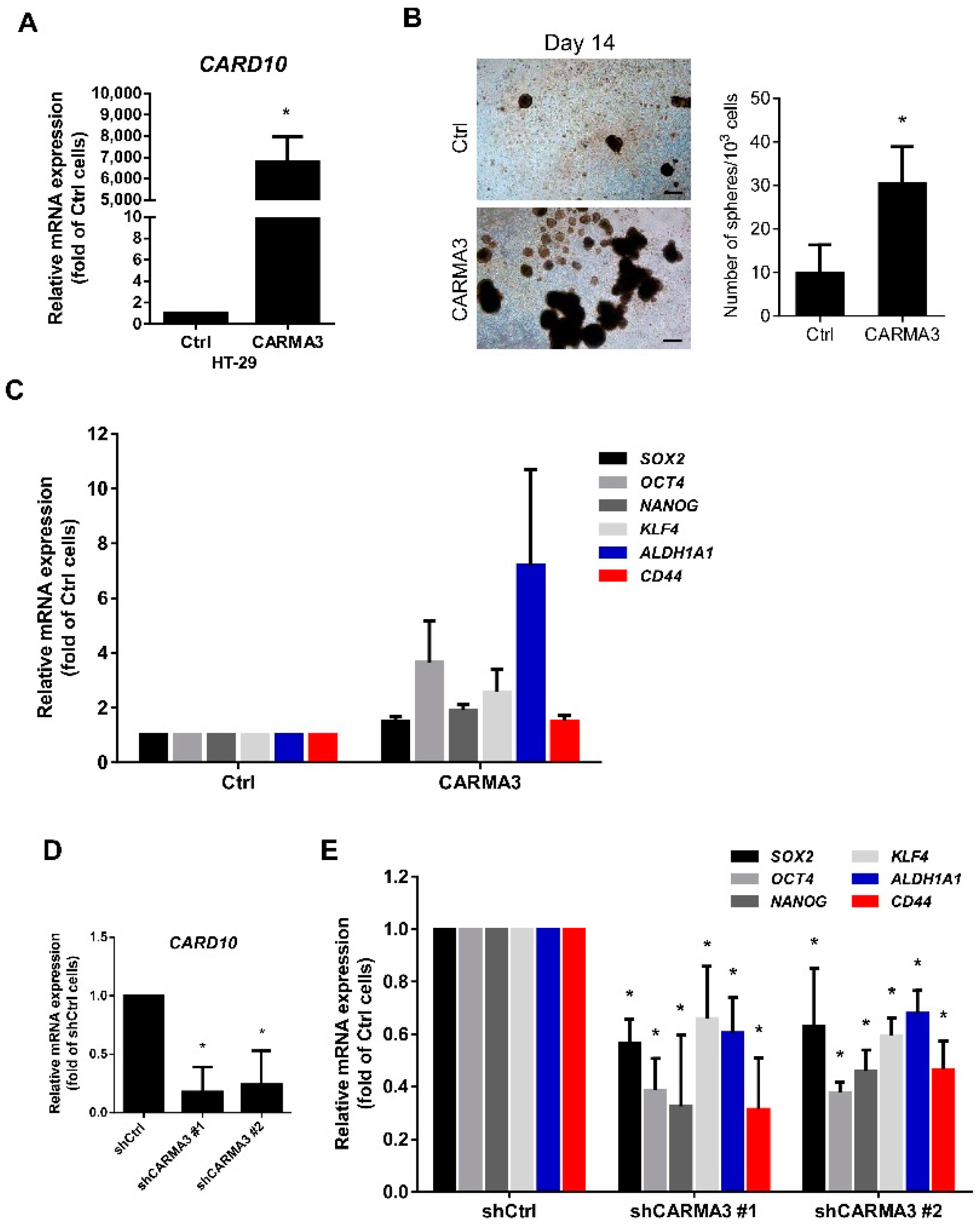
3.3. CARMA3 Expression Modulates Cancer Stemness
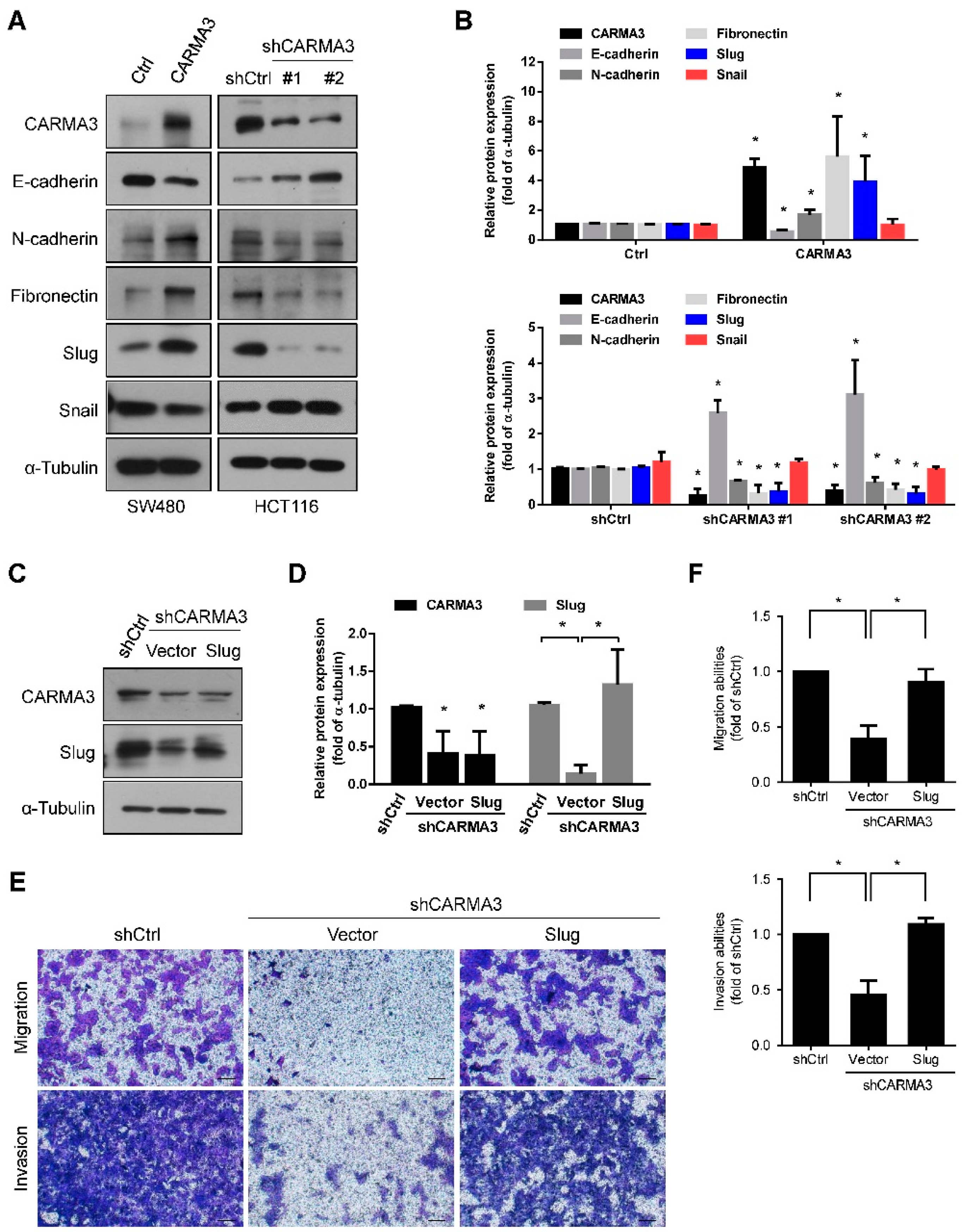
3.4. CARMA3 Enhanced EMT by Upregulating of Slug
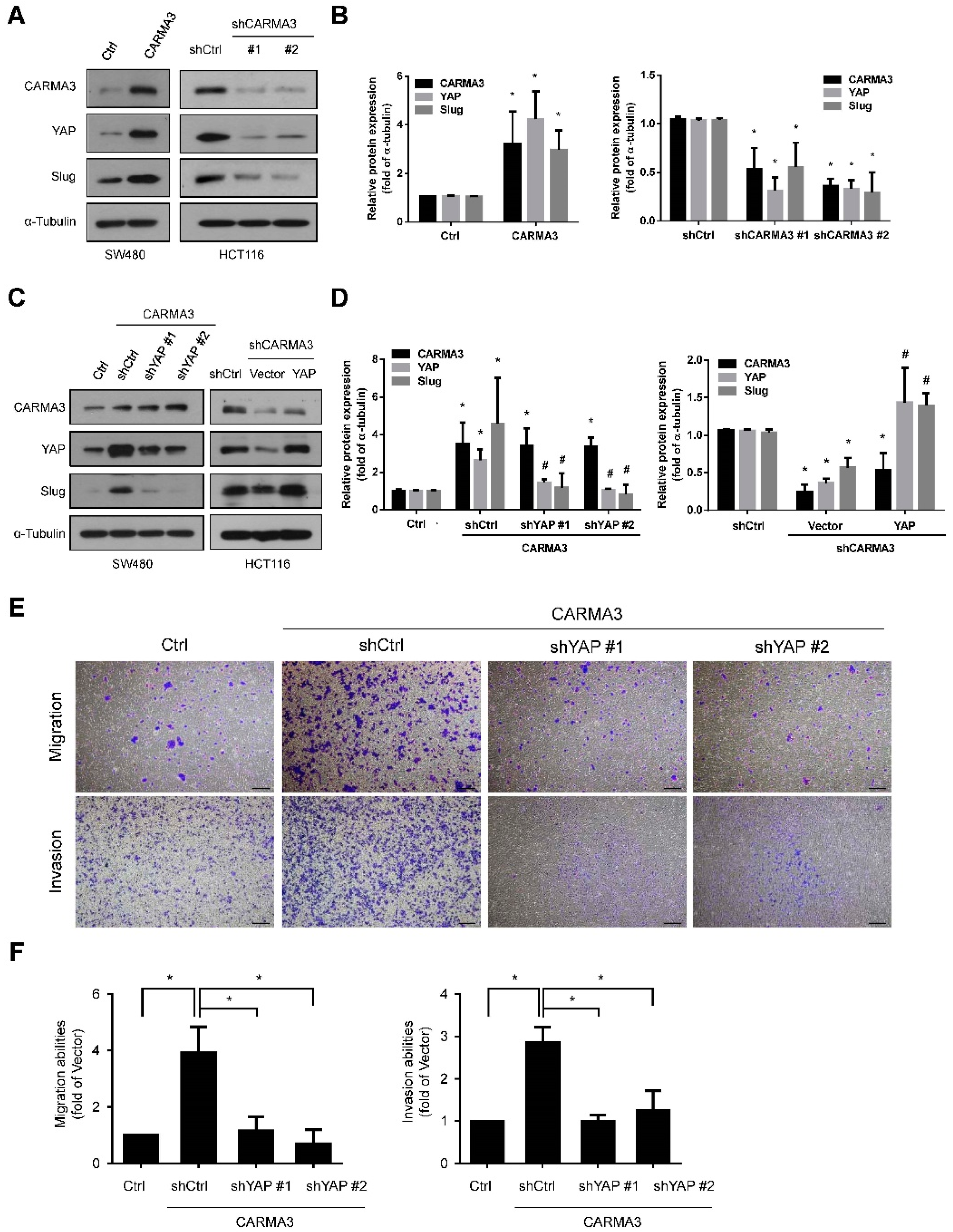
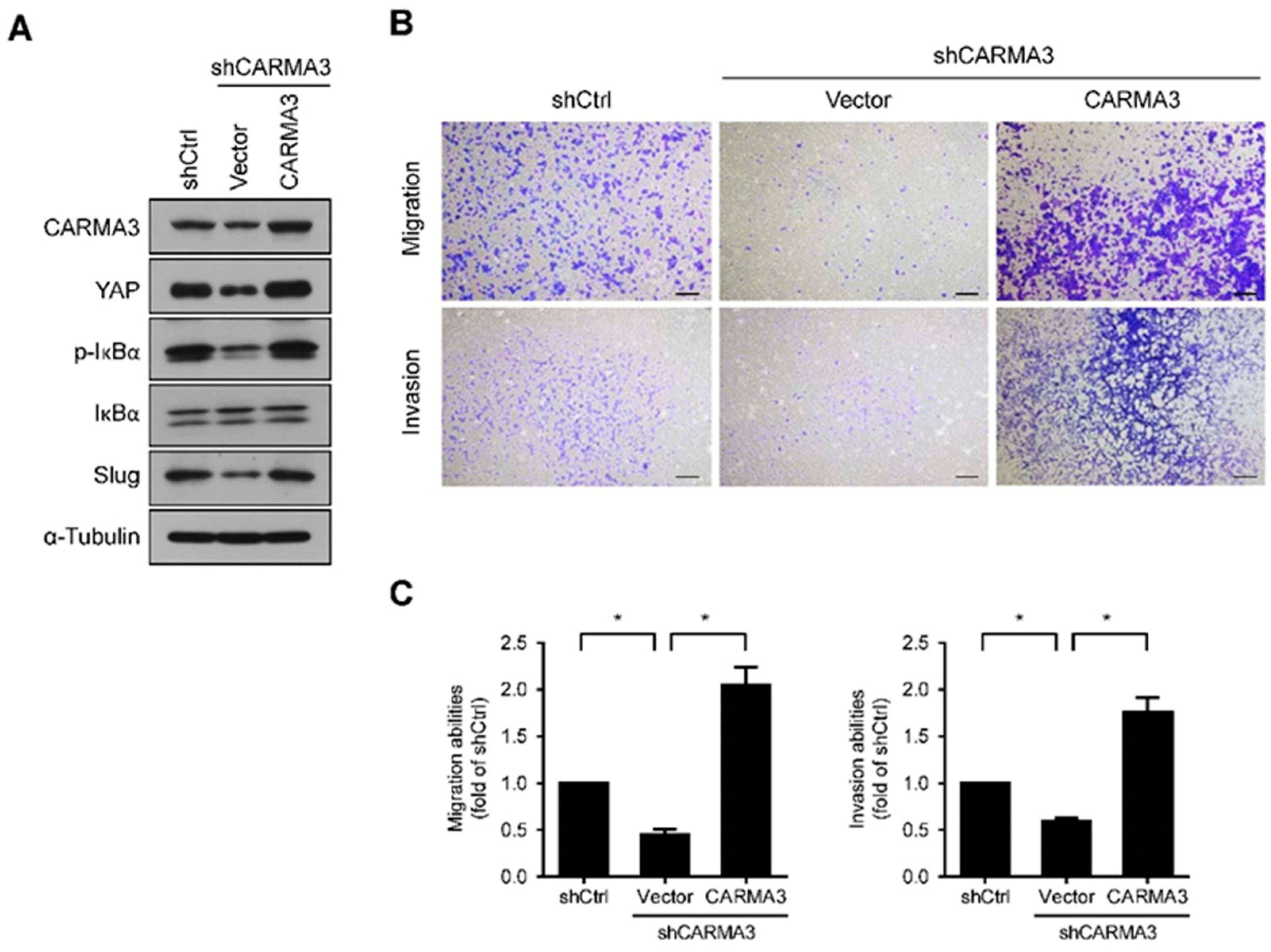
3.5. CARMA3 Mediated CRC Metastasis through YAP/Slug Regulation
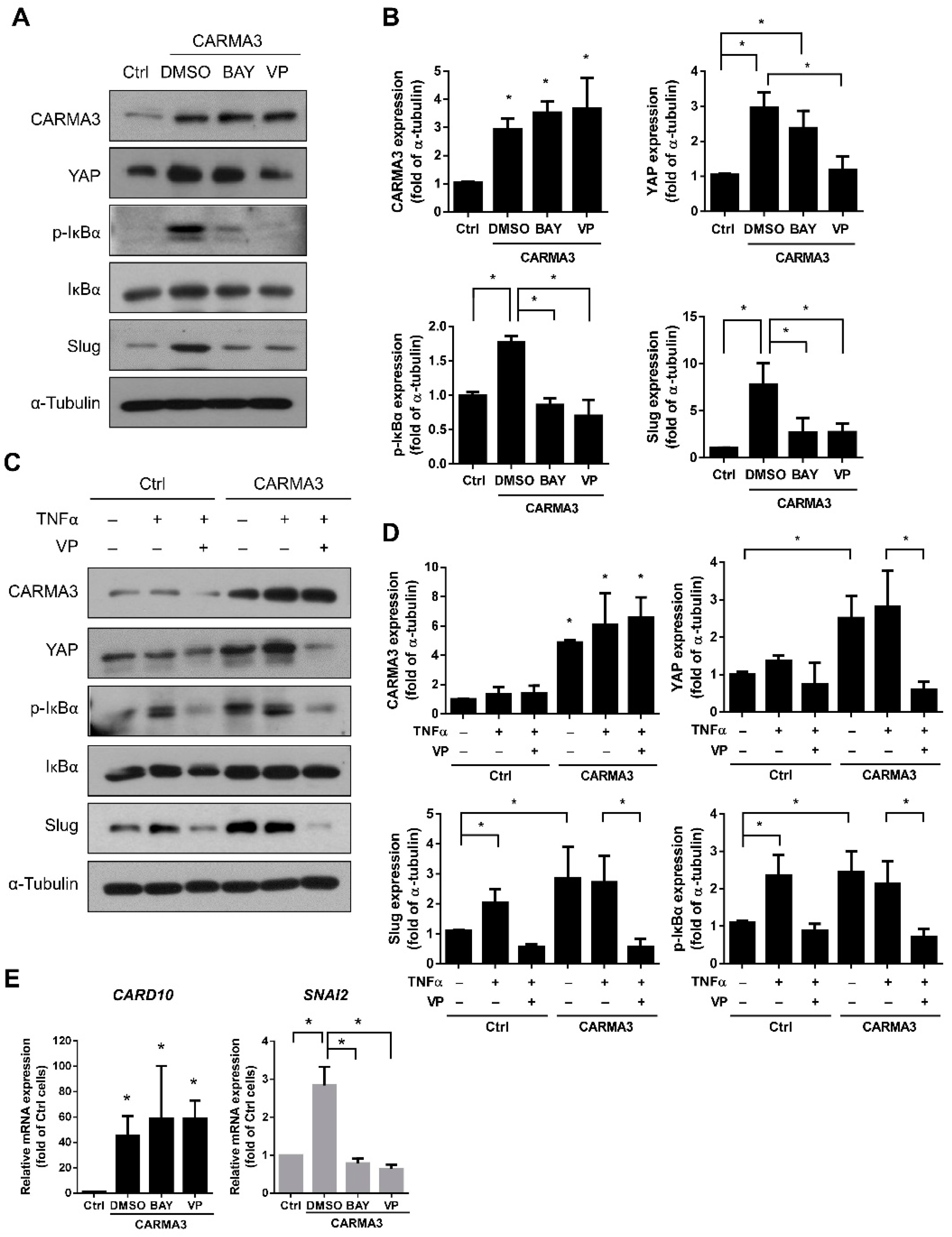
3.6. Activation of NF-κB Is Regulated by CARMA3-Induced YAP Expression
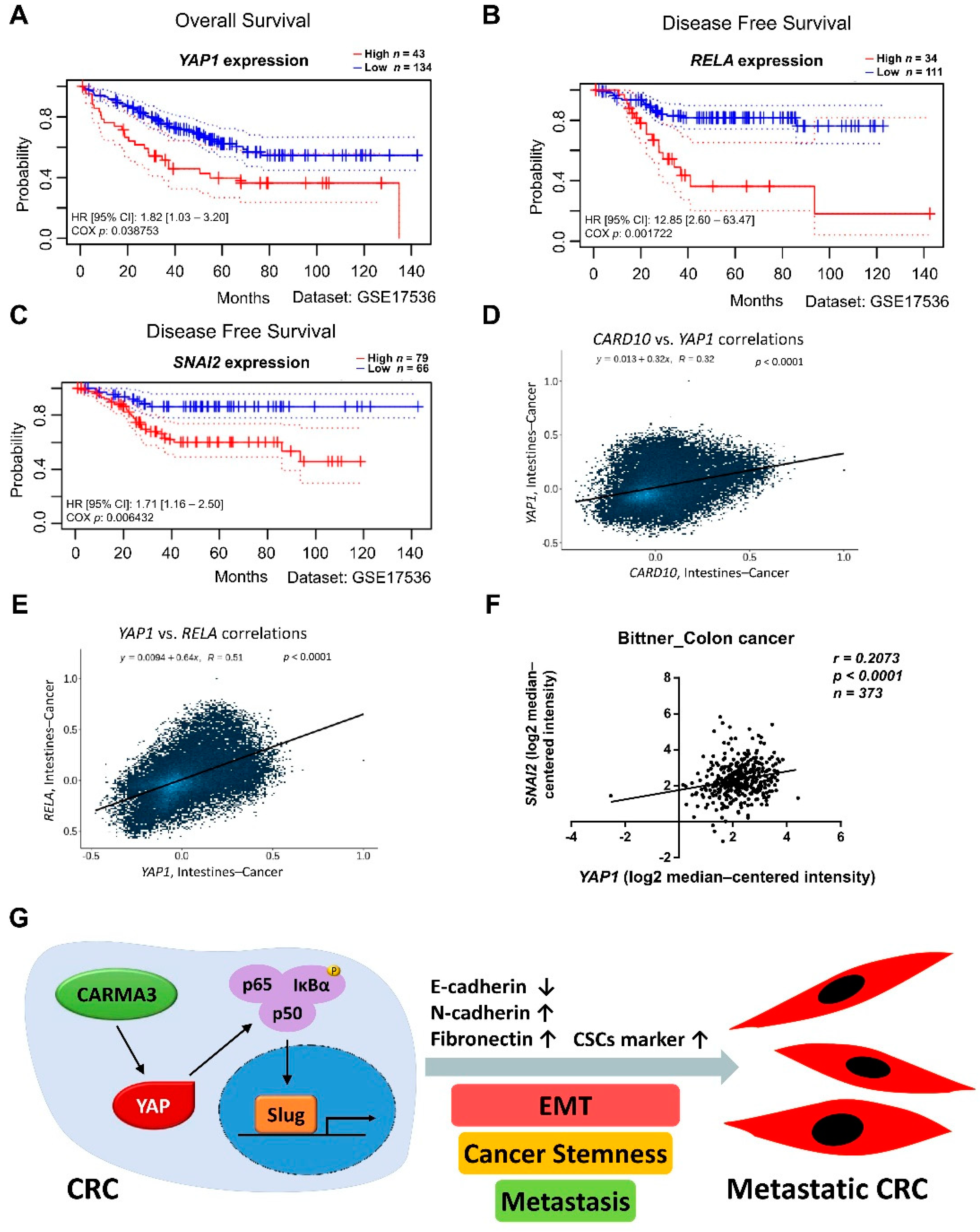
3.7. CARMA3/YAP/NF-κB/Slug Signaling Pathway Is Associated with Poor Prognosis for Human Colorectal Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- National Cancer Institute Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Colorectal Cancer. Available online: https://seer.cancer.gov/statfacts/html/colorect.html (accessed on 8 September 2021).
- Zhang, S.; Lin, X. CARMA3: Scaffold protein involved in NF-kappaB signaling. Front. Immunol. 2019, 10, 176. [Google Scholar] [CrossRef]
- Xia, Z.X.; Li, Z.X.; Zhang, M.; Sun, L.M.; Zhang, Q.F.; Qiu, X.S. CARMA3 regulates the invasion, migration, and apoptosis of non-small cell lung cancer cells by activating NF-small ka, CyrillicB and suppressing the P38 MAPK signaling pathway. Exp. Mol. Pathol. 2016, 100, 353–360. [Google Scholar] [CrossRef]
- Man, X.; Liu, T.; Jiang, Y.; Zhang, Z.; Zhu, Y.; Li, Z.; Kong, C.; He, J. Silencing of CARMA3 inhibits bladder cancer cell migration and invasion via deactivating beta-catenin signaling pathway. Onco Targets Ther. 2019, 12, 6309–6322. [Google Scholar] [CrossRef]
- Hou, H.; Li, W.X.; Cui, X.; Zhou, D.C.; Zhang, B.; Geng, X.P. CARMA3/NF-kappaB signaling contributes to tumorigenesis of hepatocellular carcinoma and is inhibited by sodium aescinate. World J. Gastroenterol. 2019, 25, 5483–5493. [Google Scholar] [CrossRef]
- Shi, D.; Zhang, Z.; Kong, C. CARMA3 transcriptional regulation of STMN1 by NF-kappaB promotes renal cell carcinoma proliferation and invasion. Technol. Cancer Res. Treat. 2021, 20, 15330338211027915. [Google Scholar] [CrossRef]
- Wang, L.; Qian, L.; Li, X.; Yan, J. MicroRNA-195 inhibits colorectal cancer cell proliferation, colony-formation and invasion through targeting CARMA3. Mol. Med. Rep. 2014, 10, 473–478. [Google Scholar] [CrossRef]
- Grabiner, B.C.; Blonska, M.; Lin, P.C.; You, Y.; Wang, D.; Sun, J.; Darnay, B.G.; Dong, C.; Lin, X. CARMA3 deficiency abrogates G protein-coupled receptor-induced NF-κB activation. Genes Dev. 2007, 21, 984–996. [Google Scholar] [CrossRef]
- Jiang, T.; Grabiner, B.; Zhu, Y.; Jiang, C.; Li, H.; You, Y.; Lang, J.; Hung, M.C.; Lin, X. CARMA3 is crucial for EGFR-Induced activation of NF-κB and tumor progression. Cancer Res. 2011, 71, 2183–2192. [Google Scholar] [CrossRef]
- McAllister-Lucas, L.M.; Ruland, J.; Siu, K.; Jin, X.; Gu, S.; Kim, D.S.; Kuffa, P.; Kohrt, D.; Mak, T.W.; Nunez, G.; et al. CARMA3/Bcl10/MALT1-dependent NF-κB activation mediates angiotensin II-responsive inflammatory signaling in nonimmune cells. Proc. Natl. Acad. Sci. USA 2007, 104, 139–144. [Google Scholar] [CrossRef]
- Ekambaram, P.; Lee, J.L.; Hubel, N.E.; Hu, D.; Yerneni, S.; Campbell, P.G.; Pollock, N.; Klei, L.R.; Concel, V.J.; Delekta, P.C.; et al. The CARMA3-Bcl10-MALT1 signalosome drives NFκB activation and promotes aggressiveness in angiotensin II receptor-positive breast cancer. Cancer Res. 2018, 78, 1225–1240. [Google Scholar] [CrossRef] [PubMed]
- Halder, G.; Johnson, R.L. Hippo signaling: Growth control and beyond. Development 2011, 138, 9–22. [Google Scholar] [CrossRef]
- Harvey, K.F.; Zhang, X.; Thomas, D.M. The Hippo pathway and human cancer. Nat. Rev. Cancer 2013, 13, 246–257. [Google Scholar] [CrossRef]
- Mao, B.; Hu, F.; Cheng, J.; Wang, P.; Xu, M.; Yuan, F.; Meng, S.; Wang, Y.; Yuan, Z.; Bi, W. SIRT1 regulates YAP2-mediated cell proliferation and chemoresistance in hepatocellular carcinoma. Oncogene 2014, 33, 1468–1474. [Google Scholar] [CrossRef] [PubMed]
- Zanconato, F.; Piccolo, S. Eradicating tumor drug resistance at its YAP-biomechanical roots. EMBO J. 2016, 35, 459–461. [Google Scholar] [CrossRef] [PubMed]
- Basu-Roy, U.; Bayin, N.S.; Rattanakorn, K.; Han, E.; Placantonakis, D.G.; Mansukhani, A.; Basilico, C. Sox2 antagonizes the Hippo pathway to maintain stemness in cancer cells. Nat. Commun. 2015, 6, 6411. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Ajani, J.A.; Honjo, S.; Maru, D.M.; Chen, Q.; Scott, A.W.; Heallen, T.R.; Xiao, L.; Hofstetter, W.L.; Weston, B.; et al. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 2014, 74, 4170–4182. [Google Scholar] [CrossRef] [PubMed]
- Cheung, P.; Xiol, J.; Dill, M.T.; Yuan, W.C.; Panero, R.; Roper, J.; Osorio, F.G.; Maglic, D.; Li, Q.; Gurung, B.; et al. Regenerative reprogramming of the intestinal stem cell state via Hippo signaling suppresses metastatic colorectal cancer. Cell Stem Cell 2020, 27, 590–604.e599. [Google Scholar] [CrossRef]
- Lee, K.W.; Lee, S.S.; Kim, S.B.; Sohn, B.H.; Lee, H.S.; Jang, H.J.; Park, Y.Y.; Kopetz, S.; Kim, S.S.; Oh, S.C.; et al. Significant association of oncogene YAP1 with poor prognosis and cetuximab resistance in colorectal cancer patients. Clin. Cancer Res. 2015, 21, 357–364. [Google Scholar] [CrossRef]
- Chen, Y.C.; Ingram, P.N.; Fouladdel, S.; McDermott, S.P.; Azizi, E.; Wicha, M.S.; Yoou, E. High-throughput single-cell derived sphere formation for cancer stem-like cell identification and analysis. Sci. Rep. 2016, 6, 27301–27312. [Google Scholar] [CrossRef] [PubMed]
- Varghese, F.; Bukhari, A.B.; Malhotra, R.; De, A. IHC Profiler: An open source plugin for the quantitative evaluation and automated scoring of immunohistochemistry images of human tissue samples. PLoS ONE 2014, 9, e96801. [Google Scholar]
- Bartha, A.; Gyorffy, B. TNMplot.com: A web tool for the comparison of gene expression in normal, tumor and metastatic tissues. Int. J. Mol. Sci. 2021, 22, 2622. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Dash, A.B.; Vo, A.P.; Sullivan, A.; Brooks, M.W.; Bell, G.W.; Richardson, A.L.; Polyak, K.; Tubo, R.; Weinberg, R.A. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature 2007, 449, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Shiozawa, Y.; Nie, B.; Pienta, K.J.; Morgan, T.M.; Taichman, R.S. Cancer stem cells and their role in metastasis. Pharmacol. Ther. 2013, 138, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Xu, E.; Liu, H.; Wan, L.; Lai, M. Epithelial-mesenchymal transition in colorectal cancer metastasis: A system review. Pathol. Res. Pract. 2015, 211, 557–569. [Google Scholar] [CrossRef]
- Zanconato, F.; Cordenonsi, M.; Piccolo, S. YAP/TAZ at the roots of cancer. Cancer Cell 2016, 29, 783–803. [Google Scholar] [CrossRef]
- Cheng, D.; Jin, L.; Chen, Y.; Xi, X.; Guo, Y. YAP promotes epithelial mesenchymal transition by upregulating Slug expression in human colorectal cancer cells. Int. J. Clin. Exp. Pathol. 2020, 13, 701–710. [Google Scholar]
- Zhou, P.; Wang, C.; Hu, Z.; Chen, W.; Qi, W.; Li, A. Genistein induces apoptosis of colon cancer cells by reversal of epithelial-to-mesenchymal via a Notch1/NF- κ B/slug/E-cadherin pathway. BMC Cancer 2017, 17, 813–820. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yue, B.; Yu, X.; Wang, Z.; Wang, M. SLUG is activated by nuclear factor kappa B and confers human alveolar epithelial A549 cells resistance to tumor necrosis factor-alpha-induced apoptosis. World J. Surg. Oncol. 2013, 11, 12–21. [Google Scholar] [CrossRef][Green Version]
- Storci, G.; Sansone, P.; Mari, S.; D’Uva, G.; Tavolari, S.; Guarnieri, T.; Taffurelli, M.; Ceccarelli, C.; Santini, D.; Chieco, P.; et al. TNFalpha up-regulates SLUG via the NF-kappaB/HIF1alpha axis, which imparts breast cancer cells with a stem cell-like phenotype. J. Cell Physiol. 2010, 225, 682–691. [Google Scholar] [CrossRef]
- Wu, D.W.; Lee, M.C.; Hsu, N.Y.; Wu, T.C.; Wu, J.Y.; Wang, Y.C.; Cheng, Y.W.; Chen, C.Y.; Lee, H. FHIT loss confers cisplatin resistance in lung cancer via the AKT/NF-κB/Slug-mediated PUMA reduction. Oncogene 2015, 34, 2505–2515. [Google Scholar] [CrossRef] [PubMed]
- Miao, Z.; Zhao, T.; Wang, Z.; Xu, Y.; Song, Y.; Wu, J.; Xu, H. CARMA3 is overexpressed in colon cancer and regulates NF-kappaB activity and cyclin D1 expression. Biochem. Biophys. Res. Commun. 2012, 425, 781–787. [Google Scholar] [CrossRef]
- Xie, C.; Han, Y.; Fu, L.; Li, Q.; Qiu, X.; Wang, E. Overexpression of CARMA3 is associated with advanced tumor stage, cell cycle progression, and cisplatin resistance in human epithelial ovarian cancer. Tumour. Biol. 2014, 35, 7957–7964. [Google Scholar] [CrossRef]
- Zhang, S.; Pan, D.; Jia, X.M.; Lin, X.; Zhao, X. The CARMA3-BCL10-MALT1 (CBM) complex contributes to DNA damage-induced NF-kappaB activation and cell survival. Protein Cell 2017, 8, 856–860. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jiang, C.; Zhou, Z.; Quan, Y.; Zhang, S.; Wang, T.; Zhao, X.; Morrison, C.; Heise, M.T.; He, W.; Miller, M.S.; et al. CARMA3 is a host factor regulating the balance of inflammatory and antiviral responses against viral infection. Cell Rep. 2016, 14, 2389–2401. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Shi, S.; Guo, Z.; Zhang, X.; Han, S.; Yang, A.; Wen, W.; Zhu, Q. Overexpression of YAP and TAZ is an independent predictor of prognosis in colorectal cancer and related to the proliferation and metastasis of colon cancer cells. PLoS ONE 2013, 8, e65539. [Google Scholar]
- Touil, Y.; Igoudjil, W.; Corvaisier, M.; Dessein, A.F.; Vandomme, J.; Monte, D.; Stechly, L.; Skrypek, N.; Langlois, C.; Grard, G.; et al. Colon cancer cells escape 5FU chemotherapy-induced cell death by entering stemness and quiescence associated with the c-Yes/YAP axis. Clin. Cancer Res. 2014, 20, 837–846. [Google Scholar] [CrossRef]
- Wang, Q.; Gao, X.; Yu, T.; Yuan, L.; Dai, J.; Wang, W.; Chen, G.; Jiao, C.; Zhou, W.; Huang, Q.; et al. REGgamma controls hippo signaling and reciprocal NF-kappaB-YAP regulation to promote colon cancer. Clin. Cancer Res. 2018, 24, 2015–2025. [Google Scholar] [CrossRef]
- Gregorieff, A.; Liu, Y.; Inanlou, M.R.; Khomchuk, Y.; Wrana, J.L. Yap-dependent reprogramming of Lgr5+ stem cells drives intestinal regeneration and cancer. Nature 2015, 526, 715–718. [Google Scholar] [CrossRef]
- Bora-Singhal, N.; Nguyen, J.; Schaal, C.; Perumal, D.; Singh, S.; Coppola, D.; Chellappan, S. YAP1 regulates OCT4 activity and SOX2 expression to facilitate self-renewal and vascular mimicry of stem-like cells. Stem Cells 2015, 33, 1705–1718. [Google Scholar] [CrossRef]
- Horst, D.; Kriegl, L.; Engel, J.; Kirchner, T.; Jung, A. Prognostic significance of the cancer stem cell markers CD133, CD44, and CD166 in colorectal cancer. Cancer Investig. 2009, 27, 844–850. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Lee, C.J.; Choi, J.H.; Kim, J.H.; Kim, J.W.; Kim, J.Y.; Nam, J.S. The JAK2/STAT3/CCND2 Axis promotes colorectal Cancer stem cell persistence and radioresistance. J. Exp. Clin. Cancer Res. 2019, 38, 399. [Google Scholar] [CrossRef]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Morel, A.P.; Lievre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef]
- Dai, X.; Ge, J.; Wang, X.; Qian, X.; Zhang, C.; Li, X. OCT4 regulates epithelial-mesenchymal transition and its knockdown inhibits colorectal cancer cell migration and invasion. Oncol. Rep. 2013, 29, 155–160. [Google Scholar] [CrossRef]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Leng, Z.; Tao, K.; Xia, Q.; Tan, J.; Yue, Z.; Chen, J.; Xi, H.; Li, J.; Zheng, H. Kruppel-like factor 4 acts as an oncogene in colon cancer stem cell-enriched spheroid cells. PLoS ONE 2013, 8, e56082. [Google Scholar] [CrossRef]
- Chang, Y.W.; Chiu, C.F.; Lee, K.Y.; Hong, C.C.; Wang, Y.Y.; Cheng, C.C.; Jan, Y.H.; Huang, M.S.; Hsiao, M.; Ma, J.T.; et al. CARMA3 represses metastasis suppressor NME2 to promote lung cancer stemness and metastasis. Am. J. Respir. Crit. Care Med. 2015, 192, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.; Horgan, P.G.; McMillan, D.C.; Edwards, J. NF-kappaB pathways in the development and progression of colorectal cancer. Transl. Res. 2018, 197, 43–56. [Google Scholar] [CrossRef] [PubMed]
- Gupta, S.C.; Sundaram, C.; Reuter, S.; Aggarwal, B.B. Inhibiting NF-kappaB activation by small molecules as a therapeutic strategy. Biochim. Biophys. Acta 2010, 1799, 775–787. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, T.-Y.; Wu, C.-T.; Sheu, M.-L.; Yang, R.-S.; Liu, S.-H. CARMA3 Promotes Colorectal Cancer Cell Motility and Cancer Stemness via YAP-Mediated NF-κB Activation. Cancers 2021, 13, 5946. https://doi.org/10.3390/cancers13235946
Chang T-Y, Wu C-T, Sheu M-L, Yang R-S, Liu S-H. CARMA3 Promotes Colorectal Cancer Cell Motility and Cancer Stemness via YAP-Mediated NF-κB Activation. Cancers. 2021; 13(23):5946. https://doi.org/10.3390/cancers13235946
Chicago/Turabian StyleChang, Ting-Yu, Cheng-Tien Wu, Meei-Ling Sheu, Rong-Sen Yang, and Shing-Hwa Liu. 2021. "CARMA3 Promotes Colorectal Cancer Cell Motility and Cancer Stemness via YAP-Mediated NF-κB Activation" Cancers 13, no. 23: 5946. https://doi.org/10.3390/cancers13235946
APA StyleChang, T.-Y., Wu, C.-T., Sheu, M.-L., Yang, R.-S., & Liu, S.-H. (2021). CARMA3 Promotes Colorectal Cancer Cell Motility and Cancer Stemness via YAP-Mediated NF-κB Activation. Cancers, 13(23), 5946. https://doi.org/10.3390/cancers13235946





