CYCLON and NPM1 Cooperate within an Oncogenic Network Predictive of R-CHOP Response in DLBCL
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. Nuclei Isolation
2.3. Solubilization of Nucleic-Acid Bound Complexes Using Micrococcal Nuclease (MNase)
2.4. Immunoprecipitation (IP) of GFP-Tagged CYCLON
2.5. Immunoblotting
2.6. Silver Staining
2.7. Mass Spectrometry Analysis
2.8. Bioinformatics Analysis
2.9. Immunofluorescence and Confocal Microscopy
2.10. Patient Samples
2.11. Tissue Microarray (TMA) and Immunohistochemistry (IHC)
2.12. Fluorescence In Situ Hybridization (FISH)
2.13. Targeted NGS Sequencing
2.14. Statistics
3. Results
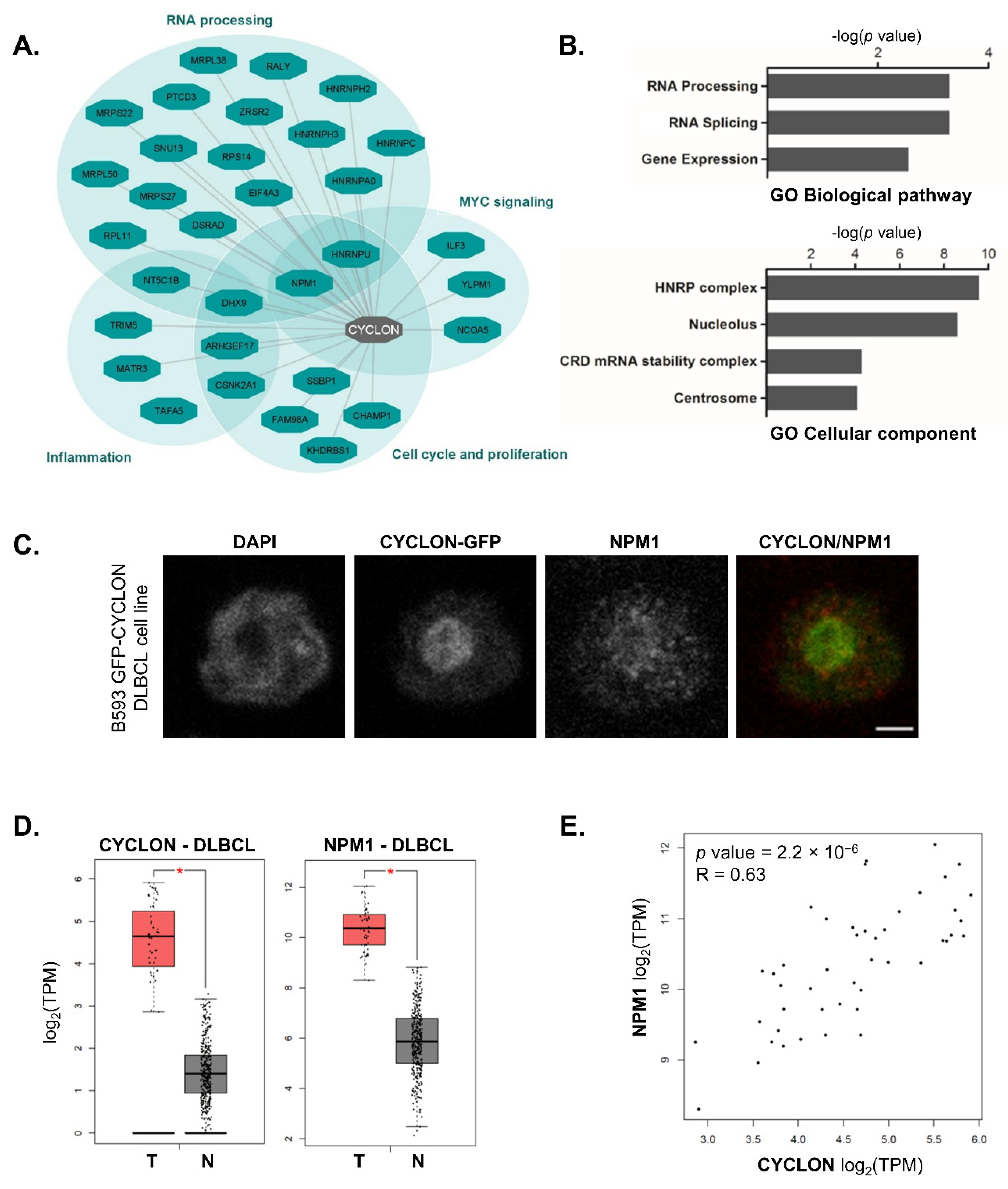
3.1. CYCLON Network Is Enriched in Nucleolar Proteins, Notably NPM1
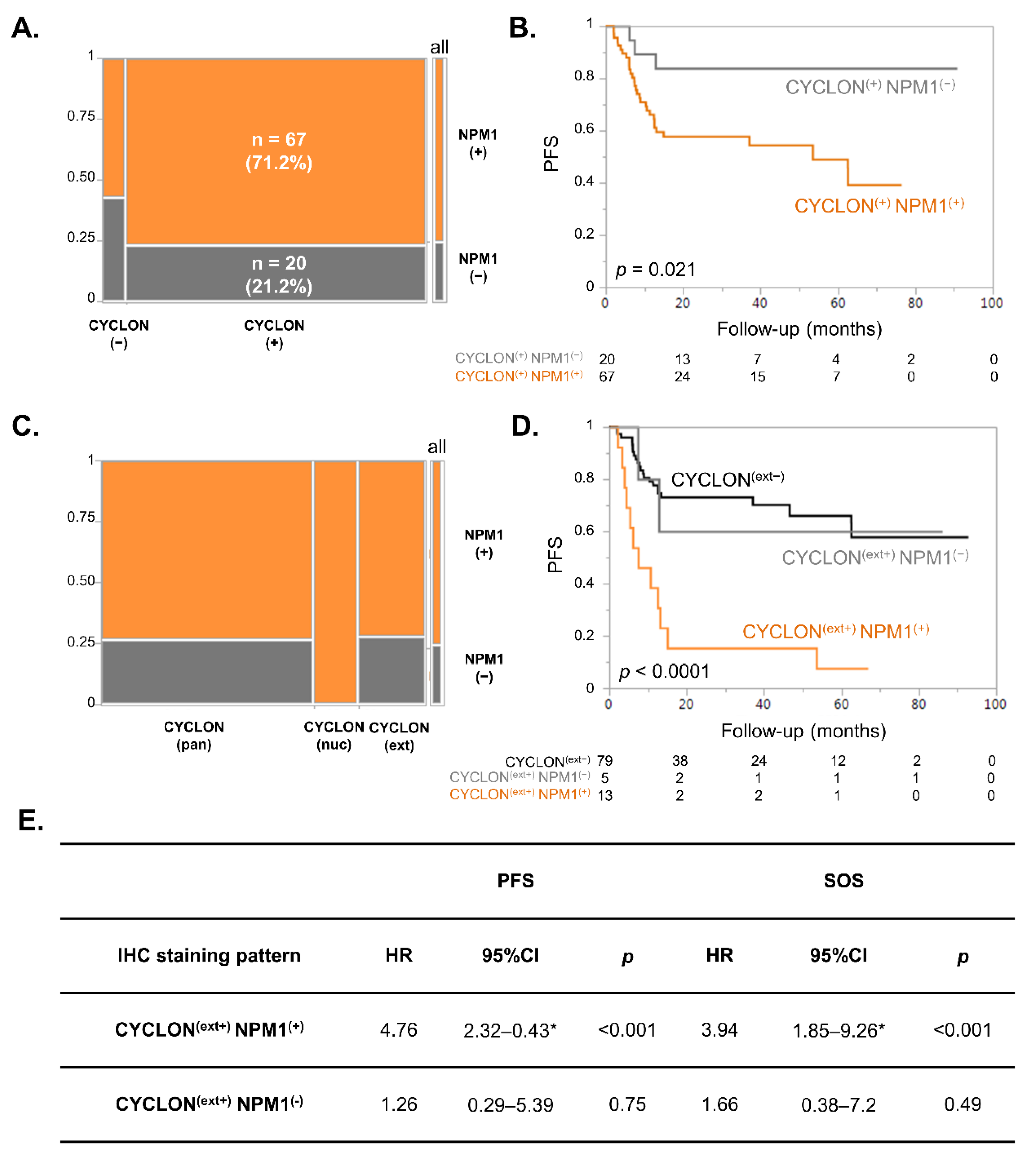
3.2. NPM1 and CYCLON Co-Expression Is Predictive of R-CHOP Response in DLBCL Patients
3.3. NPM1 Subcellular Localization Is Associated with Prognosis in DLBCL
3.4. Multivariate Analysis of CYCLON, NPM1 and R-IPI on DLBCL Prognosis
3.5. Multivariate Competing Risk Models Analyses Show That CYCLON, NPM1 and R-IPI Are Prognostic Markers of Refractory Disease-Related Death in DLBCL
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Teras, L.R.; DeSantis, C.E.; Cerhan, J.R.; Morton, L.M.; Jemal, A.; Flowers, C.R. 2016 US lymphoid malignancy statistics by World Health Organization subtypes. CA Cancer J. Clin. 2016, 66, 443–459. [Google Scholar] [CrossRef] [PubMed]
- Coiffier, B.; Thieblemont, C.; Neste, E.V.D.; Lepeu, G.; Plantier, I.; Castaigne, S.; Lefort, S.; Marit, G.; Macro, M.; Sebban, C.; et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: A study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood 2010, 116, 2040–2045. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Alizadeh, A.A.; Eisen, M.B.; Davis, R.E.; Ma, C.; Lossos, I.S.; Rosenwald, A.; Boldrick, J.C.; Sabet, H.; Tran, T.; Yu, X.; et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nat. Cell Biol. 2000, 403, 503–511. [Google Scholar] [CrossRef] [PubMed]
- Rosenwald, A.; Wright, G.; Chan, W.C.; Connors, J.M.; Campo, E.; Fisher, R.I.; Gascoyne, R.D.; Muller-Hermelink, H.K.; Smeland, E.B.; Giltnane, J.M.; et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N. Engl. J. Med. 2002, 346, 1937–1947. [Google Scholar] [CrossRef]
- Reddy, A.; Zhang, J.; Davis, N.S.; Moffitt, A.; Love, C.L.; Waldrop, A.; Leppä, S.; Pasanen, A.; Meriranta, L.; Karjalainen-Lindsberg, M.-L.; et al. Genetic and functional drivers of diffuse large B cell lymphoma. Cell 2017, 171, 481–494. [Google Scholar] [CrossRef]
- Schmitz, R.; Wright, G.W.; Huang, D.W.; Johnson, C.A.; Phelan, J.D.; Wang, J.Q.; Roulland, S.; Kasbekar, M.; Young, R.M.; Shaffer, A.L.; et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N. Engl. J. Med. 2018, 378, 1396–1407. [Google Scholar] [CrossRef]
- Wright, G.W.; Huang, D.W.; Phelan, J.D.; Coulibaly, Z.A.; Roulland, S.; Young, R.M.; Wang, J.Q.; Schmitz, R.; Morin, R.; Tang, J.; et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell 2020, 37, 551–568. [Google Scholar] [CrossRef]
- Sehn, L.H.; Berry, B.; Chhanabhai, M.; Fitzgerald, C.; Gill, K.; Hoskins, P.; Klasa, R.; Savage, K.J.; Shenkier, T.; Sutherland, J.; et al. The revised International Prognostic Index (R-IPI) is a better predictor of outcome than the standard IPI for patients with diffuse large B-cell lymphoma treated with R-CHOP. Blood 2007, 109, 1857–1861. [Google Scholar] [CrossRef]
- Emadali, A.; Rousseaux, S.; Bruder-Costa, J.; Rome, C.; Duley, S.; Hamaidia, S.; Betton, P.; Debernardi, A.; Leroux, D.; Bernay, B.; et al. Identification of a novel BET bromodomain inhibitor-sensitive, gene regulatory circuit that controls Rituximab response and tumour growth in aggressive lymphoid cancers. EMBO Mol. Med. 2013, 5, 1180–1195. [Google Scholar] [CrossRef]
- Bouroumeau, A.; Bussot, L.; Sartelet, H.; Fournier, C.; Betton-Fraisse, P.; Col, E.; David-Boudet, L.; McLeer, A.; Lefebvre, C.; Raskovalova, T.; et al. Extranucleolar CYCLON staining pattern is strongly associated to relapse/refractory disease in R-CHOP–treated DLBCL. HemaSphere 2021, 5, e598. [Google Scholar] [CrossRef]
- Hoshino, A.; Fujii, H. Redundant promoter elements mediate IL-3-induced expression of a novel cytokine-inducible gene, cyclon. FEBS Lett. 2007, 581, 975–980. [Google Scholar] [CrossRef] [PubMed]
- Fleur, S.S.; Hoshino, A.; Kondo, K.; Egawa, T.; Fujii, H. Regulation of Fas-mediated immune homeostasis by an activation-induced protein, Cyclon. Blood 2009, 114, 1355–1365. [Google Scholar] [CrossRef]
- Dominguez-Sola, D.; Victora, G.; Ying, C.Y.; Phan, R.T.; Saito, M.; Nussenzweig, M.C.; Dalla-Favera, R. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat. Immunol. 2012, 13, 1083–1091. [Google Scholar] [CrossRef] [PubMed]
- Callanan, M.B.; Le Baccon, P.; Mossuz, P.; Duley, S.; Bastard, C.; Hamoudi, R.; Dyer, M.; Klobeck, G.; Rimokh, R.; Sotto, J.J.; et al. The IgG Fc receptor, Fcgamma RIIB, is a target for deregulation by chromosomal translocation in malignant lymphoma. Proc. Natl. Acad. Sci. USA 2000, 97, 309–314. [Google Scholar] [CrossRef] [PubMed]
- Casabona, M.G.; Vandenbrouck, Y.; Attree, I.; Couté, Y. Proteomic characterization of Pseudomonas aeruginosa PAO1 inner membrane. Proteomics 2013, 13, 2419–2423. [Google Scholar] [CrossRef] [PubMed]
- Bouyssié, D.; Hesse, A.-M.; Mouton-Barbosa, E.; Rompais, M.; Macron, C.; Carapito, C.; De Peredo, A.G.; Couté, Y.; Dupierris, V.; Burel, A.; et al. Proline: An efficient and user-friendly software suite for large-scale proteomics. Bioinformatics 2020, 36, 3148–3155. [Google Scholar] [CrossRef] [PubMed]
- Wieczorek, S.; Combes, F.; Lazar, C.; Gianetto, Q.G.; Gatto, L.; Dorffer, A.; Hesse, A.-M.; Couté, Y.; Ferro, M.; Bruley, C.; et al. DAPAR & ProStaR: Software to perform statistical analyses in quantitative discovery proteomics. Bioinformatics 2017, 33, 135–136. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Pathan, M.; Keerthikumar, S.; Ang, C.-S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Dubois, S.; Viailly, P.-J.; Mareschal, S.; Bohers, E.; Bertrand, P.; Ruminy, P.; Maingonnat, C.; Jais, J.-P.; Peyrouze, P.; Figeac, M.; et al. Next-generation sequencing in diffuse large B-cell lymphoma highlights molecular divergence and therapeutic opportunities: A LYSA study. Clin. Cancer Res. 2016, 22, 2919–2928. [Google Scholar] [CrossRef]
- Sujobert, P.; Le Bris, Y.; De Leval, L.; Gros, A.; Merlio, J.P.; Pastoret, C.; Huet, S.; Sarkozy, C.; Davi, F.; Callanan, M.; et al. The need for a consensus next-generation sequencing panel for mature lymphoid malignancies. HemaSphere 2019, 3, e169. [Google Scholar] [CrossRef] [PubMed]
- Burlet, B.; Ramla, S.; Fournier, C.; Abrey-Recalde, M.J.; Sauter, C.; Chrétien, M.-L.; Rossi, C.; Duffourd, Y.; Ragot, S.; Buriller, C.; et al. Identification of novel, clonally stable, somatic mutations targeting transcription factors PAX5 and NKX2–3, the epigenetic regulator LRIF1, and BRAF in a case of atypical B-cell chronic lymphocytic leukemia harboring a t(14;18) (q32;q21). Mol. Case Stud. 2021, 7, a005934. [Google Scholar] [CrossRef] [PubMed]
- Fine, J.P.; Gray, R.J. A proportional hazards model for the subdistribution of a competing risk. J. Am. Stat. Assoc. 1999, 94, 496. [Google Scholar] [CrossRef]
- Xu, L.-H.; Fang, J.-P.; Liu, Y.-C.; Jones, A.I.; Chai, L. Nucleophosmin mutations confer an independent favorable prognostic impact in 869 pediatric patients with acute myeloid leukemia. Blood Cancer J. 2020, 10, 1–10. [Google Scholar] [CrossRef]
- Morris, S.W.; Kirstein, M.N.; Valentine, M.B.; Dittmer, K.G.; Shapiro, D.N.; Saltman, D.L.; Look, A.T. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in Non-Hodgkin’s Lymphoma. Science 1994, 263, 1281–1284. [Google Scholar] [CrossRef]
- Falini, B.; Mecucci, C.; Tiacci, E.; Alcalay, M.; Rosati, R.; Pasqualucci, L.; La Starza, R.; Diverio, D.; Colombo, E.; Santucci, A.; et al. Cytoplasmic Nucleophosmin in acute myelogenous leukemia with a normal karyotype. N. Engl. J. Med. 2005, 352, 254–266. [Google Scholar] [CrossRef]
- Verhaak, R.G.W.; Goudswaard, C.S.; Van Putten, W.; Bijl, M.A.; Sanders, M.A.; Hugens, W.; Uitterlinden, A.G.; Erpelinck, C.A.J.; Delwel, R.; Löwenberg, B.; et al. Mutations in nucleophosmin (NPM1) in acute myeloid leukemia (AML): Association with other gene abnormalities and previously established gene expression signatures and their favorable prognostic significance. Blood 2005, 106, 3747–3754. [Google Scholar] [CrossRef]
- Moy, T.I.; Boettner, D.; Rhodes, J.C.; Silver, P.A.; Askew, D.S. Identification of a role for Saccharomyces cerevisiae Cgr1p in pre-rRNA processing and 60S ribosome subunit synthesis. Microbiology 2002, 148, 1081–1090. [Google Scholar] [CrossRef][Green Version]
- López, D.J.; Rodríguez, J.A.; Bañuelos, S. Nucleophosmin, a multifunctional nucleolar organizer with a role in DNA repair. Biochim. Biophys. Acta Proteins Proteom. 2020, 1868, 140532. [Google Scholar] [CrossRef] [PubMed]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Körner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef] [PubMed]
- Colombo, E.; Marine, J.-C.; Danovi, D.; Falini, B.; Pelicci, P.G. Nucleophosmin regulates the stability and transcriptional activity of p53. Nat. Cell Biol. 2002, 4, 529–533. [Google Scholar] [CrossRef]
- Chen, Y.; Hu, J. Nucleophosmin1 (NPM1) abnormality in hematologic malignancies, and therapeutic targeting of mutant NPM1 in acute myeloid leukemia. Ther. Adv. Hematol. 2020, 11, 2040620719899818. [Google Scholar] [CrossRef] [PubMed]
- Vassiliou, G.S.; Cooper, J.L.; Rad, R.; Li, J.; Rice, S.; Uren, A.; Rad, L.; Ellis, P.; Andrews, R.; Banerjee, R.; et al. Mutant nucleophosmin and cooperating pathways drive leukemia initiation and progression in mice. Nat. Genet. 2011, 43, 470–475. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Dermani, F.K.; Khoei, S.G.; Afshar, S.; Amini, R. The potential role of nucleophosmin (NPM1) in the development of cancer. J. Cell. Physiol. 2021, 236, 7832–7852. [Google Scholar] [CrossRef]
- Wang, X.; Li, S. Protein mislocalization: Mechanisms, functions and clinical applications in cancer. Biochim. Biophys. Acta Bioenerg. 2014, 1846, 13–25. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 2016, 5, e13571. [Google Scholar] [CrossRef]
- Papageorgiou, S.G.; Thomopoulos, T.P.; Katagas, I.; Bouchla, A.; Pappa, V. Prognostic molecular biomarkers in diffuse large B-cell lymphoma in the rituximab era and their therapeutic implications. Ther. Adv. Hematol. 2021, 12, 20406207211013987. [Google Scholar] [CrossRef]
- Etchin, J.; Sanda, T.; Mansour, M.; Kentsis, A.; Montero, J.; Le, B.T.; Christie, A.L.; McCauley, D.; Rodig, S.J.; Kauffman, M.; et al. KPT-330 inhibitor of CRM1 (XPO1)-mediated nuclear export has selective anti-leukaemic activity in preclinical models of T-cell acute lymphoblastic leukaemia and acute myeloid leukaemia. Br. J. Haematol. 2013, 161, 117–127. [Google Scholar] [CrossRef]
- Garzon, R.; Savona, M.; Baz, R.; Andreeff, M.; Gabrail, N.; Gutierrez, M.; Savoie, L.; Mau-Sørensen, M.; Wagner-Johnston, N.; Yee, K.; et al. A phase 1 clinical trial of single-agent selinexor in acute myeloid leukemia. Blood 2017, 129, 3165–3174. [Google Scholar] [CrossRef]
- Kuruvilla, J.; Savona, M.; Baz, R.; Mau-Sørensen, M.; Gabrail, N.; Garzon, R.; Stone, R.; Wang, M.; Savoie, L.; Martin, P.; et al. Selective inhibition of nuclear export with selinexor in patients with non-Hodgkin lymphoma. Blood 2017, 129, 3175–3183. [Google Scholar] [CrossRef] [PubMed]
- Balusu, R.; Fiskus, W.; Rao, R.; Chong, D.G.; Nalluri, S.; Mudunuru, U.; Ma, H.; Chen, L.; Venkannagari, S.; Ha, K.; et al. Targeting levels or oligomerization of nucleophosmin 1 induces differentiation and loss of survival of human AML cells with mutant NPM1. Blood 2011, 118, 3096–3106. [Google Scholar] [CrossRef] [PubMed]
- Urbaneja, M.A.; Skjærven, L.; Aubi, O.; Underhaug, J.; López, D.J.; Arregi, I.; Alonso, M.; Cuevas, A.; Rodriguez, J.A.; Martinez, A.; et al. Conformational stabilization as a strategy to prevent nucleophosmin mislocalization in leukemia. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [PubMed]

| Parameters | Value (Range)/% [Proportion] |
|---|---|
| Median age at diagnosis (years) | 67 (29–94) |
| >60 years | 69.1% [67/97] |
| Gender | |
| Male | 55.7% [54/97] |
| Female | 44.3% [43/97] |
| Median follow up time (months) | 41.1 (2–93, 95% CI: 29.0–53.6) |
| First-line therapy | |
| R-CHOP | 80.4% [78/97] |
| R-CHOP like | 19.6% [19/97] |
| Mini R-CHOP | 8.2% [8/97] |
| R-CVP | 9.3% [9/97] |
| R-CHVP | 1.0% [1/97] |
| R-COP | 1.0% [1/97] |
| Complete response rate | 78.3% [76/97] |
| Overall response rate | 83.5% [81/97] |
| Primary refractory cases | 20.6% [20/97] |
| Relapse cases | 18.5% [18/97] |
| Specific overall survival (SOS) | 62.0% (95% CI: 49.4–72.4) |
| Progression-free survival (PFS) | 51.1% (95% CI: 36.2–64.1) |
| Ann Arbor staging classification (diagnosis) | |
| I (Single lymph node (LN) involved) | 11.3% [11/97] |
| II (2 or more LN ipsilateral to the diaphragm) | 20.6% [20/97] |
| III (LN on both sides of the diaphragm) | 21.6% [21/97] |
| IV (extralymphatic organs or tissues involvment) | 46.4% [45/97] |
| LDH > upper limit of normal | 75.0% [73/97] |
| NPM1 (IHC) | |
| Negative | 23.7% [23/97] |
| Cytoplasmic | 10.3% [10/97] |
| Nuclear | 48.4% [47/97] |
| Pan-cellular | 14.4% [14/97] |
| Uninterpretable | 3.2% [3/97] |
| Variable | Category | HR | 95% CI | p-Value | SE | HR | 95% CI | p-Value | SE |
|---|---|---|---|---|---|---|---|---|---|
| NPM1 | pan-cellular(+) versus pan-cellular(−) | 5.2 | 2.1–12.6 | <0.001 | 2.24 | 5.8 | 2.5–13.3 | <0.001 | 1.07 |
| CYCLON | extra-nucleolar(+) versus extra-nucleolar(−) | 2.9 | 1.4–6.2 | 0.002 | 1.03 | 2.8 | 1.2–6.8 | 0.007 | 2.67 |
| R-IPI | R-IPI(high) versus R-IPI (low) | 3.7 | 1.5–8.9 | 0.001 | 1.46 | 5.2 | 2.0–13.6 | <0.001 | 2.38 |
| Competing Risk (CR) | Refractory 1 | Relapse 2 | |||||
|---|---|---|---|---|---|---|---|
| sHR 3 | 95%CI 4 | p | Bs-95%CI 5 | sHR | 95%CI 4 | p | |
| CYCLON 6 | 4.04 | 1.62–10.05 | 0.003 | 1.32–12.09 | 1.08 | 0.24–4.92 | 0.92 |
| NPM1 7 | 4.64 | 1.79–12.04 | 0.002 | 1.08–14.46 | 1.63 | 0.29–9.29 | 0.58 |
| R-IPI 8 | 2.8 | 1.03–7.59 | 0.043 | 1.008–10.24 | 10.03 | 1.07–94.6 | 0.043 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bouroumeau, A.; Bussot, L.; Hamaidia, S.; Garcìa-Sandoval, A.; Bergan-Dahl, A.; Betton-Fraisse, P.; Duley, S.; Fournier, C.; Aucagne, R.; Adrait, A.; et al. CYCLON and NPM1 Cooperate within an Oncogenic Network Predictive of R-CHOP Response in DLBCL. Cancers 2021, 13, 5900. https://doi.org/10.3390/cancers13235900
Bouroumeau A, Bussot L, Hamaidia S, Garcìa-Sandoval A, Bergan-Dahl A, Betton-Fraisse P, Duley S, Fournier C, Aucagne R, Adrait A, et al. CYCLON and NPM1 Cooperate within an Oncogenic Network Predictive of R-CHOP Response in DLBCL. Cancers. 2021; 13(23):5900. https://doi.org/10.3390/cancers13235900
Chicago/Turabian StyleBouroumeau, Antonin, Lucile Bussot, Sieme Hamaidia, Andrea Garcìa-Sandoval, Anna Bergan-Dahl, Patricia Betton-Fraisse, Samuel Duley, Cyril Fournier, Romain Aucagne, Annie Adrait, and et al. 2021. "CYCLON and NPM1 Cooperate within an Oncogenic Network Predictive of R-CHOP Response in DLBCL" Cancers 13, no. 23: 5900. https://doi.org/10.3390/cancers13235900
APA StyleBouroumeau, A., Bussot, L., Hamaidia, S., Garcìa-Sandoval, A., Bergan-Dahl, A., Betton-Fraisse, P., Duley, S., Fournier, C., Aucagne, R., Adrait, A., Couté, Y., McLeer, A., Col, E., David-Boudet, L., Raskovalova, T., Jacob, M.-C., Vettier, C., Chevalier, S., Carras, S., ... Emadali, A. (2021). CYCLON and NPM1 Cooperate within an Oncogenic Network Predictive of R-CHOP Response in DLBCL. Cancers, 13(23), 5900. https://doi.org/10.3390/cancers13235900






