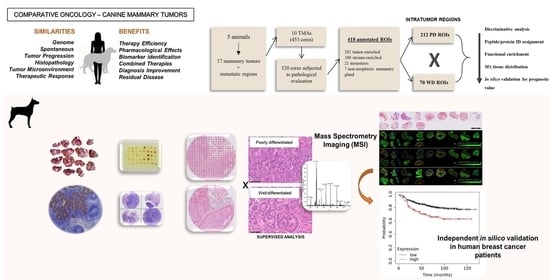
Proteomic Analysis Identifies FNDC1, A1BG, and Antigen Processing Proteins Associated with Tumor Heterogeneity and Malignancy in a Canine Model of Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. TMA Construction
2.3. Mass Spectrometry Imaging
2.4. Histopathological Staining and Annotation
2.5. MALDI-MSI Data Processing
2.6. Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS)
2.7. Statistical Analysis
2.7.1. Discriminative m/z Signals
2.7.2. Functional Enrichment Analysis
2.7.3. Independent Comparative Validation
3. Results
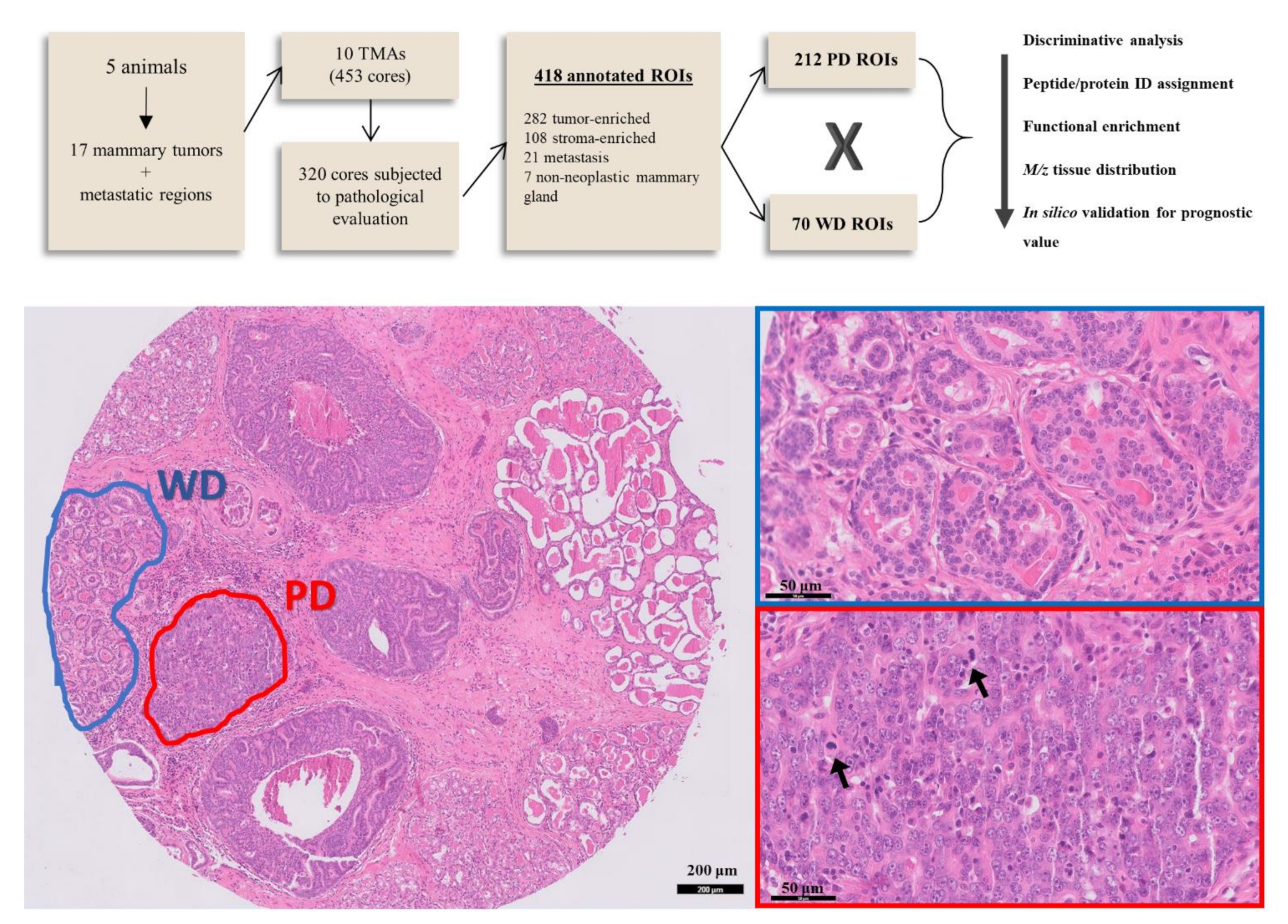
3.1. Patients, Tumor Samples and Tissue Annotation
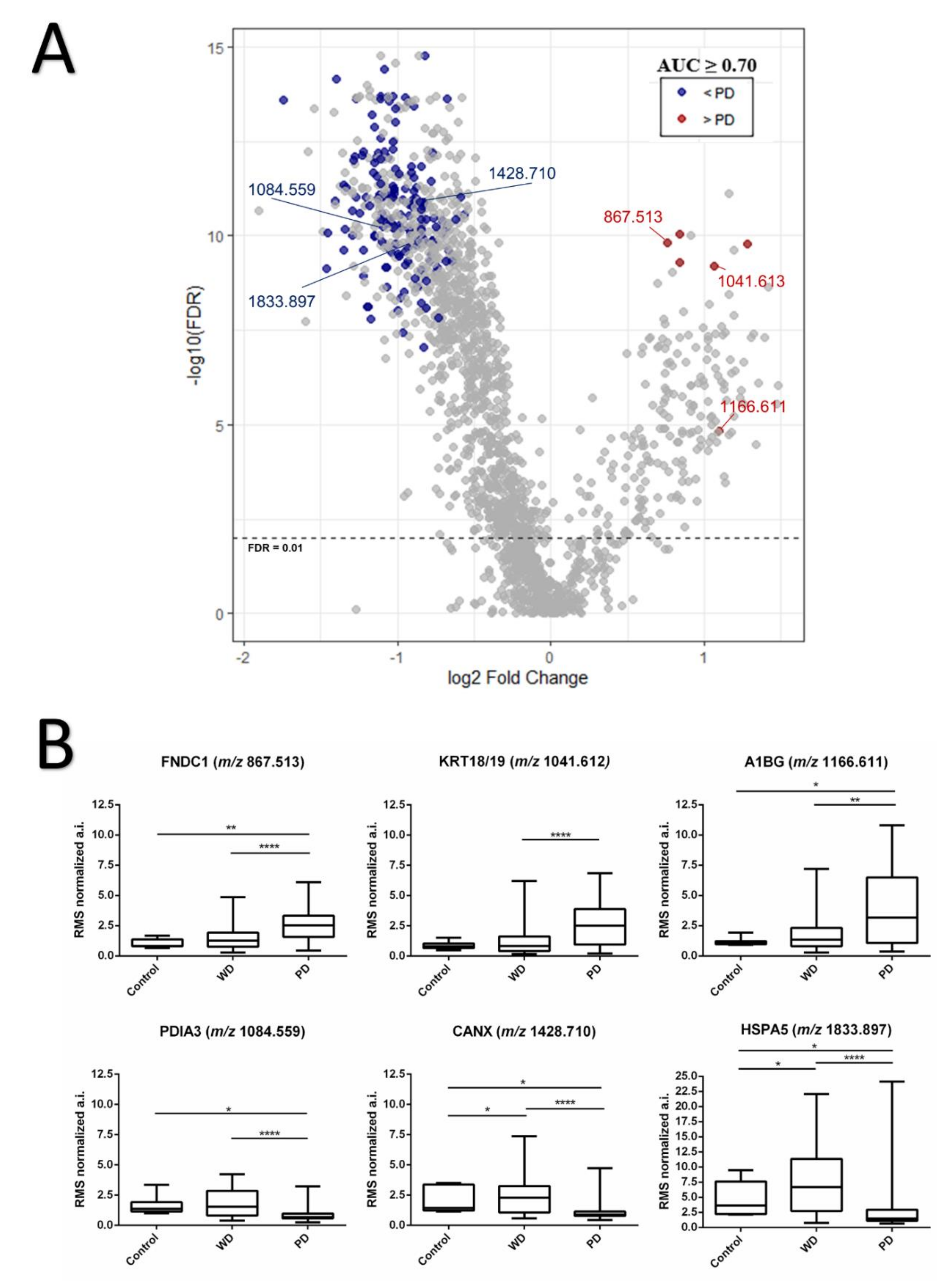
3.2. Protein Identification and Discriminative m/z Signals between Low and High Grade Intratumor Regions
3.3. Functional Enrichment Analysis of Differentially Expressed Peptides between Well and Poorly Differentiated Tumor Populations
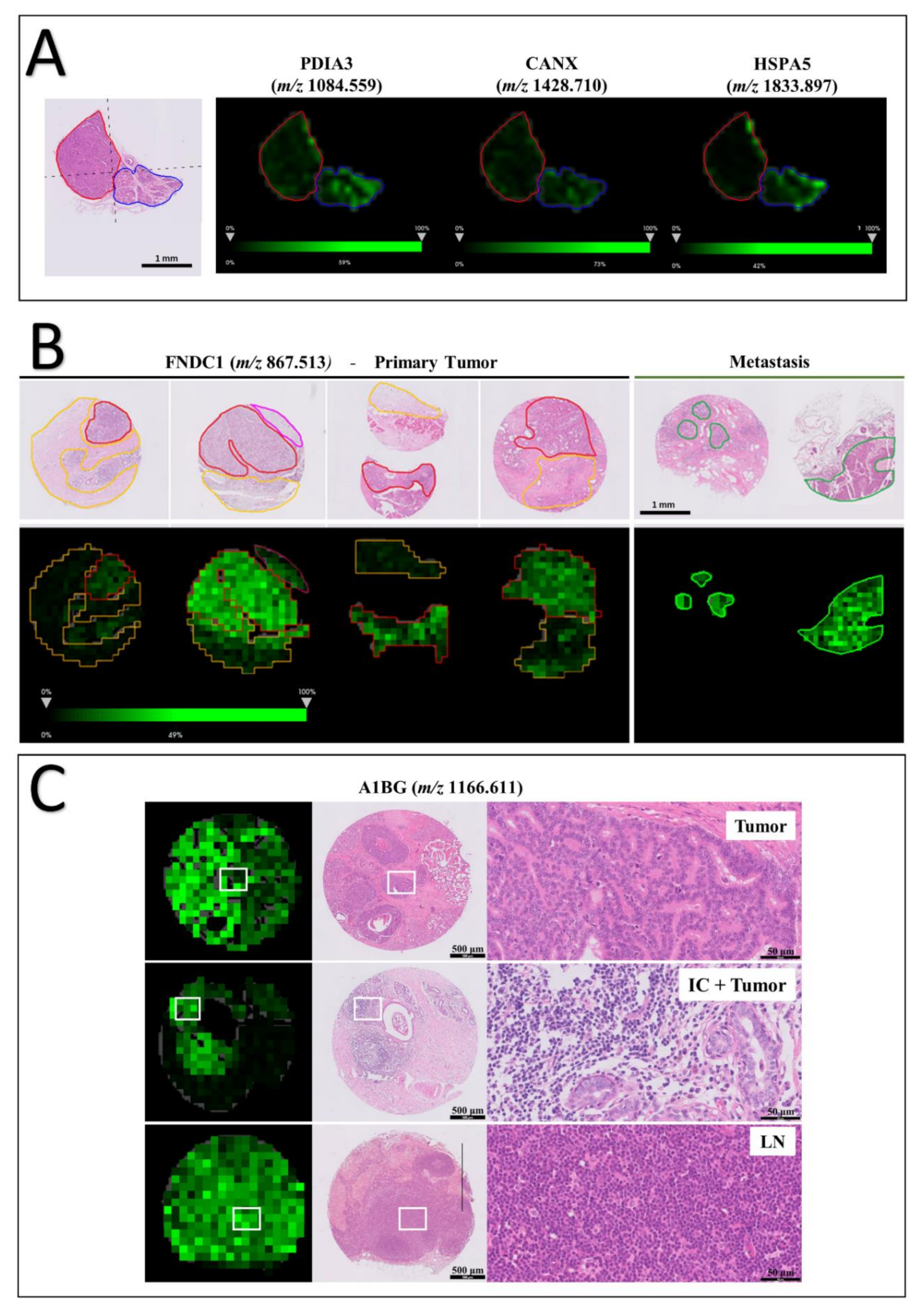
3.4. Tissue Distribution of FNDC1 and A1BG Peptides
3.5. Prognostic Value of FNDC1, A1BG, CANX, HSPA5 and PDIA3 in Human Breast Cancer Patients
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ferlay, J.; Laversanne, M.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Observatory: Cancer Tomorrow; International Agency for Research on Cancer: Lyon, France, 2020; Available online: https://gco.iarc.fr/tomorrow (accessed on 18 November 2021).
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Parker, J.S.; Mullins, M.; Cheang, M.C.U.; Leung, S.; Voduc, D.; Vickery, T.; Davies, S.; Fauron, C.; He, X.; Hu, Z.; et al. Supervised Risk Predictor of Breast Cancer Based on Intrinsic Subtypes. J. Clin. Oncol. 2009, 27, 1160–1167. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.; Kulbokas, E.J.; Zody, M.; et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef]
- Paoloni, M.; Khanna, C. Translation of new cancer treatments from pet dogs to humans. Nat. Rev. Cancer 2008, 8, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Rowell, J.L.; McCarthy, D.O.; Alvarez, C.E. Dog models of naturally occurring cancer. Trends Mol. Med. 2011, 17, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Xiong, H.; Ellis, A.E.; Northrup, N.C.; Rodriguez, C.O.; O’Regan, R.M.; Dalton, S.; Zhao, S. Molecular Homology and Difference between Spontaneous Canine Mammary Cancer and Human Breast Cancer. Cancer Res. 2014, 74, 5045–5056. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.; Keller, E.T.; Garfield, D.H.; Shen, K.; Wang, J. Stromal cells in tumor microenvironment and breast cancer. Cancer Metastasis Rev. 2013, 32, 303–315. [Google Scholar] [CrossRef]
- Swartz, M.A.; Iida, N.; Roberts, E.W.; Sangaletti, S.; Wong, M.H.; Yull, F.E.; Coussens, L.M.; DeClerck, Y.A. Tumor Microenvironment Complexity: Emerging Roles in Cancer Therapy. Cancer Res. 2012, 72, 2473–2480. [Google Scholar] [CrossRef] [PubMed]
- Balluff, B.; Hanselmann, M.; Heeren, R.M.A. Mass Spectrometry Imaging for the Investigation of Intratumor Heterogeneity, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume 134, ISBN 9780128052495. [Google Scholar]
- Vaysse, P.M.; Heeren, R.M.A.; Porta, T.; Balluff, B. Mass spectrometry imaging for clinical research—Latest developments, applications, and current limitations. Analyst 2017, 142, 2690–2712. [Google Scholar] [CrossRef] [PubMed]
- Maier, S.K.; Hahne, H.; Gholami, A.M.; Balluff, B.; Meding, S.; Schoene, C.; Walch, A.K.; Kuster, B. Comprehensive identification of proteins from MALDI imaging. Mol. Cell Proteom. 2013, 12, 2901–2910. [Google Scholar] [CrossRef]
- Dill, A.L.; Ifa, D.R.; Manicke, N.E.; Costa, A.B.; Ramos-Vara, J.A.; Knapp, D.W.; Cooks, R.G. Lipid profiles of canine invasive transitional cell carcinoma of the urinary bladder and adjacent normal tissue by desorption electrospray ionization imaging mass spectrometry. Anal. Chem. 2009, 81, 8758–8764. [Google Scholar] [CrossRef]
- Jarmusch, A.K.; Kerian, K.S.; Pirro, V.; Peat, T.; Thompson, C.A.; Ramos-Vara, J.A.; Childress, M.O.; Cooks, R.G. Characteristic lipid profiles of canine non-Hodgkin’s lymphoma from surgical biopsy tissue sections and fine needle aspirate smears by desorption electrospray ionization-mass spectrometry. Analyst 2015, 140, 6321–6329. [Google Scholar] [CrossRef]
- D’Hue, C.A.; Dhawan, D.; Peat, T.; Ramos-Vara, J.; Jarmusch, A.; Knapp, D.W.; Cooks, R.G. Fatty Acid Patterns Detected by Ambient Ionization Mass Spectrometry in Canine Invasive Urothelial Carcinoma from Dogs of Different Breeds. Bl. Cancer 2018, 4, 283–291. [Google Scholar] [CrossRef] [PubMed]
- Goldschmidt, M.; Peña, L.; Rasotto, R.; Zappulli, V. Classification and grading of canine mammary tumors. Vet. Pathol. 2011, 48, 117–131. [Google Scholar] [CrossRef] [PubMed]
- Ràfols, P.; Heijs, B.; Del Castillo, E.; Yanes, O.; McDonnell, L.A.; Brezmes, J.; Pérez-Taboada, I.; Vallejo, M.; García-Altares, M.; Correig, X. rMSIproc: An R package for mass spectrometry imaging data processing. Bioinformatics 2020, 36, 3618–3619. [Google Scholar] [CrossRef] [PubMed]
- Strohalm, M.; Hassman, M.; Košata, B.; Kodíček, M. mMass data miner: An open source alternative for mass spectrometric data analysis. Rapid Commun. Mass Spectrom. 2008, 22, 905–908. [Google Scholar] [CrossRef] [PubMed]
- Fabregat, A.; Jupe, S.; Matthews, L.; Sidiropoulos, K.; Gillespie, M.; Garapati, P.; Haw, R.; Jassal, B.; Korninger, F.; May, B.; et al. The Reactome Pathway Knowledgebase. Nucleic Acids Res. 2018, 46, D649–D655. [Google Scholar] [CrossRef] [PubMed]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1809 patients. Breast Cancer Res. Treat. 2010, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Birkbak, N.J.; Gyorffy, B.; Szallasi, Z.; Eklund, A.C. Jetset: Selecting the optimal microarray probe set to represent a gene. BMC Bioinform. 2011, 12, 474. [Google Scholar] [CrossRef] [PubMed]
- Elston, C.W.; Ellis, I.O. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: Experience from a large study with long-term follow-up. Histopathology 1991, 19, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Rakha, E.A.; Reis-Filho, J.S.; Baehner, F.; Dabbs, D.J.; Decker, T.; Eusebi, V.; Fox, S.B.; Ichihara, S.; Jacquemier, J.; Lakhani, S.R.; et al. Breast cancer prognostic classification in the molecular era: The role of histological grade. Breast Cancer Res. 2010, 12, 207. [Google Scholar] [CrossRef] [PubMed]
- Canadas, A.; França, M.; Pereira, C.; Vilaça, R.; Vilhena, H.; Tinoco, F.; Silva, M.J.; Ribeiro, J.; Medeiros, R.; Oliveira, P.; et al. Canine Mammary Tumors: Comparison of Classification and Grading Methods in a Survival Study. Vet. Pathol. 2018, 56, 208–219. [Google Scholar] [CrossRef] [PubMed]
- Kuppusamy, K.; Rajan, A.; Warrier, A.; Nadhan, R.; Patra, D.; Srinivas, P. Cytological Grading of Breast Tumors—The Human and Canine Perspective. Front. Vet. Sci. 2019, 6, 283. [Google Scholar] [CrossRef]
- Tao, L.; Huang, G.; Song, H.; Chen, Y.; Chen, L. Cancer associated fibroblasts: An essential role in the tumor microenvironment (Review). Oncol Lett. 2017, 14, 2611–2620. [Google Scholar] [CrossRef] [PubMed]
- Allinen, M.; Beroukhim, R.; Cai, L.; Brennan, C.; Lahti-Domenici, J.; Huang, H.; Porter, D.; Hu, M.; Chin, L.; Richardson, A.; et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell 2004, 6, 17–32. [Google Scholar] [CrossRef]
- Levental, K.R.; Yu, H.; Kass, L.; Lakins, J.N.; Egeblad, M.; Erler, J.T.; Fong, S.F.T.; Csiszar, K.; Giaccia, A.; Weninger, W.; et al. Matrix Crosslinking Forces Tumor Progression by Enhancing Integrin Signaling. Cell 2009, 139, 891–906. [Google Scholar] [CrossRef]
- Fang, M.; Yuan, J.; Peng, C.; Li, Y. Collagen as a double-edged sword in tumor progression. Tumour Biol. J. Int. Soc. Oncodevelopmental. Biol. Med. 2014, 35, 2871–2882. [Google Scholar] [CrossRef] [PubMed]
- Provenzano, P.P.; Eliceiri, K.W.; Campbell, J.M.; Inman, D.R.; White, J.G.; Keely, P.J. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 2006, 4, 38. [Google Scholar] [CrossRef] [PubMed]
- Arensdorf, A.M.; Diedrichs, D.; Rutkowski, D.T. Regulation of the transcriptome by ER stress: Non-canonical mechanisms and physiological consequences. Front. Genet. 2013, 4, 256. [Google Scholar] [CrossRef] [PubMed]
- Clarke, H.J.; Chambers, J.E.; Liniker, E.; Marciniak, S.J. Endoplasmic Reticulum Stress in Malignancy. Cancer Cell 2014, 25, 563–573. [Google Scholar] [CrossRef] [PubMed]
- Leone, P.; Shin, E.-C.; Perosa, F.; Vacca, A.; Dammacco, F.; Racanelli, V. MHC Class I Antigen Processing and Presenting Machinery: Organization, Function, and Defects in Tumor Cells. JNCI J. Natl. Cancer Inst. 2013, 105, 1172–1187. [Google Scholar] [CrossRef] [PubMed]
- De Charette, M.; Marabelle, A.; Houot, R. Turning tumour cells into antigen presenting cells: The next step to improve cancer immunotherapy? Eur. J. Cancer 2016, 68, 134–147. [Google Scholar] [CrossRef]
- Zhang, Y.; Baig, E.; Williams, D.B. Functions of ERp57 in the Folding and Assembly of Major Histocompatibility Complex Class I Molecules. J. Biol. Chem. 2006, 281, 14622–14631. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, H.; Chen, R.; Zhang, Y.; Sun, X.-X.; Huang, W.; Bian, H.; Chen, Z.-N. Combination of CALR and PDIA3 is a potential prognostic biomarker for non-small cell lung cancer. Oncotarget 2017, 8, 57. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, T.; Wada, R.; Kure, S.; Ishino, K.; Kudo, M.; Ohashi, R.; Fujita, I.; Uchida, E.; Yoshida, H.; Naito, Z. Expression of protein disulfide isomerase A3 and its clinicopathological association in gastric cancer. Oncol. Rep. 2019, 41, 2265–2272. [Google Scholar] [CrossRef]
- Malhotra, J.D.; Kaufman, R.J. The endoplasmic reticulum and the unfolded protein response. Semin. Cell Dev. Biol. 2007, 18, 716–731. [Google Scholar] [CrossRef]
- Faé, K.C.; da Silva Diefenbach, D.; Bilate, A.M.B.; Tanaka, A.C.; Pomerantzeff, P.M.A.; Kiss, M.H.; Silva, C.A.A.; Cunha-Neto, E.; Kalil, J.; Guilherme, L. PDIA3, HSPA5 and vimentin, proteins identified by 2-DE in the valvular tissue, are the target antigens of peripheral and heart infiltrating T cells from chronic rheumatic heart disease patients. J. Autoimmun. 2008, 31, 136–141. [Google Scholar] [CrossRef]
- Aebi, M.; Bernasconi, R.; Clerc, S.; Molinari, M. N-glycan structures: Recognition and processing in the ER. Trends Biochem. Sci. 2010, 35, 74–82. [Google Scholar] [CrossRef] [PubMed]
- Shibata, Y.; Shemesh, T.; Prinz, W.A.; Palazzo, A.F.; Kozlov, M.M.; Rapoport, T.A. Mechanisms determining the morphology of the peripheral ER. Cell 2010, 143, 774–788. [Google Scholar] [CrossRef] [PubMed]
- Molinari, M.; Helenius, A. Chaperone Selection During Glycoprotein Translocation into the Endoplasmic Reticulum. Science 2000, 288, 331–333. [Google Scholar] [CrossRef]
- Molinari, M.; Galli, C.; Vanoni, O.; Arnold, S.M.; Kaufman, R.J. Persistent Glycoprotein Misfolding Activates the Glucosidase II/UGT1-Driven Calnexin Cycle to Delay Aggregation and Loss of Folding Competence. Mol. Cell 2005, 20, 503–512. [Google Scholar] [CrossRef]
- Hammond, C.; Helenius, A. Folding of VSV G protein: Sequential interaction with BiP and calnexin. Science 1994, 266, 456–458. [Google Scholar] [CrossRef] [PubMed]
- Lakkaraju, A.K.K.; van der Goot, F.G. Calnexin Controls the STAT3-Mediated Transcriptional Response to EGF. Mol. Cell 2013, 51, 386–396. [Google Scholar] [CrossRef] [PubMed]
- Alam, A.; Taye, N.; Patel, S.; Thube, M.; Mullick, J.; Shah, V.K.; Pant, R.; Roychowdhury, T.; Banerjee, N.; Chatterjee, S.; et al. SMAR1 favors immunosurveillance of cancer cells by modulating calnexin and MHC I expression. Neoplasia 2019, 21, 945–962. [Google Scholar] [CrossRef]
- Ryan, D.; Carberry, S.; Murphy, Á.C.; Lindner, A.U.; Fay, J.; Hector, S.; McCawley, N.; Bacon, O.; Concannon, C.G.; Kay, E.W.; et al. Calnexin, an ER stress-induced protein, is a prognostic marker and potential therapeutic target in colorectal cancer. J. Transl. Med. 2016, 14, 196. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, M.; Nagashio, R.; Jiang, S.-X.; Saito, K.; Tsuchiya, B.; Ryuge, S.; Katono, K.; Nakashima, H.; Fukuda, E.; Goshima, N.; et al. Calnexin is a novel sero-diagnostic marker for lung cancer. Lung Cancer 2015, 90, 342–345. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Komohara, Y.; Domenick, N.; Ohno, M.; Ikeura, M.; Hamilton, R.L.; Horbinski, C.; Wang, X.; Ferrone, S.; Okada, H. Expression of antigen processing and presenting molecules in brain metastasis of breast cancer. Cancer Immunol. Immunother. 2012, 61, 789–801. [Google Scholar] [CrossRef] [PubMed]
- Bork, P.; Doolittle, R.F. Proposed acquisition of an animal protein domain by bacteria. Proc. Natl. Acad. Sci. USA 1992, 89, 8990–8994. [Google Scholar] [CrossRef] [PubMed]
- Erickson, H.P. Reversible unfolding of fibronectin type III and immunoglobulin domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc. Natl. Acad. Sci. USA 1994, 91, 10114–10118. [Google Scholar] [CrossRef] [PubMed]
- Bian, T.; Zheng, L.; Jiang, D.; Liu, J.; Zhang, J.; Feng, J.; Zhang, Q.; Qian, L.; Qiu, H.; Liu, Y.; et al. Overexpression of fibronectin type III domain containing 3B is correlated with epithelial-mesenchymal transition and predicts poor prognosis in lung adenocarcinoma. Exp. Med. 2019, 17, 3317–3326. [Google Scholar] [CrossRef] [PubMed]
- Obara, M.; Sakuma, T.; Fujikawa, K. The third type III module of human fibronectin mediates cell adhesion and migration. J. Biochem. 2010, 147, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Jiao, Q.; Honda, T.; Kurotani, R.; Toyota, E.; Okumura, S.; Takeya, T.; Minamisawa, S.; Lanier, S.M.; Ishikawa, Y. Activator of G Protein Signaling 8 (AGS8) Is Required for Hypoxia-induced Apoptosis of Cardiomyocytes: ROLE OF Gβγ AND CONNEXIN 43 (CX43). J. Biol. Chem. 2009, 284, 31431–31440. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Al Mamun, A.; Sakima, M.; Sato, M. Activator of G-protein signaling 8 is involved in VEGF-mediated signal processing during angiogenesis. J. Cell Sci. 2016, 129, 1210–1222. [Google Scholar] [CrossRef] [PubMed]
- Bell, A.; Bell, D.; Weber, R.S.; El-Naggar, A.K. CpG island methylation profiling in human salivary gland adenoid cystic carcinoma. Cancer 2011, 117, 2898–2909. [Google Scholar] [CrossRef]
- Liu, Y.-P.; Chen, W.-D.; Li, W.-N.; Zhang, M. Overexpression of FNDC1 Relates to Poor Prognosis and Its Knockdown Impairs Cell Invasion and Migration in Gastric Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819869928. [Google Scholar] [CrossRef]
- Das, D.K.; Naidoo, M.; Ilboudo, A.; Park, J.Y.; Ali, T.; Krampis, K.; Robinson, B.D.; Osborne, J.R.; Ogunwobi, O.O. miR-1207-3p regulates the androgen receptor in prostate cancer via FNDC1/fibronectin. Exp. Cell Res. 2016, 348, 190–200. [Google Scholar] [CrossRef] [PubMed]
- Cubedo, J.; Padró, T.; Badimon, L. Coordinated proteomic signature changes in immune response and complement proteins in acute myocardial infarction: The implication of serum amyloid P-component. Int. J. Cardiol. 2013, 168, 5196–5204. [Google Scholar] [CrossRef] [PubMed]
- Ishioka, N.; Takahashi, N.; Putnam, F.W. Amino acid sequence of human plasma alpha 1B-glycoprotein: Homology to the immunoglobulin supergene family. Proc. Natl. Acad. Sci. USA 1986, 83, 2363–2367. [Google Scholar] [CrossRef]
- Cortelazzo, A.; de Felice, C.; Leoncini, S.; Signorini, C.; Guerranti, R.; Leoncini, R.; Armini, A.; Bini, L.; Ciccoli, L.; Hayek, J. Inflammatory protein response in CDKL5-Rett syndrome: Evidence of a subclinical smouldering inflammation. Inflamm. Res. 2017, 66, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cao, J.; Li, L.; Liu, Y.; Zhao, H.; Li, N.; Li, B.; Zhang, A.; Huang, H.; Chen, S.; et al. Identification of urine protein biomarkers with the potential for early detection of lung cancer. Sci. Rep. 2015, 5, 11805. [Google Scholar] [CrossRef] [PubMed]
- Tian, M.; Cui, Y.-Z.; Song, G.-H.; Zong, M.-J.; Zhou, X.-Y.; Chen, Y.; Han, J.-X. Proteomic analysis identifies MMP-9, DJ-1 and A1BG as overexpressed proteins in pancreatic juice from pancreatic ductal adenocarcinoma patients. BMC Cancer 2008, 8, 241. [Google Scholar] [CrossRef]
- Kreunin, P.; Zhao, J.; Rosser, C.; Urquidi, V.; Lubman, D.M.; Goodison, S. Bladder Cancer Associated Glycoprotein Signatures Revealed by Urinary Proteomic Profiling. J. Proteome Res. 2007, 6, 2631–2639. [Google Scholar] [CrossRef] [PubMed]
- Kopylov, A.T.; Stepanov, A.A.; Malsagova, K.A.; Soni, D.; Kushlinsky, N.E.; Enikeev, D.V.; Potoldykova, N.V.; Lisitsa, A.V.; Kaysheva, A.L. Revelation of Proteomic Indicators for Colorectal Cancer in Initial Stages of Development. Molecules 2020, 25, 619. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Luo, X.; Hu, H.; Wang, R.; Sun, Y.; Zeng, R.; Chen, H. Integrative Proteomics and Tissue Microarray Profiling Indicate the Association between Overexpressed Serum Proteins and Non-Small Cell Lung Cancer. PLoS ONE 2012, 7, e51748. [Google Scholar]
- Canales Angélica Galicia, N.; Marina Madrid, V.; Castro Salmerón, J.; Jiménez Antúnez, A.; Mendoza-Hernández, G.; McCarron Langley, E.; Roman Bahena, M.; Castro-Romero Ivone, J. A1BG and C3 are overexpressed in patients with cervical intraepithelial neoplasia III. Oncol. Lett. 2014, 8, 939–947. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Jeong, D.H.; Kim, H.K.; Prince, A.-E.B.; Lee, D.S.; Kim, Y.N.; Han, J.; Kim, K.T. Plasma proteomic analysis of patients with squamous cell carcinoma of the uterine cervix. J. Gynecol. Oncol. 2008, 19, 173–180. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Z.; Hincapie, M.; Haab, B.B.; Hanash, S.; Pitteri, S.J.; Kluck, S.; Hogan, J.M.; Kennedy, J.; Hancock, W.S. The development of an integrated platform to identify breast cancer glycoproteome changes in human serum. J. Chromatogr. A 2010, 1217, 3307–3315. [Google Scholar] [CrossRef] [PubMed]

| Biological Process | Fold Enrichment | IDs | FDR |
|---|---|---|---|
| Protein folding in endoplasmic reticulum | >100 | CANX, HSPA5, PDIA3 | 2.7 × 10−2 |
| Extracellular matrix structural constituent conferring tensile strength | 52.16 | COL1A1, COL1A2, COL6A3, COL12A1 | 1.73 × 10−3 |
| Extracellular matrix structural constituent | 18.98 | COL1A1, COL1A2, COL6A3, COL12A1, VCAN, EMILIN2 | 1.3 × 10−3 |
| Structural molecule activity | 6.81 | COL1A1, COL1A2, COL6A3, COL12A1, VCAN, EMILIN2, LMNA, EPB41L2, CTNNA1 | 4.95 × 10−3 |
| Cadherin binding Cell adhesion molecule binding | 17.75 11.01 | LIMA1, EEF1D, CALD1, TNKS1BP1, HSPA5, FLNA, TAGLN2, SERBP1, DDX3X, CTNNA1, BAG3 | 1.07 × 10−7 9.50 × 10−6 |
| Pathway Name | Fold Enrichment | IDs | FDR |
|---|---|---|---|
| GP1b-IX-V activation signaling | >100 | COL1A1, COL1A2, FLNA | 2.47 × 10−3 |
| Platelet activation signaling and aggregation | 12.39 | COL1A1, COL1A2, FLNA, A1BG, HSPA5, TAGLN2 | 4.13 × 10−3 |
| Antigen presentation: folding, assembly and peptide loading of class I MHC | 61.69 | CANX, HSPA5, PDIA3 | 4.85 × 10−3 |
| Collagen chain trimerization | 48.60 | COL1A1, COL1A2, COL6A3, COL12A1 | 4.34 × 10−3 |
| Collagen biosynthesis and modifying enzymes | 31.92 | COL1A1, COL1A2, COL6A3, COL12A1 | 3.51 × 10−3 |
| Collagen formation | 24.03 | COL1A1, COL1A2, COL6A3, COL12A1 | 5.58 × 10−3 |
| Assembly of collagen fibrils and other multimeric structures | 35.64 | COL1A1, COL1A2, COL6A3, COL12A1 | 4.63 × 10−3 |
| Collagen degradation | 33.42 | COL1A1, COL1A2, COL6A3, COL12A1 | 4.43 × 10−3 |
| Degradation of the extracellular matrix | 15.28 | COL1A1, COL1A2, COL6A3, COL12A1 | 2.41 × 10−2 |
| ECM proteoglycans | 28.14 | COL1A1, COL1A2, COL6A3, VCAN | 4.83 × 10−3 |
| Platelet degranulation | 16.84 | A1BG, HSPA5, FLNA, TAGLN2 | 1.95 × 10−2 |
| Response to elevated platelet cutosolic Ca2+ | 16.20 | A1BG, HSPA5, FLNA, TAGLN2 | 2.08 × 10−2 |
| Signaling by receptor tyrosine kinases | 8.19 | COL1A1, COL1A2, COL6A3, STMN1, HNRNPM, CSN2, CTNNA1 | 5.38 × 10−3 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cordeiro, Y.G.; Mulder, L.M.; van Zeijl, R.J.M.; Paskoski, L.B.; van Veelen, P.; de Ru, A.; Strefezzi, R.F.; Heijs, B.; Fukumasu, H. Proteomic Analysis Identifies FNDC1, A1BG, and Antigen Processing Proteins Associated with Tumor Heterogeneity and Malignancy in a Canine Model of Breast Cancer. Cancers 2021, 13, 5901. https://doi.org/10.3390/cancers13235901
Cordeiro YG, Mulder LM, van Zeijl RJM, Paskoski LB, van Veelen P, de Ru A, Strefezzi RF, Heijs B, Fukumasu H. Proteomic Analysis Identifies FNDC1, A1BG, and Antigen Processing Proteins Associated with Tumor Heterogeneity and Malignancy in a Canine Model of Breast Cancer. Cancers. 2021; 13(23):5901. https://doi.org/10.3390/cancers13235901
Chicago/Turabian StyleCordeiro, Yonara G., Leandra M. Mulder, René J. M. van Zeijl, Lindsay B. Paskoski, Peter van Veelen, Arnoud de Ru, Ricardo F. Strefezzi, Bram Heijs, and Heidge Fukumasu. 2021. "Proteomic Analysis Identifies FNDC1, A1BG, and Antigen Processing Proteins Associated with Tumor Heterogeneity and Malignancy in a Canine Model of Breast Cancer" Cancers 13, no. 23: 5901. https://doi.org/10.3390/cancers13235901
APA StyleCordeiro, Y. G., Mulder, L. M., van Zeijl, R. J. M., Paskoski, L. B., van Veelen, P., de Ru, A., Strefezzi, R. F., Heijs, B., & Fukumasu, H. (2021). Proteomic Analysis Identifies FNDC1, A1BG, and Antigen Processing Proteins Associated with Tumor Heterogeneity and Malignancy in a Canine Model of Breast Cancer. Cancers, 13(23), 5901. https://doi.org/10.3390/cancers13235901